Abstract
The survival of all microbes depends upon their ability to respond to environmental challenges. To establish infection, pathogens such as Candida albicans must mount effective stress responses to counter host defences while adapting to dynamic changes in nutrient status within host niches. Studies of C. albicans stress adaptation have generally been performed on glucose-grown cells, leaving the effects of alternative carbon sources upon stress resistance largely unexplored. We have shown that growth on alternative carbon sources, such as lactate, strongly influence the resistance of C. albicans to antifungal drugs, osmotic and cell wall stresses. Similar trends were observed in clinical isolates and other pathogenic Candida species. The increased stress resistance of C. albicans was not dependent on key stress (Hog1) and cell integrity (Mkc1) signalling pathways. Instead, increased stress resistance was promoted by major changes in the architecture and biophysical properties of the cell wall. Glucose- and lactate-grown cells displayed significant differences in cell wall mass, ultrastructure, elasticity and adhesion. Changes in carbon source also altered the virulence of C. albicans in models of systemic candidiasis and vaginitis, confirming the importance of alternative carbon sources within host niches during C. albicans infections.
Introduction
Most microbes inhabit microenvironments that are in a constant state of flux. This is particularly true for clinically important microbial pathogens which must counter the biochemical and immunological insults imposed by their host. Pathogens must activate appropriate responses to these local environmental stresses while adapting to and assimilating the available nutrients within that niche.
Candida albicans is a major fungal pathogen of humans that can cause a variety of infections (Calderone and Clancy, 2011). C. albicans is carried as a relatively harmless commensal in the oral, gastrointestinal and urogenital microflora of 40–80% of the population. Mucosal infections (thrush) can arise when immunological defects or pharmacological interventions affect host–fungus interactions, and a significant proportion of these infections can be recurrent (Calderone, 2002). In severely immunocompromised patients C. albicans can thrive in the bloodstream, ultimately colonizing and forming lesions on internal organs (Perlroth et al., 2007; Pfaller and Diekema, 2010). Up to half of these systemic infections are fatal (Perlroth et al., 2007). Therefore, C. albicans can thrive within diverse niches in its human host.
Clearly to cause infection, C. albicans must grow and divide in these diverse niches. Cells must assimilate locally available carbon sources which can include fermentable sugars and non-fermentable carbon sources (Lorenz and Fink, 2001; Lorenz et al., 2004; Piekarska et al., 2006; Vieira et al., 2010; Ueno et al., 2011). Physiologically relevant sugars include glucose, fructose and galactose. However, these sugars are only present at low levels and are even absent in many host niches. Consequently, other non-fermentable carbon sources become essential to support yeast growth and metabolism in vivo (Piekarska et al., 2006; Vieira et al., 2010; Ueno et al., 2011). These alternative carbon sources include amino acids and organic acids. For example, lactic acid is present in ingested foods, produced via host metabolic activity, generated by endogenous lactic acid bacteria in the urogenital and gastrointestinal tracts, and is an important carbon source for Candida in the intestine (Ueno et al., 2011).
Thinking about carbon source utilization in yeasts is strongly influenced by the Saccharomyces cerevisiae paradigm. When this relatively benign model yeast is faced with a mixture of carbon sources it preferentially utilizes sugars such as glucose before assimilating alternative carbon sources (Johnston, 1999). This hierarchical metabolic activity is regulated by a complex glucose signalling network involving the co-ordinated action of AMP kinase, protein kinase A and sugar receptor repressor signalling pathways (Kim and Johnston, 2006). This signalling network downregulates metabolic pathways required for the assimilation of alternative carbon sources at both transcriptional and post-transcriptional levels (Yin et al., 1996; Sexton et al., 2007; Gancedo, 2008; Hedbacker and Carlson, 2008; Zaman et al., 2008; Askew et al., 2009). To some extent this signalling network and the downstream regulatory mechanisms have been conserved in C. albicans (Sabina and Brown, 2009). However, recent work has shown that there has been major transcriptional rewiring as well as significant divergence in the components that regulate carbohydrate and lipid metabolism in C. albicans (Martchenko et al., ; Lavoie et al., 2009). Indeed, unlike S. cerevisiae, C. albicans is classified as a glucose-negative yeast because it continues to respire in the presence of glucose (Niimi et al., 1988). Clearly, benign and pathogenic yeasts have evolved to respond differentially to glucose, reflecting their contrasting niches. S. cerevisiae is thought to have evolved under conditions of ‘feast and famine’, rapidly exploiting fermentable sugars before switching to alternative carbon sources (Johnston, 1999). In contrast, C. albicans often inhabits niches that are glucose-limited but rich in alternative carbon sources. The physiological robustness of this pathogen in vivo is probably enhanced by its ability to assimilate multiple carbon sources simultaneously, rather than the sequential use of ‘preferred’ and then ‘non-preferred’ carbon sources (Brown et al., 2007).
Genome-wide expression profiling studies have confirmed the ability of C. albicans to undergo rapid metabolic transitions (Lorenz et al., 2004; Rodaki et al., 2009). For several fungi, as well as some bacteria (e.g. Mycobacterium tuberculosis), the glyoxylate cycle plays an important role in pathogenesis and is required for full virulence in the host (Lorenz and Fink, 2001). Recent work has reinforced the essentiality of carbon metabolism for fungal pathogenicity (Price et al., 2011; Vylkova et al., 2011). In particular, the induction of lactate transporters and metabolic enzymes upon macrophage internalization (Lorenz et al., 2004) suggests that high concentrations of lactate are present in the phagosome. As peroxisomal fatty acid β-oxidation is not essential for virulence (Piekarska et al., 2006), it is conceivable that lactate assimilation and utilization might support flux through the glyoxylate cycle thereby contributing to the survival of the pathogen under glucose-limiting conditions. The dependence of Candida glabrata upon lactate assimilation in certain host niches further strengthens this view (Ueno et al., 2011). However, most virulence studies have been performed using cells grown in rich glucose-containing media, and thus the impact of carbon source on infection development has yet to be addressed.
How does growth on different carbon sources affect the ability of yeasts to adapt to dynamic changes in their environment? In S. cerevisiae, glucose downregulates the core stress response via cAMP signalling, rendering cells more sensitive to environmental insults. Following the diauxic shift to alternative carbon sources, this yeast becomes more stress-resistant (Gounalaki and Thireos, 1994; Stanhill et al., 1999; Görner et al., 2002). The situation is less clear in C. albicans as most studies of stress adaptation have been performed on glucose-grown cells. The core stress response has diverged significantly in C. albicans (Enjalbert et al., 2003; 2006; Ramsdale et al., 2008), and while glucose exposure and cAMP signalling do influence stress resistance in C. albicans (Wilson et al., 2007; Rodaki et al., 2009), this is by unknown mechanisms. The effects of alternative carbon sources upon stress resistance of C. albicans remain unexplored despite the fact that this yeast depends upon such nutrients in many host niches (Lorenz and Fink, 2001; Barelle et al., 2006).
