Polycomb repressive complexes (PRCs) establish and maintain silenced gene states through chromatin modifications, but their role in specifying cell types during vertebrate development is not known. Golden and Dasen show that the PRC1 component Bmi1 exerts dose-dependent effects on Hox gene expression in the spinal cord, defining the anterior expression domain of some Hox genes and specifying motor neuron columns and pool identities.
Keywords: Hox, epigenetics, motor neuron, polycomb, spinal cord
Abstract
Polycomb repressive complexes (PRCs) establish and maintain gene repression through chromatin modifications, but their specific roles in cell fate determination events are poorly understood. Here we show an essential role for the PRC1 component Bmi1 in motor neuron (MN) subtype differentiation through dose-dependent effects on Hox gene expression. While Bmi1 is dispensable for generating MNs as a class, it has an essential role in specifying and determining the position of Hox-dependent MN columnar and pool subtypes. These actions are mediated through limiting anterior Hox expression boundaries, functions deployed in post-mitotic MNs, temporally downstream from morphogen gradients. Within the HoxC gene cluster, we found a progressive depletion of PRC-associated marks from rostral to caudal levels of the spinal cord, corresponding to major demarcations of MN subtypes. Selective ablation of Bmi1 elicits a derepression of more posterior Hox genes, leading to a switch in MN fates. Unexpectedly, Hox patterns and MN fates appear to be sensitive to absolute PRC1 activity levels; while reducing Bmi1 switches forelimb lateral motor column (LMC) MNs to a thoracic preganglionic (PGC) identity, elevating Bmi1 expression at thoracic levels converts PGC to LMC MNs. These results suggest that graded PRC1 activities are essential in determining MN topographic organization.
The Polycomb group (PcG) is a large protein family that plays a variety of crucial roles in animal development. The depth of its involvement is highlighted by the copious number of genes that are bound by PcG members or bear histone modifications indicative of PcG-mediated repression (Boyer et al. 2006; Bracken et al. 2006; Lee et al. 2006). The function of the PcG in CNS maturation, however, remains unclear and may be dependent on temporal and spatial context. PcG complexes play somewhat disparate roles regulating cell fate decisions in several proliferating CNS populations. The PcG member Bmi1 in particular has been examined for its multifaceted role in modulating stem cell identity during development. The self-renewal capacity of neural stem cell populations, but not progenitor populations, is dependent on Bmi1, indicating that PcG pathways are key in distinguishing stem-like neural proliferation from developmental proliferation (Molofsky et al. 2003). Furthermore, overexpression of Bmi1 only facilitates proliferation in stem cell populations expressing FoxG1 in the forebrain, where it specifies production of neurons rather than glia (Fasano et al. 2009). PcG complexes have been found to have an inverse function in neocortical progenitors, where PcG members facilitate the transition from neuron to astrocyte production (Hirabayashi et al. 2009). Similarly, PcG activity can influence the generation of spinal dorsal interneurons via regulation of BMP signaling (Akizu et al. 2010).
Outside the nervous system, PcG complexes have been shown to contribute to developmental patterning via their interaction with Hox genes, which broadly govern regional morphology and cellular identity over the body axis (Mallo et al. 2010). PcG genes were first discovered as regulators of body plan and Hox expression in Drosophila (Lewis 1978; Struhl 1981; Jurgens 1985). Drosophila larvae with loss of function for the Bmi1 homolog Posterior sex combs (Psc) express AbdA and AbdB rostral to their normal boundaries (Simon et al. 1992; Martin and Adler 1993), leading to the transformation of several body segments into more posterior segments (Jurgens 1985; Brunk et al. 1991; van Lohuizen et al. 1991). Later work has demonstrated conservation of this role in mammals. Removal of Bmi1 leads to rostral shifts in the expression of Hox genes within the mesoderm as well rostral shifts in vertebral morphology (van der Lugt et al. 1994, 1996), while overexpression of Bmi1 results in the inverse phenotype (Alkema et al. 1995).
The central role that PcG complexes play in the establishment of cellular identity is significant due to the heritability of PcG-induced repression over multiple cellular divisions. While the precise mechanism of PcG-mediated silencing is unclear, the histone modifications deposited by PcG complexes are associated with compacted, silent chromatin. Classically, PcG repression in Drosophila is thought to be mediated by a two-step process (Simon and Kingston 2009). Polycomb reprssive complex 2 (PRC2) initiates repression by binding to a Polycomb response element (PRE), where it trimethylates Lys 27 on histone 3. This modification (H3K27me3) can be bound by the chromodomain found in PRC1, a complex that maintains repression of the locus and monoubiquitinates Lys 119 of histone H2A. While this model of two-step recruitment of PRC2 and PRC1 complexes is relevant in vertebrates, the behavior of PcG complexes is much more diverse. It is unclear how initial recruitment of PcG complexes occurs at mammalian loci, as only a handful of PREs have been found (Sing et al. 2009; Woo et al. 2010); however, PRC binding at several mammalian loci is regulated by noncoding RNA-binding partners (Rinn et al. 2007; Zhao et al. 2008; Khalil et al. 2009; Yap et al. 2010). There is also some evidence that PRC1 can function independently of PRC2 or that the two complexes costabilize the binding of one another (Simon and Kingston 2009). Within mammalian Hox loci, it seems likely that PRC1 and PRC2 are both involved in mediating correct patterns of repression via either a classical hierarchical recruitment model or costabilization. Work characterizing the distribution of PcG members and associated histone marks across the genome in embryonic stem (ES) and fibroblast cells has shown concomitant binding of PRC1, H3K27me3, and PRC2 at Hox loci (Boyer et al. 2006; Bracken et al. 2006; Woo et al. 2010).
While the role of PRCs in controlling the expression of developmental regulatory factors is well described, surprisingly little is known about their function in neuronal subtype diversification. In the embryonic spinal cord, Hox transcription factors determine the regional identity of motor neurons (MNs), raising the possibility that PcG proteins have an instructive role in their specification. In this context, Hox activities parse an initially uniform neuronal class into many subgroups with distinct postsynaptic targets. The largest subgroups are known as columns, three of which are distinguished by Hox genes expressed at specific levels of the neuraxis. At thoracic levels, Hoxc9 defines preganglionic (PGC) motor column MNs that innervate the sympathetic chain ganglia. Hoxc6 defines the lateral motor column (LMC) found at brachial levels that innervates the muscles of the forelimb, while Hoxc10 defines the LMC found at lumbar levels (Dasen et al. 2003). LMC neurons can be further subdivided into smaller Hox-dependent groups known as motor pools, each of which innervates a distinct limb muscle (Dasen et al. 2005). Thus, the patterning of Hox gene expression must be precisely regulated to ensure the establishment of a topographic registry between MN subtypes and their peripheral synaptic targets.
The factors governing this meticulous Hox gene code are currently not well defined. In spinal MNs, Hox patterns are thought to be regulated over two distinct phases of development. During tail bud regression and closure of the neural tube, the progenitor zone of the spinal cord is colonized by a stem cell-like population surrounding the node. These cells are exposed to retinoic acid (RA) from the rostral somitic mesoderm and fibroblast growth factors (FGFs) from the caudal presomitic mesoderm and node (Dasen and Jessell 2009). A cell remaining in the stem-like population for a longer period of time sees increased exposure to FGFs and decreased exposure to RA, resulting in a more caudal pattern of Hox gene expression upon the cell's eventual maturation as an MN.
This initial pattern of Hox expression is later refined after cells have undergone their final mitosis. At this stage, specific Hox factors display cross-repressive interactions that sharpen and maintain their expression borders (Dasen et al. 2003). Cross-repressive interactions appear to be critical to, and possibly restricted to, establishing the posterior Hox expression boundaries. For instance, in mice lacking the thoracic Hox gene Hoxc9, the more rostrally expressed Hox4–Hox8 genes are derepressed, and PGC neurons acquire an LMC fate. Analysis of histone modifications at thoracic levels indicates that Hoxc9-mediated repression is independent of the H3K27me3 mark associated with PcG-mediated repression (Jung et al. 2010). In contrast, H3K27me3 marks are found on Hox genes expressed more posteriorly, suggesting that PcG repression has a selective role in defining anterior Hox boundaries.
We show here an essential role for PRC1 function in controlling MN organization through regulation of Hox gene expression. In differentiated MNs, we observed a progressive decrease in the occupancy of PRC marks within the HoxC locus along the rostrocaudal axis. We found that the activities of the PRC1 component Bmi1 are essential in controlling the anterior limit of Hox gene expression and delineating the position of MN columnar subtypes. Attenuation or elevation of Bmi1 elicits a repositioning of Hox boundaries and motor columns, suggesting that MNs are sensitive to the overall levels of PRC1 activity. These actions appear to be deployed subsequent to the signaling gradients that determine the early positional identity of MN progenitors. Collectively, our results indicate that a key mechanism through which Hox gene patterns are controlled in MNs relies on the net level of PRC1 activity in early post-mitotic cells.
Results
Distribution of Polycomb-associated proteins at HoxC loci in the spinal cord
To assess a possible role for PRC activity in MNs, we first examined chromatin modifications and binding of PRC components at motor column-defining Hox loci at distinct rostrocaudal levels of the spinal cord. The H3K27me3 mark is a histone modification associated with PcG-mediated repression (Simon and Kingston 2009). Our previous studies provided evidence that at brachial and thoracic levels, Hox genes carry the H3K27me3 modification selectively at segments more rostral to where they are expressed (Jung et al. 2010). To assess whether this pattern of histone modifications applies generally to column-defining Hox genes at all spinal levels, we examined the distribution of H3K27me3 at cervical, brachial, thoracic, and lumbar levels (Fig. 1A,B). We also analyzed the chromatin state of Hox loci at these four spinal levels by interrogating the binding pattern of the PRC1 component Bmi1 as well as H3K4me3, a mark associated with transcriptionally active genes.