Similarly, the impact of growth on alternative carbon sources on the C. albicans cell wall remains largely unexplored, although the cell wall is a key effector of cell morphology and robustness, the major point of contact with the host, an active modulator of host immune defences and a prime target for antifungal drugs. Furthermore, a large proportion of assimilated carbon is invested in cell wall biogenesis as the cell wall comprises ∼ 30% of the yeast cell dry weight. The C. albicans yeast cell wall contains three main components: mannoproteins (∼ 39%), β-glucans (∼ 59%) and chitin (∼ 2%) (Aguilar-Uscanga and François, 2003). At a gross level, the composition of the S. cerevisiae cell wall has been shown to vary considerably in response to changes in carbon source, temperature, pH and aeration (Aguilar-Uscanga and François, 2003). Several observations imply mutual interactions between stress adaptation and cell wall architecture in C. albicans. Mannan chain length and complexity is altered when cells are grown in blood or serum rather than standard laboratory growth media (Kruppa et al., 2011). The stress-activated protein kinase Hog1 (Smith et al., 2004) influences cell wall biosynthesis (Eisman et al., 2006), partly by regulating chitin synthesis (Munro et al., 2007; Walker et al., 2008). Also, in response to certain stresses, the cell integrity pathway activates cell wall biogenesis (Navarro-Garcia et al., 1998; Eisman et al., 2006; Munro et al., 2007; Blankenship et al., 2010). In addition, stressors have recently been shown to influence mannan structures in C. albicans (Koyama et al., 2009). However, few studies have examined the influence of growth conditions on the cell wall (Kulkarni et al., 1980; Aguilar-Uscanga and François, 2003; Kawahata et al., 2006; Kruppa et al., 2011), and none has examined this in any detail despite its relevance to disease progression.
In this study we first characterized the impact of carbon source on the C. albicans cell wall, because of the critical role of the fungal cell wall during infection. We compared cells grown on glucose versus the physiologically relevant carboxylic acid, lactate, with a view to comparing this gluconeogenic carbon source with the carbon source classically used for experimental dissection of stress adaptation in C. albicans. We chose sodium lactate for these detailed cell wall analyses because lactate has been used previously to study metabolic flexibility (Vieira et al., 2010) and stress resistance (Rodaki et al., 2009). Lactate is an abundant carbon source in the intestine and vaginal mucosa and is produced at high rates by red blood cells, brain and muscle (Buchalter et al., 1989; Ueno et al., 2011). It is also found clinically in isotonic solutions commonly used after trauma, surgery or burn injury (lactated Ringer's solutions, Hartmann’ solution), all of which increase the risk of systemic candidiasis (Pfaller and Diekema, 2010). Having demonstrated the impact of lactate, we then confirmed that C. albicans is affected by other carbon sources that are commonly found in the host, including galactose, fructose, oleic acid, pyruvate, sorbitol and amino acids.
Our analyses revealed the major effects of carbon source on the architecture as well as the biochemical and biophysical properties of the cell wall. Alternative carbon sources strongly influenced adhesion, stress adaptation and drug resistance in C. albicans in vitro, as well as the virulence of this major pathogen in in vivo models of systemic candidiasis and vaginitis.
Results
Carbon source affects cell wall architecture
The cell wall architecture of yeast cells grown on glucose, lactate or a combination of these two carbon sources was examined by high-resolution freeze substitution transmission electron microscopy (FS-TEM). This revealed major differences in cell wall structure, thickness and density (Fig. 1A), which were confirmed by measuring the thickness of β-glucan and mannan layers (Fig. 1B). The cell walls of lactate-grown cells were thinner, with the β-glucan and chitin layer dramatically reduced. Furthermore, the outer mannan fibrils displayed less ordered structures compared with those of glucose-grown cells.
Fig. 1.
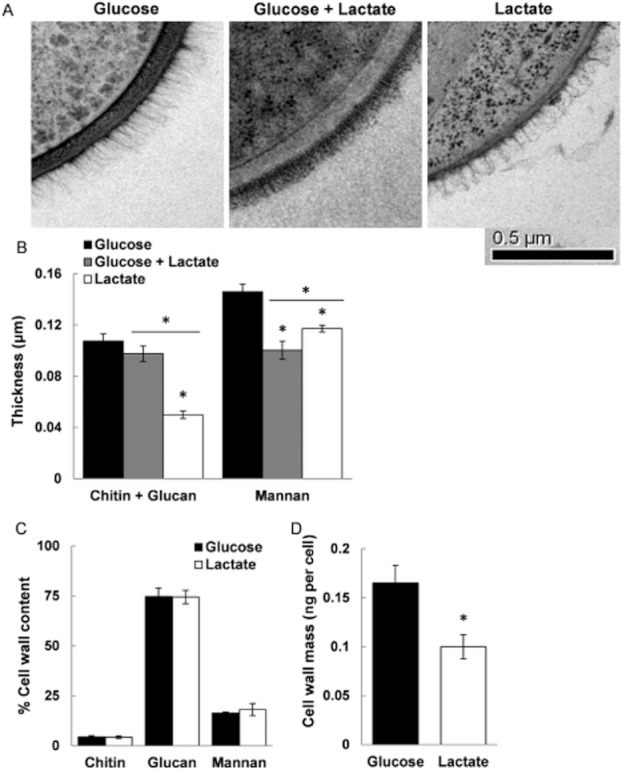
Carbon source influences cell wall architecture.
A. TEM images of the C. albicans RM1000 cell walls grown on glucose, lactate or a mixture of both (scale bar = 0.5 μm).
B. The thicknesses of the chitin plus β-glucan and mannan layers were quantified from TEM images of individual cells. Means ± SEM for n > 20 cells for each growth condition are shown.
C and D. Biochemical content (C) and cell wall biomass (D) of exponential C. albicans cells grown on glucose or lactate. Means ± SEM for three independent experiments are shown.
Relative to glucose-grown cells: *P < 0.05.
We also examined the effects of carbon source upon C. albicans cell wall composition. Lactate- and glucose-grown cells were similar with regard to the overall proportions of chitin, β-glucan and mannan in their cell walls (Fig. 1C). However, there were dramatic differences in cell wall biomass. The cell wall dry mass of lactate-grown cells was 50% less than that of glucose-grown cells (Fig. 1D). Clearly, carbon source strongly influences the cell wall architecture of C. albicans.
Carbon source affects biophysical properties of the cell wall
Given the major impact on cell wall architecture, we reasoned that changes in carbon source might also affect C. albicans cell wall properties. Cell wall porosity was investigated using an assay based on the polycation-induced leakage of UV-absorbing compounds from cells (De Nobel et al., 1990). This assay compares leakage induced by small polycations (e.g. poly-l-lysine, which induces cell leakage independent of cell wall porosity) with the release caused by large polycations (such as DEAE-dextrans, which cause limited leakage depending upon the degree of porosity of the cell wall). The cell wall porosity of lactate-grown cells was twofold higher than of glucose-grown cells (Fig. 2A), providing the evidence that carbon source has a major impact upon biophysical properties of the C. albicans cell wall. The increased porosity of lactate-grown cells was consistent with their reduced mannan fibrils (Fig. 1B), as increased porosity has been previously associated with shorter mannan side-chains (De Nobel et al., 1990).
Fig. 2.
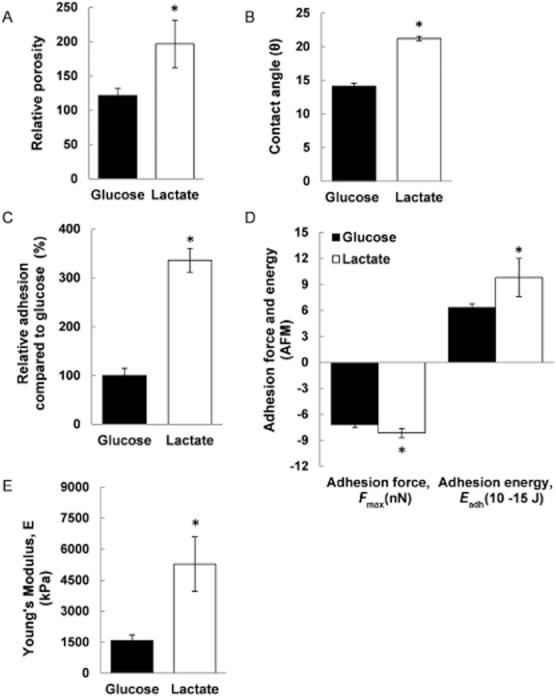
Carbon source affects biophysical properties of the cell wall. C. albicans RM1000 cells were grown to mid-exponential phase (OD600 0.4–0.6) prior to the analyses.
A. The relative cell wall porosity of glucose or lactate-grown cells assayed via polycation-induced leakage of UV-absorbing compounds (De Nobel et al., 1990).
B. Cell surface hydrophobicity of glucose- and lactate-grown C. albicans RM1000 cells assayed via the water contact angles between a liquid surface and these cells (n > 20 drops).
C. Adhesion of C. albicans cells to a non-treated polystyrene plastic surface. Adhered cells were scraped from the surface, serially diluted, plated on agar and their numbers determined as CFUs (shown as percentage CFUs compared to glucose-grown controls).
D. Adhesion force and adhesion energy of glucose- and lactate-grown cells examined by AFM (n = 6).
E. Cell surface flexibility (indicated by Young's modulus) measured by AFM (n = 6). Results are presented as means ± SEM for at least three independent experiments.
Relative to glucose-grown cells: *P < 0.05.
Cell surface hydrophobicity was examined using a biophysical assay that defines contact angles between cell surfaces and aqueous solutions (Amaral et al., 2006; Henriques et al., 2007). Significant differences were observed in these contact angles (Fig. 2B) suggesting that the surface of lactate-grown cells is more hydrophobic than that of glucose-grown cells.
Hydrophobicity has been recently linked with adhesion to abiotic surfaces in C. albicans clinical isolates (Yoshijima et al., 2010; Sardi et al., 2011). Adhesion is a key virulence trait, playing major roles in tissue colonization and biofilm formation on indwelling devices in hospital patients (Calderone, 2002). Adhesion of C. albicans cells is influenced by growth on different sugars (Willis et al., 2000; Bain et al., 2001) and therefore we predicted that growth on non-fermentable carbon sources might also influence C. albicans adherence. As expected, lactate-grown C. albicans cells adhered more strongly to plastic surfaces than cells grown on glucose (Fig. 2C). Furthermore, we confirmed that the adhesive force of lactate-grown cells was significantly greater than for glucose-grown cells using atomic force microscopy in force-mapping mode (AFM-FM) (Müller and Dufrêne, 2007) (Figs 2D and S1). Moreover, the adhesion energy displayed by lactate-grown cells was also significantly elevated (> 50%) (Fig. 2D).
The elasticity of the C. albicans cell wall was also examined by AFM. This nanomechanical analysis revealed a threefold increase in Young's modulus (Dague et al., 2010) for lactate-grown cells (Fig. 2E), indicating that their cell walls were less elastic than those of glucose-grown cells. Therefore, increased mechanical pressure exerted by lactate-grown cells might promote epithelial penetration during host invasion (Dalle et al., 2009).
Taken together, these data indicate that changes in carbon source have a major impact upon the biophysical properties of the cell wall and upon the adherence of C. albicans cells, a key virulence trait of this pathogen.
Osmotic stress resistance and adaptation are affected by carbon source
As the cell wall physically protects fungal cells from environmental insults, we reasoned that carbon source might also exert effects upon stress resistance in C. albicans. First we examined the impact on osmotic stress resistance, showing that lactate-grown cells were much more resistant to high concentrations of salt (2 M NaCl) than glucose-grown cells (Fig. 3A). Blood glucose and lactate levels are maintained within homeostatic limits. Glucose levels are generally about 3–5 mM (∼ 0.1%) and lactate concentrations are about 1–20 mM (up to 0.2%). Lactate levels vary in mucosal niches with concentrations in the vaginal fluid reaching 110 mM (∼ 1.2%) (O'Hanlon et al., 2011). Therefore, we tested a wide range of lactate and glucose concentrations (0.1% to 4%). This revealed that differential osmotic stress sensitivities are observed for cells grown at more physiological levels of these carbon sources (Fig. S2).
Fig. 3.
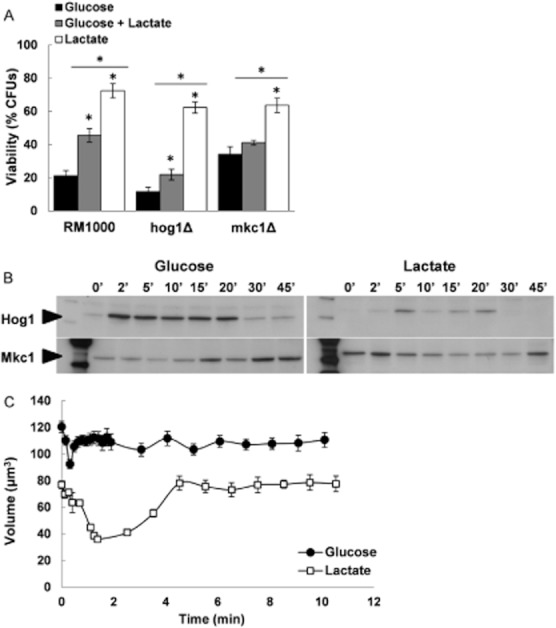
Osmotic stress resistance and adaptation are affected by carbon source.
A. Resistance of wild-type (RM1000), hog1Δ and mkc1Δ C. albicans cells (Table S1) to osmotic stress (2 M NaCl) during exponential growth on glucose, lactate or glucose plus lactate. Means ± SEM for four independent experiments are presented. Relative to glucose-grown cells: *P < 0.05.
B. Phosphorylation and activation of Hog1 and Mkc1 in glucose- and lactate-grown cells following exposure to 1 M NaCl as revealed by Western blotting with phospho-specific antibodies. Similar results were obtained in four independent experiments.
C. Dynamic changes in cell volume following hyperosmotic shock (1 M NaCl; n > 25 cells).
This increased resistance might reflect increased signalling via the Hog1 stress-activated protein kinase, because this MAP kinase pathway is critical for osmoadaptation in C. albicans (San José et al., 1996; Smith et al., 2004). This was tested by examining the dynamics of Hog1 phosphorylation during hyperosmotic stress (1 M NaCl; Fig. 3B). Hog1 was activated to a lesser extent in lactate-grown cells, although they were more resistant to 2 M NaCl. Furthermore, this increased resistance was retained in a hog1Δ mutant (Fig. 3A). These observations indicate that these effects of carbon source upon osmotic stress resistance were not dependent upon Hog1 signalling.
The cell integrity (Mkc1) signalling pathway also plays a role in cell wall biogenesis and osmoadaptation in C. albicans (Navarro-Garcia et al., 1998; 2005). Therefore, we examined the dynamics of Mkc1 phosphorylation (Fig. 3C). Higher basal levels of Mkc1 phosphorylation were observed in lactate-grown cells, suggesting active cell wall remodelling even in the absence of stress. While Mkc1 phosphorylation was delayed in glucose-grown cells, Mkc1 activation was observed immediately after NaCl addition to lactate-grown cells (Fig. 3C). However, MKC1 deletion did not block the effects of lactate on osmotic stress resistance (Fig. 3A). We conclude that the effects of lactate are not dependent on signalling via the cell integrity pathway.