Figure 1.
Chromatin marks and expression of Hox genes along the rostrocaudal axis. (A) Schematic of Hox protein expression and Hox-dependent MN columnar subtypes. Rostral is to the left. LMC MNs innervate the limbs, and PGC MNs project to sympathetic chain ganglia. For simplicity, Hox-independent motor columns are not shown. (B) H3K27me3 status of Hoxc6, Hoxc9, and Hoxc10 promoter regions at cervical, brachial, thoracic, and lumbar levels. Fold enrichment is shown over IgG controls for each level. (C) Distribution of Bmi1 binding at columnar Hox loci shows distribution similar to that of H3K27me3. The absence of Bmi1 and H3K27me3 binding to lumbar chromatin indicates that antibodies specifically recognize marks at other levels. (D) Distribution of H3K4me3 marks. (E) Levels of Hoxc6, Hoxc9, and Hoxc10 mRNA at each spinal level.
We used chromatin immunoprecipitation (ChIP) assays to monitor the levels of H3K27me3, H3K4me3, and Bmi1 found near the promoter regions of Hoxc6, Hoxc9, and Hoxc10, three column-defining Hox genes expressed at brachial, thoracic, and lumbar levels of the spinal cord, respectively. To determine the functional outcome of the chromatin state of Hoxc6, Hoxc9, and Hoxc10, we also quantified mRNA levels of these genes. Spinal cords were isolated from Hamburger and Hamilton (HH) stage 25–27 chick embryos, a stage at which all MN columnar subtypes have been generated, and processed by collecting either chromatin or total RNA from each level. Samples were then subjected to either first strand synthesis using oligo(dT) or ChIP for H3K27me3, H3K4me3, or Bmi1.
At cervical levels, none of the column-defining Hox genes were expressed, and all three Hox genes bore high levels of H3K27me3. At brachial levels, only Hoxc6 was expressed, and H3K27me3 was found at Hoxc9 and Hoxc10. At thoracic levels, Hoxc9 was expressed, and H3K27me3 was only found at Hoxc10. Finally, at lumbar levels, only Hoxc10 was expressed, and H3K27me3 was not observed at any Hox gene examined (Fig. 1A,B,E). Thus, H3K27me3 is associated with Hox genes expressed more caudally than the level examined, and furthermore, Hox genes bearing high levels of H3K27me3 show low levels of transcription.
The pattern of Bmi1 binding to Hox loci closely follows that of H3K27me3. At cervical levels, Bmi1 was bound to all three of the silenced Hox genes. At brachial levels, significant levels of Bmi1 were only found at the silenced Hox genes Hoxc9 and Hoxc10. Levels of Bmi1 binding to Hoxc9 relative to Hoxc10 appeared to be lower, however, whereas H3K27me3 was found at both loci at roughly the same level. At thoracic levels, Bmi1was bound only to the more caudally expressed Hox gene, Hoxc10. Finally, at lumbar levels, Bmi1 was not found in significant amounts at any Hox gene examined (Fig. 1C).
The binding patterns of H3K4me3 to Hox loci suggest a complex interplay between chromatin modifications associated with active and repressed genes. At each level, H3K4me3 was found at the active Hox gene: At brachial levels, it was found at Hoxc6; at thoracic levels, it was found at Hoxc9; and at lumbar levels, it was found at Hoxc10 (Fig. 1D). However, H3K4me3 was also present at Hoxc6 at cervical levels and at Hoxc9 at brachial levels. These two loci also carry high levels of H3K27me3 and Bmi1, indicating that they may carry bivalent marks (Fig. 1D). The H3K4me3 mark was also observed at Hoxc9 at lumbar levels (Fig. 1D), while Hoxc9 mRNAs and PRC marks were not detected, suggesting a distinct mechanism of repression. Nevertheless, these data indicate that there is a progressive depletion of PRC-associated marks at Hox loci along the rostrocaudal axis and that all column-defining Hox genes bearing PRC marks are repressed.
Bmi1 is dispensable for generating MNs as a class
To determine the role of PcG complexes in specifying Hox-dependent MN subtypes, we assessed the function of Bmi1, a well-defined PRC1 component. Bmi1 has been shown to regulate rostrocaudal positional identity in the mesoderm of mice, and mutants display a one- to two-segment rostral shift in Hox expression (van der Lugt et al. 1994, 1996; Alkema et al. 1995). We hypothesized that similar shifts within the spinal cord of Bmi1 mutant mice might disrupt the specification of MN columnar subtypes. Analysis of Bmi1 mutants at embryonic day 12.5 (E12.5) showed that spinal morphology and expression of the transcription factors Hb9 and Isl1/2 were normal, indicating Bmi1 is not required for MN generation. We also observed a rostral shift in Hoxc9 expression in the dorsal spinal cord, but this did not translate to a significant change in MN identity (Supplemental Fig. S1). These results indicate that in mice, the loss of Bmi1 alone does not impact the specification of MN subtypes.
In mammals, several homologs of Bmi1 have been identified, including Mel-18 (Schuettengruber and Cavalli 2009). Mel-18 knockout mice display a phenotype similar to Bmi1 mutants, while Bmi1 and Mel-18 double-knockout mice are not viable beyond E9.5, indicating that these homologs compensate for each other (Akasaka et al. 2001). The embryonic chick provides a potential system to circumvent the limitations of studying Bmi1/Mel-18 function for several reasons. First, the accessibility of the chick spinal cord allows acute and selective depletion of genes of interest via in ovo electroporation, avoiding the confounding side effects of global morphological distortions stemming from segmentation defects in the mesoderm and skeleton. Second, Mel-18 has not been described in the chick, and the potential lack of a functional Bmi1 homolog in the spinal cord avoids the issue of compensation.
We depleted Bmi1 via in ovo electroporation of dsRNAs at HH stage 13/14, a stage in which the neural tube contains exclusively progenitors, and analyzed embryos 3 d later (around stage 27). A constitutive nuclear lacZ construct was included to label the region that incorporated dsRNA. Electroporation of Bmi1 dsRNA led to a marked depletion of Bmi1 protein within electroporated neurons (Fig. 2A). In contrast, electroporation of a scrambled dsRNA had no effect on Bmi1 or Hox expression patterns (Supplemental Fig. S2). To ensure that phenotypes seen after Bmi1 depletion were specific to PRC1 function, we assessed expression levels of PRC2-associated marks. Neither global H3K27me3 nor embryonic ectoderm development (Eed) levels were altered (Fig. 2B), indicating that Bmi1 depletion selectively affects PRC1.
Figure 2.
Bmi1 depletion does not affect generation of MNs. (A) Expression of Bmi1 is diminished upon electroporation of Bmi1 dsRNA. The electroporated half of the cord is indicated by a bolt. A nuclear LacZ construct was coelectroporated to grossly mark the electroporated region. (B) Expression of H3K27me3 and Eed is not affected by Bmi1 dsRNA. (C) Expression of post-mitotic MN markers Hb9 and Isl1/2 is also not affected. (D) Expression of Olig2, Pax6, and Nkx6.1 in MN progenitors is unaffected. (E) Time line of Bmi1 depletion upon electroporation of dsRNA at HH stage 13. Embryos were fixed and analyzed for Bmi1 expression at different time points following electroporation. Expression of Isl1/2 marks post-mitotic MNs.
We further characterized the temporal dynamics of Bmi1 depletion by analyzing different time points following dsRNA electroporation. At 8 h following electroporation (around stage 16/17), there was no depletion of Bmi1 protein in the electroporated spinal cord (Fig. 2E). However, after 28 h (around stage 18/19, as post-mitotic MNs appear), Bmi1 protein expression was reduced by 75%–80%. After 60 h, we observed some recovery in Bmi1 levels (Fig. 2E). Nevertheless, electroporation of Bmi1 dsRNA results in the depletion of Bmi1 protein during the key period of MN specification.
To characterize the effect of Bmi1 loss, we first assessed expression of markers for general features of MN identity. MNs originate in the central progenitor zone of the spinal cord at a specific dorso–ventral position defined by expression of Olig2, Nkx6.1, and Pax6 (Jessell 2000; Shirasaki and Pfaff 2002). Patterning of MN progenitors was unaffected after depletion of Bmi1 (Fig. 2D). Post-mitotic MNs can be distinguished by the expression of Hb9, Isl1/2, and Lhx3. Expression of these factors is unchanged in the absence of Bmi1 (Fig. 2C). PRC1 function therefore does not appear to be necessary for generating MNs as a class.
Bmi1 is required for proper Hox boundaries in the spinal cord
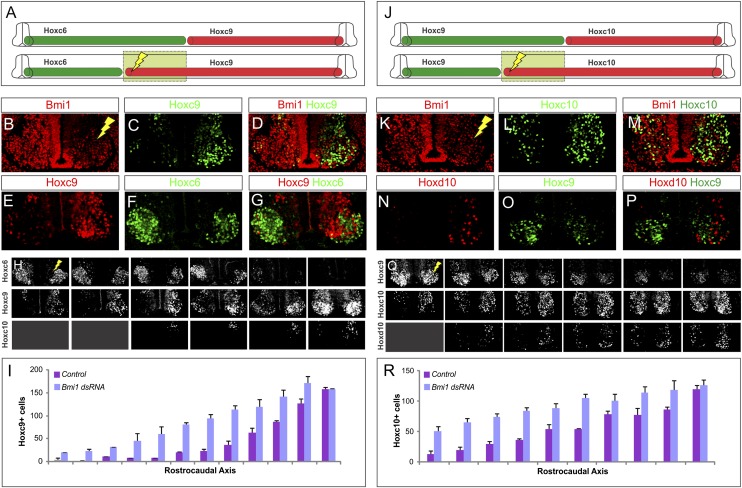
We next analyzed the effect of Bmi1 depletion on Hox expression. Hoxc9 is normally expressed at thoracic levels and restricted from brachial MNs. In embryos electroporated with Bmi1 dsRNA, we observed a marked derepression of Hoxc9 at brachial levels (Fig. 3A–D). Hoxc9 was ectopically expressed rostral to its endogenous domain, extending to mid-brachial levels (Fig. 3A,H,I; data not shown). As a central regulator of Hox patterning, Hoxc9 represses the expression of Hoxc6 (Dasen et al. 2003). Consistent with this repressive function, Hoxc6 was extinguished in 96% of ectopic Hoxc9 MNs such that the overall Hox topography of the thoracic region was extended rostrally (Fig. 3E–G; Supplemental Table S1).
Figure 3.
Effect of Bmi1 loss on Hox patterns in spinal MNs. (A) Summary of the effects of Bmi1 depletion at brachial levels. (B–D) Hoxc9 extends to brachial levels after Bmi1 depletion. (E–G) Hoxc6 is repressed in cells that express Hoxc9. (H) Serial sections showing extension of Hoxc9 and loss of Hoxc6. There is no change in Hoxc10 in MNs, although some Hoxc10+ cells are observed in dorsal regions. Results shown are typical of electroporations where >80% of cells have lost Bmi1. (I) Quantification of Hoxc9+ cells along the rostrocaudal axis of the electroporated and nonelectroporated halves of chick spinal cord HH stages 25–28. Results show averaged cell counts from two embryos. Error bars show standard error. (J) Effects of Bmi1 loss at thoracic/lumbar levels. (K–N) Hoxc10 and Hoxd10 are derepressed. (O,P) Ectopic Hoxd10 cells do not express Hoxc9. (Q) Serial section showing extension of Hox10 proteins and loss of Hoxc9. (R) Quantification of Hoxc10+ cells along the rostrocaudal axis of the electroporated and nonelectroporated halves of the spinal cord. Results show averaged cell counts from four embryos.
We next determined whether Bmi1 was required for regulation of Hox expression at other levels of the spinal cord. We first examined the effect of Bmi1 depletion on Hox expression at the thoracic–lumbar interface. Here we observed a derepression of Hoxc10 and Hoxd10, two Hox genes normally restricted to lumbar levels (Fig. 3J–M,Q,R). Similar to the effects of Hoxc9 derepression, ectopic Hoxc10 and Hoxd10 resulted in the repression of Hoxc9 (Fig. 3N–P). However, although we found similar levels of Bmi1 depletion, Hox10 genes were less effectively derepressed in MNs, with most ectopic cells found in the dorsal spinal cord (Fig. 3Q). Bmi1 depletion at the cervical–brachial interface was ineffective at producing a rostral shift in Hoxc6 expression (data not shown), likely due to relatively high PRC activities at this level. Nevertheless, these results suggest that PRC1 functions to repress posterior Hox genes generally, with the most pronounced effects on Hoxc9 expression.
Bmi1 is necessary for the development of MN topography
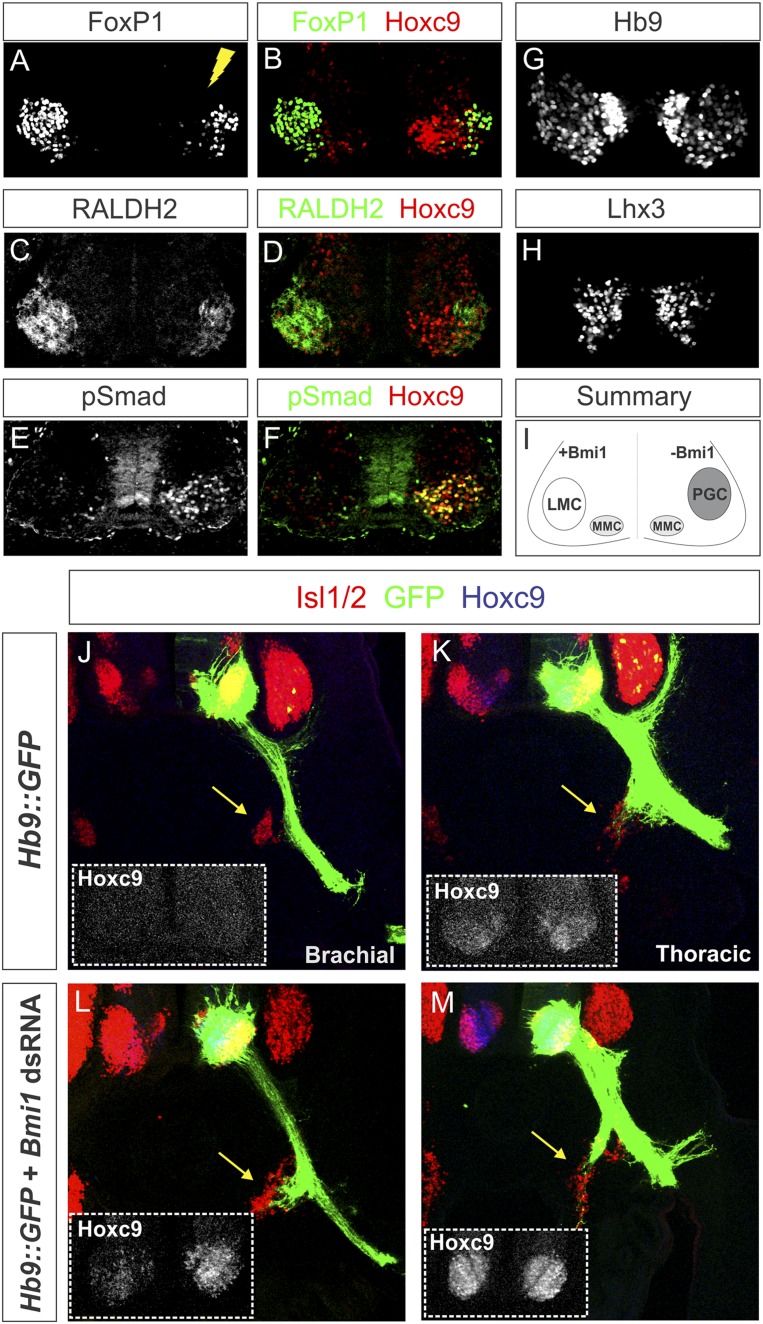
We next assessed whether changes in Hox patterns induced by Bmi1 depletion resulted in transformations of MN columnar identity. Brachial MNs can be defined by expression of Raldh2 and high FoxP1 levels, while thoracic PGC MNs are selectively labeled by pSmad1/5/8 (Dasen et al. 2008). After Bmi1 depletion, we observed an ablation of the LMC, as assessed by loss of Raldh2 and FoxP1 expression (Fig. 4A–D). In addition, ectopic pSmad+ MNs were detected in 25% of the Hoxc9+ MNs at brachial levels, demonstrating a transformation to a PGC fate (Fig. 4E,F; Supplemental Table S1). In contrast, expression of Lhx3 and Hb9 in axial projecting medial motor column (MMC) neurons was unchanged (Fig. 4G,H). Loss of Bmi1 at brachial levels therefore results in the derepression of Hoxc9, leading to a loss of LMC identity and the respecification to a PGC fate, indicating a selective effect on Hox-dependent MN subtypes (Fig. 4I).
Figure 4.
Bmi1 depletion converts forelimb LMC MNs to a PGC fate. (A–D) Expression of the LMC markers FoxP1 and Raldh2 is diminished after depletion of Bmi1 at brachial levels. (E,F) Expression of the PGC marker pSmad is detected at brachial levels and coexpresses Hoxc9. Not all ectopic Hoxc9 cells label with pSmad, as Hoxc9 is derepressed in non-MN cells. (G,H) Expression of pan-MN markers Hb9 and Lhx3 is not affected at brachial levels. (I) Summary showing changes in MN molecular identity following Bmi1 depletion. (J,K) Vibratome sections of chick embryos electroporated with a Hb9∷GFP construct to label motor axons (green). Sympathetic chain ganglia are indicated by yellow arrows and are stained by Isl1/2. (L,M) MNs coelectroporated with Bmi1 dsRNA and Hb9∷GFP express ectopic Hoxc9, and their axons are redirected to the sympathetic chain ganglia at brachial levels.
The brachial LMC can be further subdivided into smaller MN groups, known as motor pools. Hox genes are differentially expressed within the LMCs and define codes for pool fates (Dasen et al. 2005). We asked whether the transformations in MN identity elicited by Bmi1 depletion also affected motor pool organization. Staining for the brachial Hox genes Hoxa5, Hoxc6, and Hoxc8 showed that all Hox genes underwent a rostral shift in their expression (Supplemental Fig. S3). We also assessed two Hox-dependent motor pools, identified by their expression of the transcription factors Scip and Pea3 (Dasen et al. 2005). These pools were eroded within their normal domains, while cells in the rostral portion of the brachial compartment (which retain an LMC fate) acquired Scip+ or Pea3+ identities (Supplemental Fig. S3). Thus, Bmi1 actions are not restricted to motor column-defining Hox genes.