As signalling through these key pathways was not essential for the elevated osmotic stress resistance of lactate-grown cells (Fig. 3A) and given the dramatic changes observed in cell wall architecture (Fig. 1), we reasoned that the increased osmotic stress resistance of lactate-grown cells might be mediated through alterations in the biophysical properties of the cell wall. To test this we examined the dynamic changes in cell volume propagated by hyperosmotic stress (Fig. 3C). Relative to glucose-grown cells, lactate-grown cells displayed dramatic reductions in cell volume (> 50% versus 20%). Furthermore, we observed significant decreases in the rates of volume reduction and recovery in lactate-grown cells (−30.65 μm3 min−1 and 13.75 μm3 min−1 respectively) compared to glucose-grown cells (−87.21 μm3 min−1 and 38.56 μm3 min−1 respectively). Thus, although the decrease in cell volume was more dramatic for lactate-grown cells, their volume changes occurred at a slower rate, consistent with the AFM observation that the walls of lactate-grown cells are less elastic (Fig. 2E). Taken together, these observations suggest that changes in carbon source affect the architecture and flexibility of the cell wall, and that this in turn affects the dynamics of cellular adaptation and resistance to hyperosmotic stress.
Carbon source affects resistance to other stresses and antifungal drugs
Clearly, the influence of cell wall remodelling might extend to other types of stress, and cell wall stresses in particular. Calcofluor White and Congo Red interfere with the synthesis and cross-linking of chitin and glucan respectively. Lactate-grown cells displayed increased resistance to both cell wall agents (Fig. 4A). This increased resistance was also observed for hog1Δ and mkc1Δ cells grown on lactate (Fig. S3A–C).
Fig. 4.
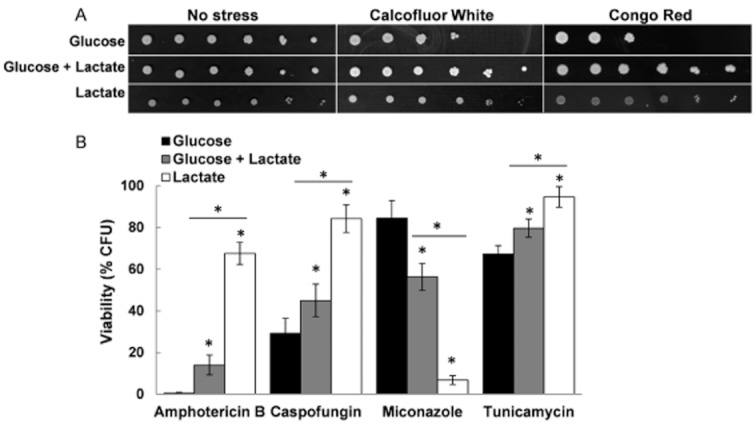
Carbon source impacts upon resistance to other stresses and antifungal drugs.
A. Sensitivity of C. albicans RM1000 to Calcofluor White (200 μg ml−1) and Congo Red (300 μg ml−1) when grown on glucose, lactate or a mixture of both.
B. Sensitivity of C. albicans RM1000 to the antifungal drugs amphotericin B (Ambisome; 10 μg ml−1), caspofungin (6.4 ng ml−1), miconazole (25 μg ml−1) and tunicamycin (4 μg ml−1).
Relative to glucose-grown cells: *P < 0.05.
Next, we compared the sensitivity of glucose- and lactate-grown cells to antifungal drugs. We tested caspofungin (an echinocandin that inhibits 1,3-β-glucan synthase), tunicamycin (a nucleoside antibiotic that blocks N-linked mannosylation), amphotericin B (a polyene that interferes with plasma membrane function by binding to ergosterol) and miconazole (an azole that inhibits ergosterol synthesis). Compared with glucose-grown cells, lactate-grown cells were resistant to caspofungin, tunicamycin and amphotericin B, but were more sensitive to miconazole (Fig. 4B). To address the contrasting effects of carbon source on resistance to membrane-targeting antifungals, we tested the sensitivity of an ergosterol biosynthetic mutant (erg11Δ) to amphotericin B and miconazole. Ergosterol levels in erg11Δ cells are below detection and consequently this strain displays increased resistance to amphotericin B and azoles (Sanglard et al., 2003). Although glucose-grown erg11Δ cells displayed increased resistance to miconazole compared to the control strain (CAF4-2) (Fig. S3E), the differential miconazole resistance of glucose- and lactate-grown cells was maintained in both mutant and control strains (Fig. S3D and E). Although sterol levels may differ between glucose- and lactate-grown cells, this does not account for their differential sensitivities to ergosterol-targeting antifungals. This suggests that differential carbon source availability in vivo, and the resultant cell wall remodelling, is likely to alter the susceptibility of C. albicans cells to therapeutic intervention.
The observed phenotypes extend to other pathogenic Candida species
We tested whether the phenotypic impact of carbon source is observed in other pathogenic Candida species. We compared the resistance of lactate- and glucose-grown cells of C. glabrata, Candida dubliniensis, Candida tropicalis, Candida krusei, Candida lusitaniae and Candida guillermondii clinical isolates (Table S1) to hyperosmotic stress and amphotericin B. In addition, we included two C. albicans clinical isolates (ATCC90028, a blood isolate, and ATCC10231, an oropharynx isolate) for comparison with the laboratory strains examined in the above experiments (MacCallum et al., 2009). In all cases, when these Candida species were grown on lactate they displayed significantly increased resistance to hyperosmotic stress and amphotericin B compared with cells grown on glucose (Fig. 5). This confirmed that carbon source has a pronounced effect upon stress and drug resistance for all of the pathogenic Candida species tested.
Fig. 5.
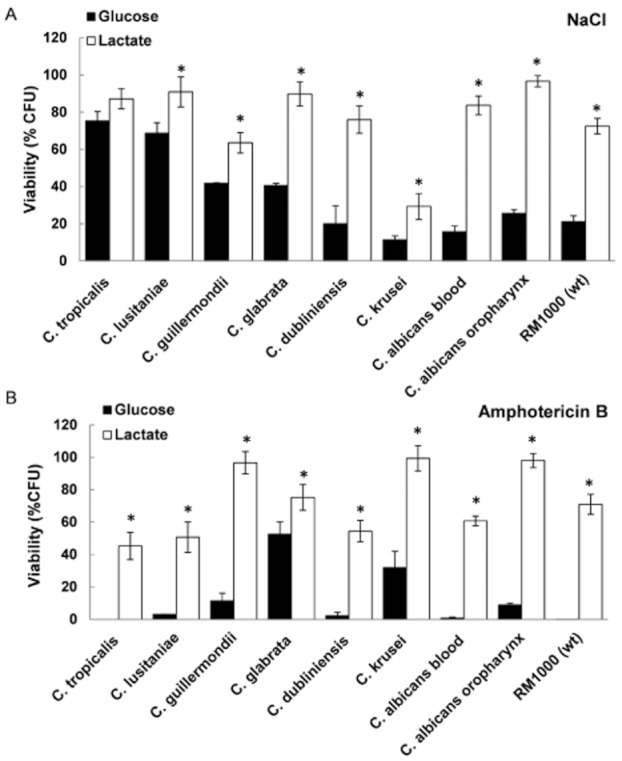
Carbon source impacts upon resistance of other pathogenic Candida species. Candida cells (Table S1) were grown to exponential phase on glucose or lactate, and their resistance to (A) hyperosmotic stress (2 M NaCl) and (B) amphotericin B (Ambisome; 10 μg ml−1) was determined. Means ± SEM for at least three independent experiments are presented.