MN fate switch after Bmi1 depletion causes a redirection of motor axons
To further assess the consequences of respecifying the identity of LMC neurons, we examined the peripheral connectivity of brachial MNs lacking Bmi1. Brachial LMC axons normally project ventrolaterally toward the limb bud and avoid the sympathetic chain ganglia. Consistent with this pattern, electroporation of a construct expressing GFP under the control of the Hb9 promoter labeled motor axons that bypass the sympathetic ganglia and project to the limb (Fig. 4J). However, when Hb9∷GFP was coelectroporated with dsRNA against Bmi1, the axons of caudal LMC MNs innervated sympathetic ganglia (Fig. 4J–M). Changes in Hox patterning following depletion of Bmi1 therefore result in the respecification of motor columns at both the level of molecular identity and peripheral connectivity.
Bmi1 regulates brachial MN identity through modulation of Hoxc9 gene expression
The effects of Bmi1 depletion on Hoxc6 and MN organization could be mediated directly by Bmi1 or indirectly by changes in Hoxc9. Bmi1 depletion phenocopies the switch in MN fates observed after brachial Hoxc9 misexpression (Dasen et al. 2003), suggesting that the observed effects are solely due to ectopic Hoxc9. To test this, embryos were coelectroporated with dsRNAs targeting both Bmi1 and Hoxc9, which effectively depleted both proteins from MNs (Supplemental Fig. S4). Concurrent depletion of Bmi1 and Hoxc9 at brachial levels had no effect on Hoxc6 expression (Supplemental Fig. S4), consistent with the idea that Hoxc9 is necessary to repress brachial Hox genes. In addition, expression of FoxP1 in LMC MNs was not affected, and no ectopic pSmad+ MNs were observed (Supplemental Fig. S4). When Bmi1 and Hoxc9 expression was reduced at thoracic levels, we observed a phenotype similar to that seen in Hoxc9 knockout mice (Jung et al. 2010); PGC MNs were lost, Hoxc6 was derepressed, and MNs acquired an LMC fate (Supplemental Fig. S4). These data indicate that the phenotypes observed in the absence of Bmi1 are due to the derepression of Hoxc9 and not to a direct effect of Bmi1 on either MN identity or other Hox genes.
Bmi1 depletion does not affect Hox patterns in spinal progenitors
The pattern of Hox expression in the spinal cord is initially controlled through opposing gradients of FGFs and RA, which act on progenitors shortly after neurogenesis (Dasen and Jessell 2009). The rostrocaudal limits of Hox expression domains are later refined post-mitotically via cross-repressive interactions with other Hox genes (Dasen et al. 2003; Jung et al. 2010). Having demonstrated that Bmi1 is necessary to delineate the correct rostral boundaries of Hox genes expressed at thoracic and lumbar levels, we next asked at which stage in development Bmi1-mediated repression of Hox genes is established.
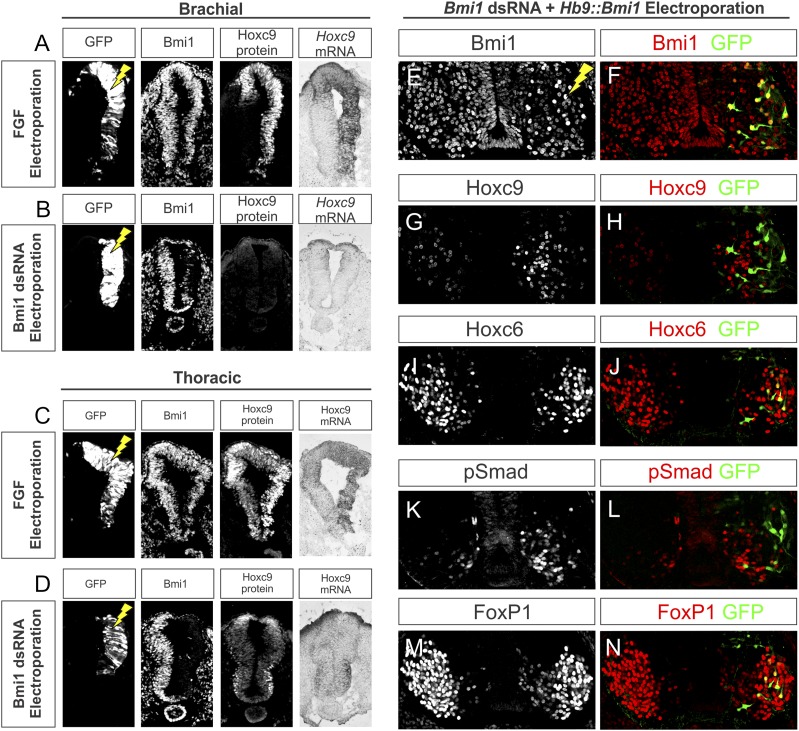
We first examined the potential involvement of Bmi1 in programming early Hox expression mediated by FGF signaling. From HH stages 10 to 14, neural tube progenitors express Hoxc9 mRNA and protein at thoracic levels, and elevation of FGF signaling at these stages induces rostral shifts in Hoxc9 expression, resulting in a loss of LMC MNs and a rostral extension of the PGC (Dasen et al. 2003). The similarity of the effects of elevated FGF8 to Bmi1 depletion led us to explore whether Bmi1 similarly regulates Hoxc9 in neural progenitors. We replicated the effects of FGF8 and analyzed embryos at a slightly later stage, demonstrating that at HH stage 12 +24 h, FGF8 overexpression induces a rostral shift of Hoxc9 mRNA and proteins in brachial neural tube progenitors but does not affect Bmi1 levels (Fig. 5A,C). Depletion of Bmi1 at HH stage 12 +24 h did not show similar changes in early Hox expression patterns, as Hoxc9 was not ectopically expressed in brachial progenitors (Fig. 5B,D). Ectopic Hoxc9 expression was observed at a slightly later stage (stage 14 +28 h), prior to the appearance of the first brachial post-mitotic MN (Supplemental Fig. S5). These results indicate that Bmi1 is not necessary at the progenitor phase of MN development for the patterning of Hox genes mediated by FGFs.
Figure 5.
The developmental timing of Bmi1-mediated control of Hox expression. (A) Elevation of FGF signaling induces Hoxc9 in brachial progenitors. HH stage 14 chick embryos were electroporated and analyzed after 24 h. A GFP expression construct was coelectroporated to grossly mark the electroporated region. Bmi1 expression is unchanged by FGF8 overexpression. (B) Effects of Bmi1 depletion on brachial progenitors. Hoxc9 immunostaining and in situ hybridization show that brachial Hoxc9 expression does not change in the absence of Bmi1. (C,D) Hoxc9 is grossly unaffected at thoracic levels after FGF elevation or Bmi1 depletion. (E,F) Rescue of Bmi1 expression at brachial levels of the spinal cord. Embryos were coelectroporated with Bmi1 dsRNA and Hb9∷Bmi1-ires-GFP. (G,H) Expression of Hoxc9 is shifted rostrally into the brachial level on the electroporated side of the cord; however, Hoxc9 is excluded from cells rescued for Bmi1. (I,J) Expression of Hoxc6 is reduced on the electroporated side of the cord but is restored in rescued cells. (K,L) pSmad is excluded from cells expressing GFP. (M,N) Expression of FoxP1 is restored in cells expressing GFP.
To gain further insight into the temporal requirement for PcG complexes in restricting Hox patterns, we examined the effects of selectively depleting PRC function from MN progenitors. Ezh2 is a core component of PRC2 and is necessary for depositing H3K27me3. To remove PRC2 function from MN progenitors, we crossed a floxed Ezh2 line (Su et al. 2003) to Olig2∷Cre mice (Dessaud et al. 2007) in which Cre is expressed in MN progenitors. Analysis of global H3K27me3 levels demonstrated that while the PRC2 mark was effectively depleted from progenitors, H3K27me3 was retained in post-mitotic MNs (Supplemental Fig. S6), likely due to compensation by the Ezh2 homolog Ezh1 (Ezhkova et al. 2011). We found no change in Hox patterns or MN organization over the rostrocaudal axis after conditional ablation of Ezh2 (Supplemental Fig. S6). We conclude that PRC2 function is not necessary within progenitors for correct expression of Hox genes, further supporting a late role for PRC function.
If PRC1 activity is deployed in early post-mitotic MNs, then selective late expression of Bmi1 should rescue the effects of early Bmi1 depletion. To test this, embryos were coelectroporated with both Bmi1 dsRNA and an Hb9∷Bmi1iresGFP construct and analyzed for changes in Hox expression and MN identity. Coelectroporation of these constructs at brachial levels effectively depleted levels of Bmi1 expression except in individual post-mitotic cells expressing Hb9∷Bmi1 (Fig. 5E,F). Because of differences in the electroporation efficiency between plasmids and dsRNAs, Bmi1-rescued MNs were typically surrounded by cells that lacked Bmi1. In cells rescued for Bmi1, 99% did not express Hoxc9, despite ectopic Hoxc9 expression in neighboring Bmi1-negative cells (Fig. 5G,H). Furthermore, 65% of Bmi1-rescued cells expressed Hoxc6 and 38% expressed FoxP1, but only 4% expressed pSmad (Fig. 5I–N; Supplemental Table S1). Thus, expression of Bmi1 at the post-mitotic stage can cell-autonomously rescue Hox patterning and MN identity.