Relative to glucose-grown cells: *P < 0.05.
Observed phenotypes extend to other physiologically relevant carbon sources
Host niches contain complex mixtures of fermentable and non-fermentable carbon sources. Therefore, we extended our analyses to test the impact of mixed carbon sources. Growing C. albicans cells on a mixed medium containing both glucose and lactate (each at 1%) resulted in intermediate levels of resistance compared to cells grown on glucose or lactate alone (Figs 3A and 4). This correlated with the intermediate architecture of the cell wall in cells grown on lactate plus glucose (Fig. 1A and B).
Next, we examined how stress and drug resistance are affected by other carbon sources, including sugars (galactose, fructose and sorbitol), a fatty acid (oleic acid), another short-chain organic acid (pyruvate) and mixed amino acids. Cells grown on these carbon sources displayed significant differences in their sensitivities to hyperosmotic stress, cell wall stress and antifungal drugs when compared to the glucose-grown cells (Fig. 6). In general, cells grown on sugars exhibited lower resistances to osmotic stress (Fig. 6A) and higher sensitivities to amphotericin B (Fig. 6B).
Fig. 6.
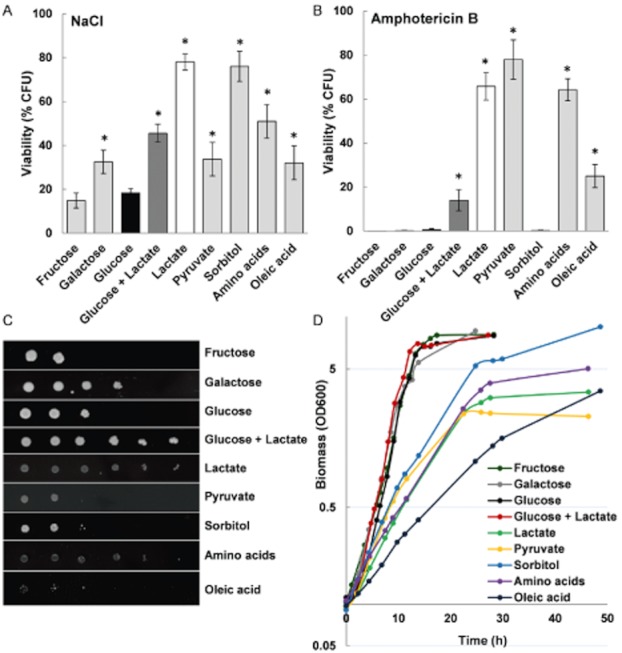
Impact of various carbon sources upon stress and drug resistance.
A–C. Sensitivity of C. albicans RM1000 to (A) 2 M NaCl, (B) 10 μg ml−1 amphotericin B and (C) 300 μg ml−1 Congo Red following growth on different carbon sources. Data represent results from at least three independent experiments (means ± SEM).
Relative to glucose-grown cells: *P < 0.05.
D. Growth curves of C. albicans RM1000 on different carbon sources. Cells were grown in either 2% or 0.2% (for oleic acid) carbon source for 4 h at 30°C and the relative levels of growth were determined by monitoring the OD600. Each curve represents the average of three biological replicates, with a maximum SEM ± 0.4.
It was conceivable that stress resistance simply correlated with a decreased growth rate. Hence, we monitored the growth of C. albicans cells on the different carbon sources during 48 h (Fig. 6D) and calculated the doubling times of these cultures during exponential phase (Fig. S4A). The doubling time was then plotted against the resistance level to hyperosmotic stress (Fig. S4B), amphotericin B (Fig. S4C) and Congo Red (Fig. S4D). This showed that stress resistance did not correlate directly with the relative doubling time on different carbon sources (Spearman correlation coefficients: sensitivities to osmotic stress P = 0.46; amphotericin B P = 0.062; Congo Red P = 0.65; Fig. S3B–D). This was most clearly illustrated by the sensitivities of cells grown in medium containing glucose plus lactate. These cells displayed similar growth rates to those grown on glucose alone, but significant differences were observed in their resistance to osmotic stress, cell wall stresses and antifungal drugs (Figs 3A and 4). Therefore, stress resistance did not correlate inversely with growth rate.
Growth on serum affects C. albicans cell wall architecture
We analysed mixed carbon sources because host microenvironments contain mixtures of nutrients (above). To reinforce the physiological relevance of our observations, we examined the impact of serum upon cell wall architecture. As expected, the cell walls of C. albicans grown in serum (< 0.1% glucose) showed marked differences compared with those of glucose-grown cells (Fig. 7A). In particular, a dramatic decrease in the length of mannan chains and a decrease in the thickness of the chitin and β-glucan layer were observed (Fig. 7B). These differences in the mannan length and structure were consistent with a recent biochemical report describing alterations in mannan complexity in cells grown on blood or serum agar (Kruppa et al., 2011). We observed that cells grown on serum supplemented with 1% glucose displayed intermediate phenotypes, marked by a significant lengthening of the outer mannan fibrils (Fig. 7A). C. albicans cells grown on serum displayed high levels of filamentation and self-adhesion, thereby precluding colony-forming units (CFUs) measurements. Therefore, we were unable to compare the stress or drug resistance of these cells with lactate- and glucose-grown cells (Figs 3 and 4). Nevertheless, it was clear that serum-grown cells displayed analogous changes in cell wall architecture to those grown on a mixed carbon source (Figs 1A and 7A).
Fig. 7.
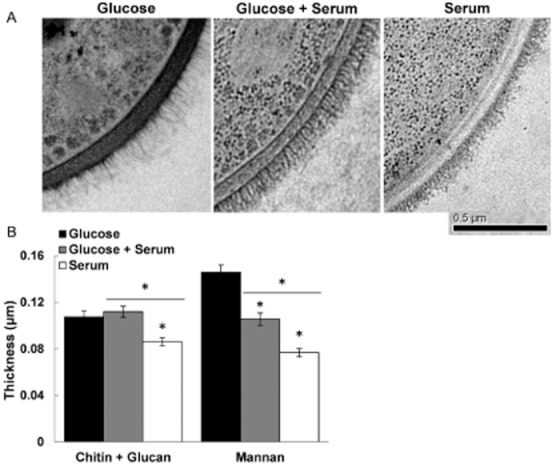
Growth on serum affects cell wall architecture.
A. TEM of the C. albicans RM1000 yeast cell walls growing on 2% glucose, serum or serum plus 1% glucose (scale bar = 0.5 μm).
B. The thicknesses of the chitin plus β-glucan and mannan layers were quantified from TEM images of individual cells. Means ± SEM for n > 20 cells for each growth condition are shown.
Relative to glucose-grown cells: *P < 0.05.
Carbon source affects the virulence of C. albicans
The above observations strongly suggested that changes in carbon source might affect the physiological fitness of C. albicans cells in vivo. To test this, we assayed the virulence of cells grown on galactose, glucose, oleic acid, lactate, glucose plus lactate or a mix of amino acids in mouse models of systemic and vaginal infection.