Elevation of Bmi1 expression converts PGC MNs to an LMC fate
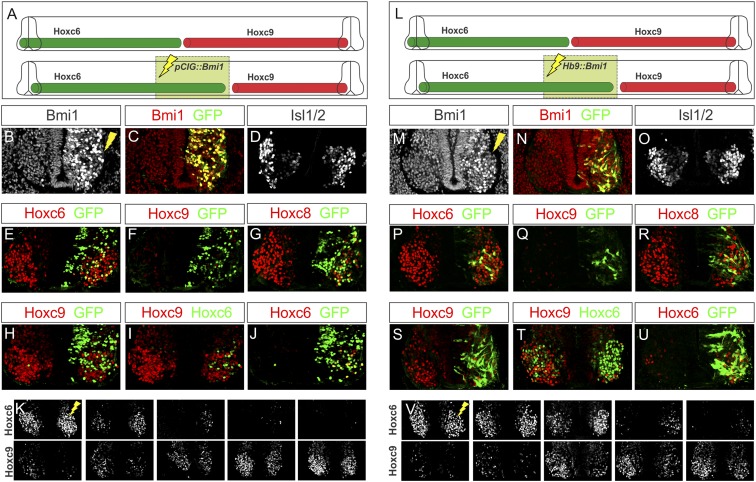
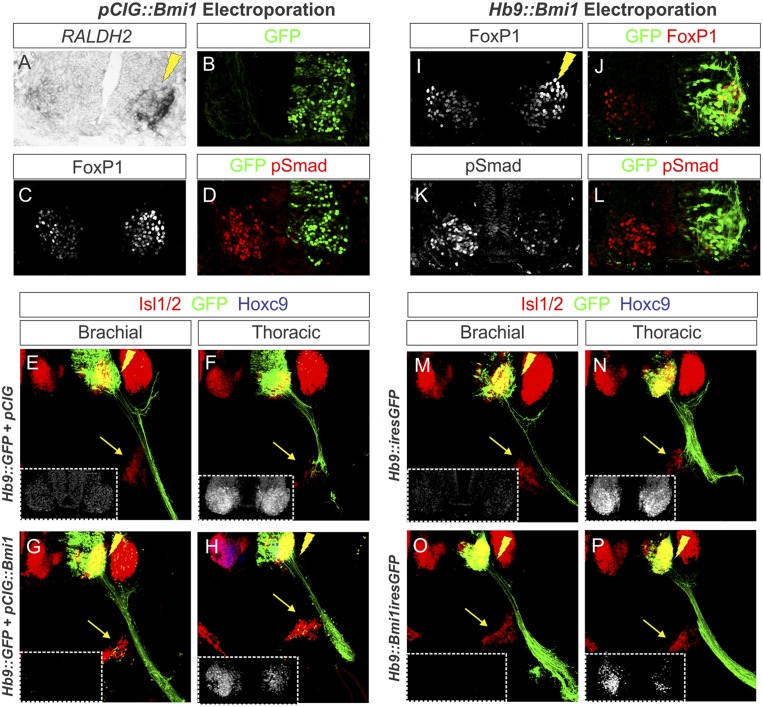
Mice overexpressing Bmi1 globally are characterized by a caudal shift in the expression of Hox genes within the mesoderm as well as an accompanying anterior transformation in vertebral identity (Alkema et al. 1995), suggesting that Hox patterning along the neuraxis may be similarly sensitive to Bmi1 levels. In order to investigate a potential role of Bmi1 dosage in controlling Hox expression in MNs, we electroporated a constitutive Bmi1-ires-nuclearGFP expression vector (pCIG∷Bmi1) into both brachial and thoracic neural tubes and assessed Hox patterning and columnar specification. Quantification of nuclear Bmi1 levels revealed an approximately fivefold elevation in protein levels in electroporated cells. Expression of Hb9 and Isl1/2 remained normal under conditions of elevated Bmi1, indicating that MN generation was unaffected (Fig. 6A–D; data not shown). In addition, expression of Hoxc6 in brachial LMC MNs was unaffected by Bmi1 elevation, and no ectopic Hoxc9 was detected (Fig. 6E,F). However, expression of Hoxc8 was diminished in caudal brachial LMC MNs, and these cells expressed ectopic Hoxa5 (Fig. 6G; data not shown). At thoracic segments, cells expressing high levels of Bmi1 displayed a marked decrease in Hoxc9, and MNs lacking Hoxc9 ectopically expressed Hoxc6 (Fig. 6H–K; Supplemental Table S1). These results, in conjunction with the effects of Bmi1 depletion, suggest that a subset of Hox genes is sensitive to Bmi1 levels.
Figure 6.
Elevation of Bmi1 expression at thoracic levels extinguished Hoxc9. (A) Schematic of the effects of Bmi1 elevation at thoracic levels of the spinal cord. (B,C) Bmi1 expression is elevated after electroporation of a pCIG∷Bmi1 expression construct. (D) Bmi1 elevation does not affect MN generation, marked by Isl1/2. (E–G) At brachial levels, Hoxc6 is unaffected, while Hoxc8 expression is diminished. No ectopic Hoxc9 is observed. (H–J) At thoracic levels, Hoxc9 expression is ablated, and ectopic Hoxc6 is detected. (K) Serial sections showing that Hoxc6 expression is not affected at brachial levels but is extended caudally. (L–V) Effects of post-mitotic overexpression of Bmi1 using an Hb9∷Bmi1-ires-EGFP expression construct. (M–O) Bmi1 expression is increased on the electroporated half of the embryo but not within the progenitor zone. Isl1/2 expression is unchanged. (P–R) Effects of Bmi1 elevation on brachial Hox expression. (S–U) Hoxc9 expression in GFP+ cells is reduced, and ectopic Hoxc6 is observed. (V) Extension of Hoxc6 to thoracic levels.
To further characterize the effects of elevated Bmi1 expression, we analyzed motor column specification and peripheral connectivity. At brachial levels, Bmi1 elevation did not affect motor column identity, as assessed by FoxP1 and Raldh2 expression, consistent with a preservation of Hoxc6 expression (data not shown). However, at thoracic levels, pSmad expression was ablated, and MNs ectopically expressed high FoxP1 levels and Raldh2, indicating a transformation of the PGC into an LMC identity (Fig. 7A–D; Supplemental Table S1). The fate switch observed after Bmi1 overexpression was reflected in the projection patterns of MNs overexpressing Bmi1, which failed to innervate the sympathetic chain ganglia (Fig. 7E–H). Thus, elevation of Bmi1 levels has the inverse effect of Bmi1 depletion, leading to a caudal shift in Hox expression and a transformation of MN identity.
Figure 7.
Effects of Bmi1 elevation on MN columnar identity. (A–D) Effects of Bmi1 elevation on MN columnar identity at thoracic levels. LMC markers Raldh2 and (high) FoxP1 are ectopically expressed, while pSmad expression is extinguished. (E,F) Vibratome sections of chick embryos electroporated with Hb9∷GFP construct to label motor axons (green). Sympathetic chain ganglia are indicated by yellow arrows. (G,H) MNs coelectroporated with pCIG∷Bmi1 have reduced Hoxc9 expression, and their axons are directed away from the sympathetic chain ganglia. (I–L) Effects of Hb9∷Bmi1-ires-EGFP electroporation at thoracic levels. FoxP1 expression is elevated, while pSmad expression is reduced. (M–P) Vibratome sections of chick embryos electroporated with Hb9-GFP construct to label motor axons (green). Sympathetic chain ganglia are indicated by yellow arrows. MNs coelectroporated with Hb9∷Bmi1-ires-EGFP have reduced Hoxc9 expression, and their axons are redirected away from the sympathetic chain ganglia.
We further reasoned that if Bmi1 acts post-mitotically to regulate MN identity, then late elevation of Bmi1 levels should recapitulate the phenotypes observed when Bmi1 is overexpressed indiscriminately within the spinal cord. We therefore electroporated Hb9∷Bmi1iresGFP and evaluated MN development. As with pCIG∷Bmi1, expression of Hb9 and Isl1/2 were unaffected, indicating normal MN generation (Fig. 6L–O; data not shown). At brachial levels, there was no change in Hoxc6, and LMC identity was retained, although Hoxc8 expression was diminished (Fig. 6P–R). At thoracic levels, electroporated cells extinguished Hoxc9 and expressed Hoxc6 (Fig. 6S–V). Additionally, post-mitotic Bmi1 overexpression ablated the PGC, thoracic MNs acquired an LMC fate, and motor axons bypassed the sympathetic ganglia (Fig. 7I–P). Thus, Hb9∷Bmi1 embryos displayed the same Hox and columnar transformation phenotypes as those expressing elevated Bmi1 throughout the spinal cord, providing supporting evidence that Bmi1 activity regulates Hox expression post-mitotically.