For systemic infection, virulence was assayed by determining infection outcome scores, which are based on measurements of weight changes and kidney burdens 3 days post challenge (MacCallum et al., 2010). In this assay a higher ‘outcome score’ reflects greater weight loss and higher fungal burdens and thus increased virulence (MacCallum et al., 2010). Carbon source was found to have a significant impact upon the virulence of C. albicans during systemic infection (Fig. 8). Indeed, significant differences were found for all parameters (kidney burden, weight change and outcome score). Mice infected with C. albicans cells grown on lactate, glucose plus lactate, amino acids all displayed increased fungal burdens and weight loss compared with glucose-grown cells (Fig. 8A and B). In contrast, mice infected with C. albicans cells grown on oleic acid displayed reduced fungal burdens in the kidney and these mice actually gained weight (Fig. 8A and B). Therefore, growth on oleic acid reduced the virulence of C. albicans in systemic infections (Fig. 8C). These observations closely paralleled the stress phenotypes induced by growth on alternative carbon sources (Fig. 6).
Fig. 8.
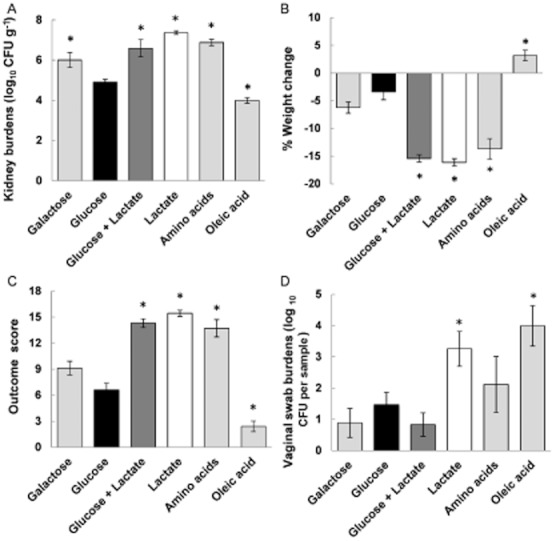
Growth on alternative carbon sources influences C. albicans virulence. C. albicans (RM1000 containing Clp20, Table S1) cells grown on different carbon sources were used to infect mice intravenously.
A and B. Kidney burdens (A) and weight change (B) were measured 72 h post infection (means ± SEM; n = 6).
C. Infection outcome scores were calculated after 72 h (means ± SEM; n = 6), higher outcome scores reflecting more severe infection (MacCallum et al., 2010).
D. Mice were infected intravaginally with C. albicans (RM1000 containing Clp20) cells grown on different carbon sources. Fungal burdens in vaginal swabs were measured 72 h post infection (means ± SEM; n = 6).
In all panels, the asterisk (*) indicates P < 0.05 relative to glucose-grown C. albicans infection.
For vaginal infection, virulence was assayed by measuring fungal burdens in mice 3 days post infection. Significantly increased virulence was observed for C. albicans cells grown on lactate or oleic acid compared to the control glucose-grown cells (Fig. 8D). Differences for the other carbon sources were not statistically significant. Interestingly, growth on oleic acid led to reduced virulence in the intravenous model but increased virulence in the vaginal model (Fig. 8C and D).
Discussion
Candida albicans occupies diverse niches in its human host and many of these microenvironments offer complex mixtures of nutrients. Some niches continuously supply low concentrations of glucose (e.g. ∼ 0.1% in the blood, during haematogenously disseminated candidiasis), while others provide transient exposure to fermentable sugars (e.g. the oral cavity and gastrointestinal tract) or probably lack glucose (e.g. on skin or nails). Nevertheless, it is often assumed that in vitro observations made with cells grown on relatively high concentrations of glucose are relevant to growth in vivo. Almost without exception, the experimental dissection of fitness attributes, such as stress adaptation, in C. albicans has been performed on glucose-grown cells (Alonso-Monge et al., 2003; Enjalbert et al., 2003; Smith et al., 2004; Navarro-Garcia et al., 2005; Eisman et al., 2006; Cheetham et al., 2007). Furthermore, antifungal drug susceptibilities are generally assayed on glucose-grown cells, the glucose concentration and the medium composition significantly influencing the minimum inhibitory concentration of drug (Bartizal and Odds, 2003).
We reveal that growth on alternative, physiologically relevant carbon sources has a major impact upon the stress, drug resistance and virulence of C. albicans. Cells grown on some non-fermentable carbon sources were more resistant to amphotericin B and caspofungin as well as to osmotic and cell wall stresses (Figs 3, 4 and 6). These observations were replicated in other pathogenic Candida species (Fig. 5). Consistent with these in vitro observations, growth of C. albicans cells on alternative carbon sources significantly altered their virulence (Fig. 8). Moreover, the impact on virulence was highly dependent on the site of infection. While growth on some carbon sources (e.g. oleic acid) promoted virulence during mucosal infection, presumably conferring a fitness advantage during disease onset in this environment, the same carbon source reduced virulence in a systemic infection (Fig. 8). Interestingly, growth on sugars such as glucose or galactose resulted in low fungal burdens in both infection models, whereas growth on amino acids or lactate promoted both systemic and vaginal infection. A recent study has also suggested that lactate utilization is required for Candida proliferation in the gastrointestinal tract (Ueno et al., 2011).
It has been reported previously that the growth medium used to prepare C. albicans inoculums affects the ability of strains to gain an initial invasive advantage immediately after injection into host, therefore influencing virulence (Odds et al., 2000). Our data now indicate that the carbon source strongly influences cell wall architecture and environmental adaptation during infection. Although C. albicans cells will adapt to the available nutrients in their local niche (Lorenz et al., 2004; Rodaki et al., 2009), our data show clearly that the initial fitness advantage conferred by growth on different carbon sources is ultimately reflected in the infection outcome in both systemic and vaginal infections (Fig. 8). Obviously differential carbon source availability within diverse host niches will also influence the virulence of C. albicans during disease progression.
It was conceivable that carbon source might influence stress sensitivity in C. albicans via key stress signalling mechanisms such as the Hog1 and the Mkc1 pathways. However, elevated stress resistance did not correlate with increased activation of the Hog1 and Mkc1 MAP kinases (Fig. 3B). Moreover, the increased resistance of lactate-grown cells was not blocked by inactivation of either kinase (Fig. 3A). Therefore, while the impact of carbon upon signalling almost certainly contributes to stress adaptation, this does not account for the major effects of carbon source upon stress resistance.
Instead, carbon source exerts its effects on stress resistance largely through alterations in the C. albicans cell wall. Growth on a non-fermentable carbon source, or mixtures of fermentable plus non-fermentable carbon sources, led to major changes in the C. albicans cell wall architecture (Fig. 1). These changes were manifested in significant alterations in the biophysical and mechanical properties of the cell wall, some of which affect stress adaptation (Fig. 2). Such properties have been predicted by mathematical modelling to play an important role in the adaptive processes of yeast cells (Schaber et al., 2010). Therefore, we suggest that the observed changes in cell wall elasticity led to decreased rates of cell volume change in C. albicans cells (Fig. 3C) thereby promoting their increased osmotic stress resistance. Presumably, the major remodelling of the cell wall in response to carbon source also underlies the altered cellular resistance to cell wall stresses and antifungal drugs.
How does the carbon source influence the cell wall? Given that remarkably little is known about how the cell wall polysaccharides are interlinked, the detailed mechanisms by which carbon source affects these linkages may take some time to define. However, the carbon source is likely to affect cell wall biogenesis by impacting directly upon metabolic fluxes as well as indirectly through regulatory networks (Ihmels et al., 2005; Martchenko et al., ; Askew et al., 2009). We reason that when glucose is present at high concentrations, excess carbon might flow via hexose phosphates into the biosynthesis of β-glucan and mannan, generating an elaborate cell wall that is relatively thick. In contrast, during growth on non-fermentable carbon sources the hexose phosphates required for cell wall biosynthesis must be generated by gluconeogenesis, an energetically demanding process. Less carbon is probably committed to cell wall biosynthesis under these more challenging conditions, leading to the construction of a leaner but stiffer cell wall.