Bmi1 expression overrides the effects of FGF signaling on MN organization
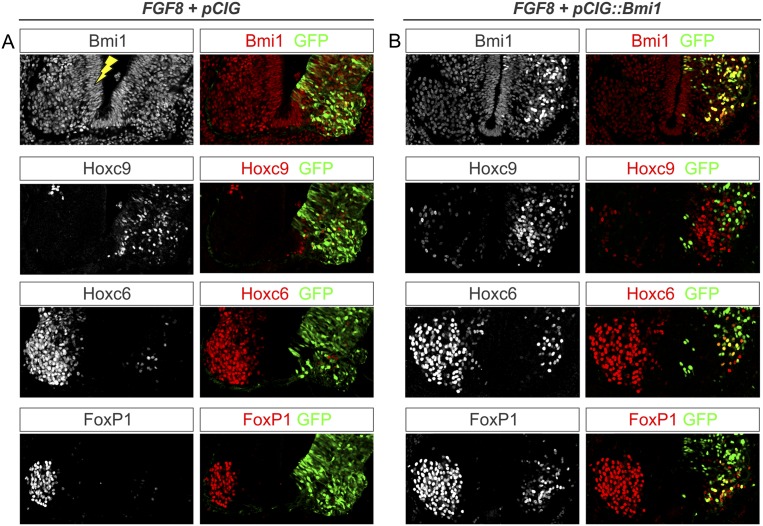
Although the levels of Bmi1 are normally homogeneous throughout the spinal cord (see Supplemental Fig. S6), Bmi1 gain and loss of function elicit complimentary phenotypes, suggesting that Hox genes may be sensitive to PRC1 activity levels at the time of MN differentiation. Because Hox patterns are initially controlled through signaling gradients that vary over the rostrocaudal axis, we next set out to further define the epistatic relationship between FGF signaling and levels of PRC1 activity. Two lines of evidence indicate that Bmi1 acts independently of FGF to control Hox patterns. First, while elevation of FGF at brachial levels induces Hoxc9 in progenitors, removal of Bmi1 fails to do so, suggesting that Bmi1 acts temporally downstream from FGF. Second, manipulation of Bmi1 levels under the Hb9 promoter is capable of switching Hox patterns in MNs, suggesting that PRC1 actions are deployed near the time of MN differentiation. These observations suggest a model in which PRC1 activities at Hox loci along the rostrocaudal axis are established subsequent to the initial patterns controlled by morphogens.
If PRC1 activities are set downstream from FGF, then elevation of Bmi1 levels might be sufficient to “reprogram” changes in Hox expression patterns elicited by elevated FGF8. We therefore tested whether the rostral shift in Hoxc9 expression induced by FGF8 could be overcome by coexpressing Bmi1. We coelectroporated CMV∷FGF8 with pCIG∷Bmi1 and assessed Hox patterns and MN fate specification. Consistent with previous studies, expression of FGF8 alone abolished Hoxc6 and FoxP1 expression and induced Hoxc9 at brachial levels (Fig. 8A). After coexpression of FGF8 and Bmi1, we found that Hoxc6 and FoxP1 were effectively rescued in ∼30% of cells that ectopically expressed Bmi1 and that these cells lacked Hoxc9 (Fig. 8B; Supplemental Table S1). Because of the non-cell-autonomous nature of the FGF8-mediated conversion, rescued cells were typically surrounded by cells that expressed ectopic Hoxc9. These results demonstrate that expression of Bmi1 is sufficient to cell-autonomously erase the positional identity imposed on progenitor cells by early signaling gradients.
Figure 8.
PRC1 reprogramming of early Hox patterning. (A) Effects of elevating FGF8 at brachial levels. Hoxc9 is expressed ectopically at brachial levels on the electroporated side of the cord, while expression of Hoxc6 is ablated. Expression of FoxP1 is also ablated. (B) Effects of pCIG∷Bmi1 and CMV∷FGF8 coelectroporation. Bmi1 expression levels are increased in the electroporated cells. Hoxc9 is expressed ectopically at brachial levels, except in cells expressing Bmi1/GFP. Expression of Hoxc6 is reduced at brachial levels, but cells expressing Bmi1/GFP are rescued for Hoxc6. Likewise, expression of FoxP1 is reduced at brachial levels but rescued in cells expressing Bmi1.
Discussion
PRCs are thought to contribute to the molecular memory of developmental programs by ensuring that certain genes remain silenced long after the disappearance of early patterning signals. While Hox genes are well-defined targets of PRC activities, the mechanisms and timing of their contributions to Hox regulation during neuronal specification are poorly defined. Our studies suggest that Hox-dependent MN subtypes are sensitive to Bmi1 levels at the time of their differentiation, and altering Bmi1 activity in vivo is sufficient to interconvert the fates of LMC and PGC neurons. The apparent sensitivity of certain Hox genes, such as Hoxc9, to Bmi1 levels indicates that one feature of PRC function is to set and maintain the anterior limits of key Hox genes at the interface between columnar subtypes. We discuss these findings in the context of the developmental regulation of Hox genes and suggest possible mechanisms of PRC actions in restricting Hox patterns during MN differentiation.
Control of Hox expression in the developing spinal cord by PRC1
Analysis of the genomic binding profiles of PRC components in ES cells suggests a central role in the control of a large repertoire of developmentally critical genes (Boyer et al. 2006; Bracken et al. 2006; Lee et al. 2006). In this study, we found a more restricted role for the core PRC1 component Bmi1 in the organization of MN subtypes along the rostrocaudal axis. Ablation of Bmi1 expression has no affect on progenitor patterning or the differentiation of MNs as a class and is not required in MN columns that are specified independent of Hox function, such as the MMC. In contrast, Bmi1 activities are necessary for the segregation of limb-innervating LMC MNs from thoracic PGC neurons. We found that manipulation of Bmi1 levels can switch the fates of MNs at brachial and thoracic levels of the spinal cord, leading to alterations in their molecular identity and peripheral connectivity. These results indicate that PRC1 has a critical but selective function in MN subtype specification.
The actions of Bmi1 in MN differentiation are mediated through their effects on Hox genes and are distinct from regulatory interactions previously shown to influence Hox expression in the spinal cord. Hox patterns are known to be set during two phases of regulation in progenitors and early post-mitotic neurons. Initially, gradients of FGFs and RA install a rough pattern of future Hox expression during formation of the neural tube, a pattern later refined through cross-repressive interactions (Dasen and Jessell 2009). At the brachial–thoracic interface, the repressive actions of Hoxc9 are known to be essential in defining the posterior boundary of Hoxc6 as well as excluding Hox4–Hox8 gene paralogs from thoracic levels (Jung et al. 2010). The absence of the PRC-associated mark at Hox genes repressed by Hoxc9, however, indicates that this repressive event does not require PRC function.
Our results indicate that PRC activities are used to selectively delimit the anterior extent of Hox expression in MNs. We found that at a given segmental level, Hox genes expressed at more caudal regions are characterized by the presence of PRC marks (Fig. 9A). At brachial levels, the Hoxc9 gene bears the H3K27me3 mark and is occupied by Bmi1, suggesting PRC1 function may be required for its exclusion. Consistent with this idea, depletion of Bmi1 at brachial levels leads to ectopic Hoxc9 rostral to its normal boundary. Under these conditions, the loss of Hoxc6 from brachial MNs appears to be a consequence of ectopic Hoxc9, since combined depletion of Bmi1 and Hoxc9 preserves Hoxc6 and LMC specification (Fig. 9A). These findings indicate that PRC1 activities are integrated with Polycomb-independent Hox cross-repression to ensure the establishment of proper Hox boundaries in MNs.
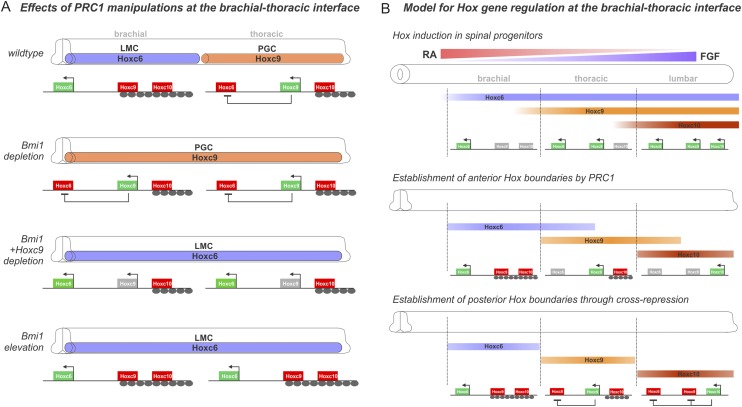
Figure 9.
Model for PRCs in controlling Hox expression and MN columnar topography. (A) Summary of the effects of Bmi1 manipulations on Hox expression and columnar position at the brachial/thoracic boundary. Bmi1 depletion leads to ectopic Hoxc9 expression at thoracic levels and converts LMC MNs to a PGC fate. Loss of Hoxc6 is due to ectopic Hoxc9, as combined Hoxc9 and Bmi1 depletion rescues the phenotype. Elevation of Bmi1 at thoracic levels represses Hoxc9 and converts PGC MNs to an LMC fate. (B) Control of Hox gene expression in MNs by signaling gradients, PRCs, and cross-repression. Hox transcription is initiated in neural progenitors through the actions of positional gradients. Anterior boundaries are reinforced through the action of PRC1. Posterior boundaries are controlled through Hox cross-repressive interactions.
Timing of PRC actions in MN development
While the chromatin marks deposited by PRC2 in all three germ layers are sequentially removed from Hox loci at early stages of development (Soshnikova and Duboule 2009), our data suggest a relatively late role for PRC1 in controlling the final pattern of Hox protein expression. We found that both elevation of FGF8 signaling and loss of Bmi1 at brachial levels induce Hoxc9 and convert LMC MNs to a PGC fate. However, the progenitor pattern of Hoxc9 is unchanged under conditions of Bmi1 depletion, and unlike the effects FGF8 exposure, ectopic Hoxc9 is observed only just before the first post-mitotic MNs appear. Moreover, loss of the H3K27me3 mark from MN progenitors fails to alter Hox patterns in MNs, while post-mitotic Bmi1 expression is sufficient to rescue early Bmi1 depletion. These observations point to a relatively late role for PRC1 to refine and maintain Hox patterns subsequent to early signaling gradients.
If Hox gene expression is initially set in neural progenitors, why might PRC1 act in early post-mitotic neurons? One possible scenario is that a temporal delay in PRC1-mediated repression might be used to ensure this restriction is developmentally linked to Hox cross-repressive activities. While the transcription of the Hoxc6, Hoxc9, and Hoxc10 genes are sequentially activated along the rostrocaudal axis at the progenitor phase (Dasen et al. 2003), Hox proteins are either weakly expressed or undetectable, with most proteins observed only in post-mitotic neurons. Thus, Hox cross-repressive interactions can only emerge as MNs differentiate. For the case of Hoxc9, the restriction of its expression to thoracic levels has an essential role in creating a permissive environment for the emergence of the Hox4–Hox8 gene network required for LMC diversification (Jung et al. 2010). PRC1 action may therefore be deployed only at the time necessary to establish the final pattern of Hoxc9 (Fig. 9B).
PcG function in vertebrate Hox gene regulation
While the roles of PcG complexes in regulating Hox expression in Drosophila have been extensively studied, it is unclear whether the mechanisms of recruitment are preserved in vertebrates. In Drosophila, PcG complexes are recruited by PREs, conserved sequence elements that are capable of conferring PRC-dependent repression. In contrast, PRC1 and PRC2 appear to be broadly distributed across Hox loci in vertebrates (Bracken et al. 2006; Lee et al. 2006; Rinn et al. 2007), raising the question of whether their recruitment relies on PREs. Recently, the presence of a PRE within the HoxD locus that confers PcG-dependent repression has been reported (Woo et al. 2010). While the identification of additional PREs will facilitate analysis of Hox gene regulation in vertebrates, one confounding factor may be varied strategies used to regulate Hox genes in different cell types, and if additional PREs exist, they will have distinct functions within a particular cellular context.
Our results suggest that that in MNs, the Hoxc9 gene is particularly sensitive to the loss of PRC1 activity. We found that Bmi1 depletion fails to derepress Hoxc6 at cervical levels, and at thoracic levels, the majority of ectopic Hox10+ cells are found in the dorsal spinal cord. The apparent bias toward Hoxc9 regulation in MNs is consistent with microarray analysis of Bmi1 mutant neurospheres, showing Hox9 genes as the most significantly derepressed, with few changes in other Hox paralogs (Molofsky et al. 2003). The regulation of Hoxc9 by Bmi1 may reflect a greater need to ensure a restricted domain of expression, as Hoxc9 has a unique role in controlling MN topography, and unlike most Hox genes, this activity is not compensated by other paralogs (Jung et al. 2010). The HoxC locus, like the HoxD cluster (Tschopp et al. 2012), may contain regulatory elements that are geared toward the regulation of only a subset of critical Hox genes in the context of neural tissue.
PRC1 activity levels control Hox expression in MNs
We observed that overexpression of Bmi1 at thoracic levels represses Hoxc9 and converts PGC MNs to an LMC fate. These effects are somewhat surprising, given that at this level of the spinal cord, the Hoxc9 gene does not carry the H3K27me3 mark thought to be necessary for PRC1 recruitment. Neighboring genes in the HoxC cluster, such as Hoxc10, do, however, contain the PRC2 mark at thoracic levels; thus, it is possible that the effects of PRC1 overexpression reflect an action of neighboring Hox genes on Hoxc9. One plausible scenario is that Bmi1 elevation retains Hoxc9 in association with transcriptionally inactive regions of the cluster. Alternatively, Bmi1 may act like a DNA-binding transcription factor, with a bias toward regulating Hoxc9. DNA-binding activities have been reported for Bmi1 and its homologs, and a Bmi1 response element has been shown to be present in the Hoxc9 gene (Meng et al. 2010).
Regardless of the precise mechanisms, the effects of Bmi1 depletion and overexpression indicate that in MNs, HoxC genes are sensitive to the overall levels of PRC1 activities. If PRC1 activity levels are critical in defining Hox patterns, how might this system operate during normal development? The level and distribution of Bmi1 expression remains constant over the rostrocaudal axis, indicating that PRC1 activities are not set through graded expression. One possibility is that Bmi1 acts as a limiting factor that can grossly raise or lower the overall activity of PRC1, while additional factors are responsible for the graded activity of the complex over the rostrocaudal axis. One candidate for such a partner is a long noncoding RNA (lncRNA), which appears to regulate PRC binding at several loci, including Hox clusters (Spitale et al. 2011). Differential transcription of such an RNA over the rostrocaudal axis could have the effect of grading PRC recruitment to Hox loci. The recent identification of thousands of lncRNAs acting both in cis and in trans (Guttman et al. 2011) will confound defining whether they contribute to Hox gene regulation, and moreover, genetic evidence supporting an instructive role for lncRNAs is currently lacking (Schorderet and Duboule 2011). Nevertheless, if such a graded component exists, it would provide an attractive candidate for linking the early positional gradients to the chromatin modifications necessary for precisely controlling Hox patterns in the CNS.
Materials and methods
ChIP assays
Cervical, brachial, thoracic, and lumbar spinal cords were dissected, and tissues were homogenized in 1.1% formaldehyde using a dounce B homogenizer. Chromatin was extracted and fragmented by sonication. Chromatin extracts (20 μg) were incubated overnight at 4°C with 4–5 μg of either specific antibodies or species-matched immunoglobulins (IgGs). Protein A/G-agarose beads (Roche) were added for 2.5 h at 4°C, and complexes were washed seven times with RIPA and eluted with 1% SDS. DNA–protein de-cross-linking was performed overnight at 65°C followed by RNase and then proteinase K treatment. DNA was purified via QIAprep spin columns (Qiagen). Hox promoter regions were amplified using SYBR Green PCR master mix (Applied Biosystems) and detected by quantitative PCR. Oligonucleotide sequences are provided in the Supplemental Material. Fold enrichments were calculated over IgG: Fold enrichment = 2−(ΔCt), where ΔCt = (CtIP − CtIgG). Fold enrichments over 10% input were similar to fold enrichment over IgG. The following antibodies were used: rabbit anti-H3K27me3 (Millipore), rabbit anti-H3K4me3 (Millipore), mouse anti-Bmi1 (Millipore), and rabbit anti-mouse IgG (Millipore).
In ovo electroporation
Electroporation of plasmids and dsRNAs was performed as described (Dasen et al. 2005). To identify electroporated neurons, siRNAs (final concentration 8.5 mg/mL) were combined with a nuclear LacZ expression plasmid (final concentration 0.5 mg/mL). The target sequence against chick Bmi1 was 5′-AGAACAGACUGGAACGAAAUU-3′. Results are representative of at least two electroporated embryos in which the electroporation efficiency in MNs was >60%. Quantification of the effects of gene manipulations is shown in Supplemental Table S1.
In situ hybridization and immunohistochemistry
In situ hybridization and immunohistochemistry were performed as described (Jung et al. 2010). GFP-labeled axons were visualized in projections of confocal Z-stacks (500–1000 μm). Antibodies against Hox proteins, LIM HD proteins, and other proteins were generated as described (Dasen et al. 2005, 2008). Additional antibodies were used as follows: mouse anti-Bmi1 (1:600; Millipore), rabbit anti-GFP (1:1000; Invitrogen), rabbit anti-H3K27me3 (Millipore), and rabbit anti-EED (1:1000; Abcam). Quantification of protein levels was performed as previously described (Dasen et al. 2008).
RNA isolation and RT–PCR
RNA was isolated from tissues via TRIzol LS reagent (Invitrogen) according to the manufacturer's instructions. First strand cDNA was synthesized from ∼100 ng of RNA using the SuperScript II First Strand Synthesis system for RT–PCR (Invitrogen). cDNA was amplified using SYBR Green PCR master mix and detected by quantitative PCR. Fold enrichments were calculated over GAPDH: Fold enrichment = 2−(ΔCt), where ΔCt = (CtHox/Bmi1 − CtGAPDH).
Acknowledgments
We thank Myungin Baek, Catarina Catela, and Denis Duboule for comments on the manuscript, and Steve Burden, Gord Fishell, and Jessica Treisman for discussions. We also thank Sean Morrison and Maarten van Lohuizen for Bmi1 mutants, and Alexander Tarakhovsky for the Ezh2 conditional allele. J.D. is supported by grants from the McKnight Foundation, Alfred P. Sloan, Project ALS, NYSTEM, HHMI and NIH R01 NS062822.
Footnotes
Supplemental material is available for this article.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.199133.112.