In conclusion, our findings show clearly that carbon source significantly influences the resistance of C. albicans and other pathogenic Candida species to environmental stresses and antifungal drugs. This infers that carbon sources such as lactate and amino acids, which are available in many human niches, modulate the fitness of C. albicans cells in vivo, and furthermore that differential nutrient availability within diverse host niches has a strong influence upon the ability of Candida cells to counteract local stresses and resist pharmacological intervention. Indeed, growth on different carbon sources strongly influenced the virulence of C. albicans in murine models of systemic and vaginal candidiasis. The next challenge is to define which local nutrients are assimilated by C. albicans cells during disease establishment and progression, and how this influences the behaviour of this pathogen in the host.
Experimental procedures
Strains and growth conditions
The Candida strains and clinical isolates used in this study are described in Table S1. Strains were grown at 30°C in minimal medium containing 2% carbon source (glucose, fructose, galactose, sodium lactate, sodium pyruvate, sorbitol or a mix of amino acids), 0.67% yeast nitrogen base without amino acids (YNB) and supplemented with 10 μg ml−1 of the appropriate auxotrophic requirements. Similar to previous analyses (Piekarska et al., 2006), a lower concentration of carbon source was used for cells grown on oleic acid (0.2% instead of 2%). For serum experiments, cells were grown in 20% fetal bovine serum in PBS (Invitrogen, Paisley, UK) with or without 1% glucose. Unless otherwise stated, RM1000 (Negredo et al., 1997) cells were grown overnight at 30°C, diluted to an OD600 of 0.1 in fresh medium, and harvested in mid-exponential phase (OD600 = 0.5) for analyses and sensitivity assays. With the exception of serum cultures (pH 6.5–7.0), all experiments were performed with yeast cells grown at a pH 5.2–5.6.
Growth was monitored on different carbon sources by measuring the biomass of cultures (OD600) during 48 h of growth. Each curve represents the average of three biological replicates, with a maximum SEM ± 0.4. The linear equation during exponential phase was calculated for each curve, and the doubling time was calculated using this equation.
Freeze substitution transmission electron microscopy
Freeze substitution transmission electron microscopy was performed as described previously (Netea et al., 2006), except that C. albicans cells were harvested by filtration rather than by centrifugation to maximize cell wall integrity and ultrathin sections were cut at a thickness of 100 nm. Samples were imaged with a Philips CM10 transmission microscope (FEI UK) equipped with a Gatan 600 W camera and images were recorded using Digital Micrograph (Gatan, Abingdon Oxon, UK). The thicknesses of the chitin plus β-glucan and mannan cell wall layers were measured using Image J and by averaging > 20 measurements for each cell (n > 20 cells).
Cell wall biochemistry
The chitin, β-glucan and mannan content of cell walls were measured using previously described procedures (Plaine et al., 2008; Lee et al., 2011). Briefly, cell walls were prepared from mid-exponential C. albicans grown on glucose or lactate. Cells were washed five times with dH2O, disrupted with glass beads (Sigma, G9268) using a Fastprep cell breakage machine (Thermo Savant, Middlesex, UK). Extracts were centrifuged at 13 000 r.p.m. for 3 min and washed five times in 1 M NaCl. Cell walls were washed by heating at 100°C for 10 min in 2% SDS, 0.3 M β-mercaptoethanol, 1 mM EDTA, 50 mM Tris-HCl, pH 6.8. Cell wall pellets were then washed and resuspended in dH2O, freeze-dried and the dry weight recorded to calculate the cell wall biomass per cell. The cell wall pellets were hydrolysed in 100% trifluoroacetic acid at 100°C for 3 h, the acid evaporated at 90°C for 1 h and samples resuspended in sterile dH2O at 10 mg ml−1. Glucosamine, glucose and mannose contents were then determined by high-performance anion-exchange chromatography with pulsed amperometric detection (HPAEC-PAD) in a carbohydrate analyser system from Dionex (Surrey, UK) (Lee et al., 2011), using hydrolysed samples from three independent biological replicates, each with technical duplicates.
Cell wall porosity
Cell wall porosity was assayed using methods adapted from an existing protocol (De Nobel et al., 1990). Briefly, mid-exponential cells were washed twice with dH2O and aliquots of 108 cells were incubated with shaking (200 r.p.m.) for 30 min at 30°C in 1 ml of either 10 mM Tris-HCl pH 7.4 (control), the same buffer containing 5 mg ml−1 DEAE-dextran (500 kDa) or buffer containing 15 μg ml−1 poly-l-lysine (50 kDa). Cells were then pelleted by centrifugation and the A260 of supernatants measured. Relative porosity was calculated using the following formula: relative porosity = 100 × (ADEAE − Abuffer) / (Apoly-l-lysine − Abuffer). Results are means ± SEM of five independent experiments, each with two technical replicates.
Surface hydrophobicity
Microscope slides were prepared with a uniform layer of 2% agar in 10% glycerol in water. C. albicans cells were harvested in mid-exponential phase, resuspended in sterile H2O to an OD600 of 2 and applied to the surface of these agar slides. This procedure was repeated 2–3 times and the slides left to dry until a uniform lawn was obtained. Water contact angles were then determined at room temperature using the sessile drop technique (Soumya et al., 2011) with a goniometer (FTA 1000 Analyzes System, First Ten Angstroms, Cambridge, UK) according to manufacturer's instructions. At least seven independent measurements were taken for each slide and three independent replicates were analysed for each growth condition.
Cell adhesion
Mid-exponential C. albicans cells were washed twice with dH2O and resuspended in PBS. About 107 cells in 2 ml of PBS were added to 12-well plates (non-treated polystyrene; Costar, Corning, Ewloe, UK) and cells allowed to adhere for 1 h at 37°C without shaking. After washing three times with PBS, adhered cells were scraped off the plastic surface into 1 ml of PBS and quantified by OD600 and counting CFUs. Results represent the average CFUs ± SEM for five independent replicate experiments for each growth condition, each with technical duplicates.
Atomic force microscopy measurements
The nanomechanical properties (i.e. adhesive force, adhesive energy and Young's modulus) of mid-exponential C. albicans cells grown in glucose or lactate were investigated by AFM force mapping (Adya and Canetta, 2011). These experiments were conducted in contact mode and a Si3N4 cantilever (Bruker UK) of nominal spring constant 0.01 N m−1 and a resonant frequency of ∼ 7 kHz was used. The tip (radius of curvature < 10 nm) microfabricated on the AFM cantilever was brought into contact with the surface of the cell. AFM experiments were performed over a grid of 10 × 10 points on an individual cell and the force spectroscopy curves recorded on each point. Measurements were again repeated on three randomly chosen cells per duplicate slide, thus giving a total of six measurements growth condition. During the force mapping experiments an average normal force of 2 nN was applied by the AFM cantilever onto the cell. The tip was withdrawn vertically away from the cell surface at a speed of 0.5 μm s−1. In each experiment, the maximum adhesive force in nanonewtons (nN) of the cell surface to the tip (Fmax), the maximum distance in micrometres (μm) of deformation of the cell (dmax) and the adhesion energy in joules (J) (Wadh) were measured. The force spectroscopy curves obtained during an AFM force mapping experiment were force–height curves. To transform these curves into force–distance (F–d) curves, the real distance, d, between the sample and the AFM tip was calculated by subtracting the deflection of the cantilever, z, from the height values that corresponded to the measured piezodisplacement, zpiezo: d = zpiezo − z (Binning et al., 1986). The Young's modulus of elasticity (E) was determined by fitting the slope of the trace curve to the Hertz model (Hertz, 1986).