References
- Akasaka T, van Lohuizen M, van der Lugt N, Mizutani-Koseki Y, Kanno M, Taniguchi M, Vidal M, Alkema M, Berns A, Koseki H 2001. Mice doubly deficient for the Polycomb Group genes Mel18 and Bmi1 reveal synergy and requirement for maintenance but not initiation of Hox gene expression. Development 128: 1587–1597 [DOI] [PubMed] [Google Scholar]
- Akizu N, Estaras C, Guerrero L, Marti E, Martinez-Balbas MA 2010. H3K27me3 regulates BMP activity in developing spinal cord. Development 137: 2915–2925 [DOI] [PubMed] [Google Scholar]
- Alkema MJ, van der Lugt NM, Bobeldijk RC, Berns A, van Lohuizen M 1995. Transformation of axial skeleton due to overexpression of bmi-1 in transgenic mice. Nature 374: 724–727 [DOI] [PubMed] [Google Scholar]
- Boyer LA, Plath K, Zeitlinger J, Brambrink T, Medeiros LA, Lee TI, Levine SS, Wernig M, Tajonar A, Ray MK, et al. 2006. Polycomb complexes repress developmental regulators in murine embryonic stem cells. Nature 441: 349–353 [DOI] [PubMed] [Google Scholar]
- Bracken AP, Dietrich N, Pasini D, Hansen KH, Helin K 2006. Genome-wide mapping of Polycomb target genes unravels their roles in cell fate transitions. Genes Dev 20: 1123–1136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunk BP, Martin EC, Adler PN 1991. Drosophila genes Posterior Sex Combs and Suppressor two of zeste encode proteins with homology to the murine bmi-1 oncogene. Nature 353: 351–353 [DOI] [PubMed] [Google Scholar]
- Dasen JS, Jessell TM 2009. Hox networks and the origins of motor neuron diversity. Curr Top Dev Biol 88: 169–200 [DOI] [PubMed] [Google Scholar]
- Dasen JS, Liu JP, Jessell TM 2003. Motor neuron columnar fate imposed by sequential phases of Hox-c activity. Nature 425: 926–933 [DOI] [PubMed] [Google Scholar]
- Dasen JS, Tice BC, Brenner-Morton S, Jessell TM 2005. A Hox regulatory network establishes motor neuron pool identity and target-muscle connectivity. Cell 123: 477–491 [DOI] [PubMed] [Google Scholar]
- Dasen JS, De Camilli A, Wang B, Tucker PW, Jessell TM 2008. Hox repertoires for motor neuron diversity and connectivity gated by a single accessory factor, FoxP1. Cell 134: 304–316 [DOI] [PubMed] [Google Scholar]
- Dessaud E, Yang LL, Hill K, Cox B, Ulloa F, Ribeiro A, Mynett A, Novitch BG, Briscoe J 2007. Interpretation of the sonic hedgehog morphogen gradient by a termporal adaptation mechanism. Nature 450: 717–721 [DOI] [PubMed] [Google Scholar]
- Ezhkova E, Lien WH, Stokes N, Pasolli HA, Silva JM, Fuchs E 2011. EZH1 and EZH2 cogovern histone H3K27 trimethylation and are essential for hair follicle homeostasis and wound repair. Genes Dev 25: 485–498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fasano CA, Phoenix TN, Kokovay E, Lowry N, Elkabetz Y, Dimos JT, Lemischka IR, Studer L, Temple S 2009. Bmi-1 cooperates with Foxg1 to maintain neural stem cell self-renewal in the forebrain. Genes Dev 23: 561–574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guttman M, Donaghey J, Carey BW, Garber M, Grenier JK, Munson G, Young G, Lucas AB, Ach R, Bruhn L, et al. 2011. lincRNAs act in the circuitry controlling pluripotency and differentiation. Nature 477: 295–300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirabayashi Y, Suzki N, Tsuboi M, Endo TA, Toyoda T, Shinga J, Koseki H, Vidal M, Gotoh Y 2009. Polycomb limits the neurogenic competence of neural precursor cells to promote astrogenic fate transition. Neuron 63: 600–613 [DOI] [PubMed] [Google Scholar]
- Jessell TM 2000. Neuronal specification in the spinal cord: Inductive signals and transcriptional codes. Nat Rev Genet 1: 20–29 [DOI] [PubMed] [Google Scholar]
- Jung H, Lacombe J, Mazzoni EO, Liem KF Jr, Grinstein J, Mahony S, Mukhopadhyay D, Gifford DK, Young RA, Anderson KV, et al. 2010. Global control of motor neuron topography mediated by the repressive actions of a single hox gene. Neuron 67: 781–796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurgens G 1985. A group of genes controlling the spatial expression of the bithorax complex in Drosophila. Nature 316: 153–155 [Google Scholar]
- Khalil AM, Guttman M, Huarte M, Garber M, Raj A, Morales DR, Thomas K, Presser A, Bernstein BE, van Oudenaarden A, et al. 2009. Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proc Natl Acad Sci 106: 11667–11672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee TI, Jenner RG, Boyer LA, Guenther MG, Levine SS, Kumar RM, Chevalier B, Johnstone SE, Cole MF, Isono K, et al. 2006. Control of developmental regulators by Polycomb in human embryonic stem cells. Cell 125: 301–313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis EB 1978. A gene complex controlling segmentation in Drosophila. Nature 276: 565–570 [DOI] [PubMed] [Google Scholar]
- Mallo M, Wellik DM, Deschamps J 2010. Hox genes and regional patterning of the vertebrate body plan. Dev Biol 344: 7–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin EC, Adler PN 1993. The Polycomb group gene Posterior Sex Combs encodes a chromosomal protein. Development 117: 641–655 [DOI] [PubMed] [Google Scholar]
- Meng S, Luo M, Sun H, Yu X, Shen M, Zhang Q, Zhou R, Ju X, Tao W, Liu D, et al. 2010. Identification and characterization of Bmi-1-responding element within the human p16 promoter. J Biol Chem 285: 33219–33229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molofsky AV, Pardal R, Iwashita T, Park IK, Clarke MF, Morrison SJ 2003. Bmi-1 dependence distinguishes neural stem cell self-renewal from progenitor proliferation. Nature 425: 962–967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinn JL, Kertesz M, Wang JK, Squazzo SL, Xu X, Brugmann SA, Goodnough LH, Helms JA, Farnham PJ, Segal E, et al. 2007. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell 129: 1311–1323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schorderet P, Duboule D 2011. Structural and functional differences in the long non-coding RNA Hotair in mouse and human. PLoS Genet 7: e1002071 doi: 10.1371/journal.pgen.1002071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuettengruber B, Cavalli G 2009. Recruitment of polycomb group complexes and their role in the dynamic regulation of cell fate choice. Development 136: 3531–3542 [DOI] [PubMed] [Google Scholar]
- Shirasaki R, Pfaff SL 2002. Transcriptional codes and the control of neuronal identity. Annu Rev Neurosci 25: 251–281 [DOI] [PubMed] [Google Scholar]
- Simon JA, Kingston RE 2009. Mechanisms of Polycomb gene silencing: Knowns and unknowns. Nat Rev Mol Cell Biol 10: 697–708 [DOI] [PubMed] [Google Scholar]
- Simon J, Chiang A, Bender W 1992. Ten different Polycomb group genes are required for spatial control of the abdA and AbdB homeotic products. Development 114: 493–505 [DOI] [PubMed] [Google Scholar]
- Sing A, Pannell D, Karaiskakis A, Sturgeon K, Djabali M, Ellis J, Lipshitz HD, Cordes SP 2009. A vertebrate polycomb response element governs segmentation of the posterior hindbrain. Cell 138: 885–897 [DOI] [PubMed] [Google Scholar]
- Soshnikova N, Duboule D 2009. Epigenetic temporal control of mouse Hox genes in vivo. Science 324: 1320–1323 [DOI] [PubMed] [Google Scholar]
- Spitale RC, Tsai M-C, Chang HY 2011. RNA templating the epigenome. Epigenetics 6: 539–543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Struhl G 1981. A gene product required for correct initiation of segmental determination in Drosophila. Nature 293: 36–41 [DOI] [PubMed] [Google Scholar]
- Su IH, Basavaraj A, Krutchinsky AN, Hobert O, Ullrich A, Chait BT, Tarakhovsky A 2003. Ezh2 controls B cell development through histone H3 methylation and Igh rearrangement. Nat Immunol 4: 124–131 [DOI] [PubMed] [Google Scholar]
- Tschopp P, Christen AJ, Duboule D 2012. Bimodal control of Hoxd gene transcription in the spinal cord defines two regulatory subclusters. Development 139: 929–939 [DOI] [PubMed] [Google Scholar]
- van der Lugt NM, Domen J, Linders K, van Roon M, Robanus-Maandag E, te Riele H, van der Valk M, Deschamps J, Sofroniew M, van Lohuizen M, et al. 1994. Posterior transformation, neurological abnormalities, and severe hematopoietic defects in mice with a targeted deletion of the bmi-1 proto-oncogene. Genes Dev 8: 757–769 [DOI] [PubMed] [Google Scholar]
- van der Lugt NM, Alkema M, Berns A, Deschamps J 1996. The Polycomb-group homolog Bmi-1 is a regulator of murine Hox gene expression. Mech Dev 58: 153–164 [DOI] [PubMed] [Google Scholar]
- van Lohuizen M, Frasch M, Wientjens E, Berns A 1991. Sequence similarity between the mammalian bmi-1 proto-oncogene and the Drosophila regulatory genes Psc and Su(z)2. Nature 353: 353–355 [DOI] [PubMed] [Google Scholar]
- Woo CJ, Kharchenko PV, Daheron L, Park PJ, Kingston RE 2010. A region of the human HOXD cluster that confers polycomb-group responsiveness. Cell 140: 99–110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yap KL, Li S, Munoz-Cabello AM, Raguz S, Zeng L, Mujtaba S, Gil J, Walsh MJ, Zhou M-M 2010. Molecular interplay of the non-coding RNA ANRIL and methylated histone H3 lysine 27 by polycomb CBX7 in transcriptional silencing of INK4a. Mol Cell 38: 662–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J, Sun BK, Erwin JA, Song J-J, Lee JT 2008. Polycomb proteins targeted by a short repeat RNA to the mouse X-chromosome. Science 322: 750–756 [DOI] [PMC free article] [PubMed] [Google Scholar]