Cell volume
Mid-exponential C. albicans cells were adhered to a poly-l-lysine-coated surface for 30–60 min and stained with 2 μg ml−1 Calcofluor White to visualize cell wall chitin. Dynamic changes in individual cell volumes were monitored microscopically using a Zeiss Observer Z.1 microscope (Zeiss, Welwyn Garden City, UK) and their initial volume was recorded using Volocity software (Cambridge, UK). Cells were exposed to 1 M NaCl and changes in cell volume monitored by taking Z-stacked images every 10 s for the first minute and then every minute for the next 30 min (n > 25 cells for each condition).
Western blotting
Protein extracts were prepared from mid-exponential C. albicans cells and subjected to Western blotting as described before (Smith et al., 2004). Hog1 activation was detected using a phospho-specific phospho-p38 MAPK (Thr180/Tyr182) antibody 9211 (New England Biolabs, Hitchin, UK). Mkc1 activation was detected using a phospho-specific phospho-p44/42 MAPK (Erk1/2; Thr202/Tyr204) antibody 4370 (New England Biolabs). In both cases the secondary antibody was HRP-labelled anti-rabbit IgG 7074 (New England Biolabs) which was detected using Pierce ECL PlusTM Western blotting reagents (Thermo Scientific, Cramlington, UK).
Stress resistance
To examine hyperosmotic stress, mid-exponential C. albicans cells were exposed to 2 M NaCl for 1 h, at 30°C and CFUs measured relative to untreated control cells. Means ± SEM for at least five independent experiments are presented. To examine cell wall stresses, serial dilutions of mid-exponential C. albicans cells were plated onto agar starting from a dilution of 5 × 107 cells per spot and diluted 1/10 thereafter. The YNB agar contained the specified carbon source and was supplemented with Congo Red (300 μg ml−1) or Calcofluor White (200 μg ml−1). Plates were incubated for 2–4 days at 30°C and then photographed. Results were compared against no stress plates. Results shown are representative of data accumulated from at least three independent experiments.
Antifungal susceptibility
Antifungal drug susceptibility was analysed by treating mid-exponential C. albicans cells grown on the specified carbon source with tunicamycin (4 μg ml−1), caspofungin (0.08 μg ml−1), amphotericin B (Ambisome 10 μg ml−1 and 20 μg ml−1 for two Candida pathogenic isolates that showed decreased sensitivity) or miconazole (25 μg ml−1) for 1 h at 30°C. Cells were then serially diluted and plated onto agar. CFUs were quantified and antifungal drug sensitivities calculated relative to those observed for untreated cells. Means ± SEM for at least three independent experiments are presented.
Candida albicans virulence assays
All animal experimentation conformed to the requirements of the UK Home Office legislation and of the Ethical Review Committee of the University of Aberdeen.
The 3 day murine intravenous challenge model of C. albicans infection (MacCallum et al., 2010) was used to determine the impact of carbon source on systemic virulence. Female BALB/c mice [6–8 weeks; 18.9 ± 0.1 g (mean ± SEM); Harlan, UK] were randomly assigned to and housed in groups of six with food and water provided ad libitum. RM1000 + Clp20 (JC21) (Smith et al., 2004) cells were grown overnight at 30°C in YNB medium with 2% carbon source with constant agitation. The cultures were diluted to an OD600 of 0.1 in fresh medium, and harvested at mid-exponential phase (OD600 = 0.5). Cells were harvested, washed with sterile saline and the cell counts adjusted by OD600 to provide a 100 μl cell suspension estimated to deliver a challenge dose of 5 × 104 CFU g−1 mouse body weight. Actual challenge doses were determined from viable counts read after 24 h and were approximately 4.8 × 104 CFU g−1 (range: 4.2–6.4 × 104 CFU g−1). Each inoculum was used to infect one group of mice, which was randomly allocated. Mice were infected intravenously via a lateral tail vein, and were monitored and weighed daily. At 72 h post challenge, mice were weighed, humanely terminated by cervical dislocation, the kidneys removed aseptically and renal fungal burdens determined. For each animal, fungal burdens were measured for both kidneys by viable count. Virulence was assessed by fungal kidney burdens and by percentage weight change at 72 h and an outcome score calculated (MacCallum et al., 2010). The outcome score is devised to allow approximately equal contributions of the two parameters and was calculated using the following formula: outcome score = log CFU g−1 kidney − (0.5 × percentage weight change) (MacCallum et al., 2010). Statistical differences between body weight changes, kidney burdens and outcome scores were determined by the Mann–Whitney and Kruskall–Wallis tests using IBM spss (version 19).
The murine vaginal infection model (Fidel and Sobel, 1999) was used to determine the effect of carbon source on the ability of C. albicans to colonize/infect vaginal tissue. Female BALB/c mice [6–8 weeks; 20.2 ± 0.1 g (mean ± SEM); Harlan, UK] were randomly allocated into groups of six mice. On day 3, relative to infection, mice were treated subcutaneously with 20 μg of oestradiol valerate (Sigma) in sesame oil (Sigma). Oestradiol was administered every 3 days during the infection. C. albicans inocula were prepared from the cultures used for the intravenous challenge model, aiming for 1 × 106 CFU per 20 μl. Actual inoculum levels were determined by viable counts and were approximately 1 × 106 CFU per mouse (range: 0.9–1.3 × 106 CFU per mouse). Inocula were administered intravaginally using a pipette. Mice were sampled using swabs on day 3 and day 7 post infection. Swabs were dispersed in 100 μl of 3% Tween80 in saline and dilutions plated out on Sabouraud dextrose plus chloramphenicol and gentamicin agar. On day 7 mice were humanely terminated by cervical dislocation.
Statistical analyses
Results from independent replicate experiments are expressed as means ± SEM. With the exception of animal experiments, results were compared using two-sample Student's t-test, with a significance cut-off of 0.05. A significance cut-off of 0.1 was used for adhesion force and adhesion energy measured by AFM. Correlation was determined using the Spearman correlation coefficient (ρ) with P-values smaller than 0.05 denoting a significant correlation.
Acknowledgments
We thank members of the Aberdeen Fungal Group and the European FINSysB Network, and particularly Dr Despoina Kaloriti, Dr Louise Walker and Ms Keunsook Lee, for insightful discussions and assistance with some experiments. We thank Susan Budge for excellent technical support and Dr Elisabeta Canetta for help with AFM experiments. This work was supported by the European Commission (FINSysB, PITN-GA-2008-214004; STRIFE, ERC-2009-AdG-249793) and by the UK Biotechnology and Biological Research Council (BB/F00513X/1) and the Wellcome Trust (080088).
Author contributions
Conceived and designed the experiments: I. V. E., D. M., N. A. R. G., A. J. P. B. Performed the experiments: I. V. E., A. K. A., D. M. Analysed the data: I. V. E., D. M., A. K. A., A. J. P. B. Contributed analysis tools: S. W., A. B. Wrote the manuscript: I. V. E., A. J. P. B.
Supporting information
Additional Supporting Information may be found in the online version of this article:
Fig. S1. Illustration of the atomic force microscopy experiment (adapted from Adya and Canetta, 2011).
A. Contact between the AFM probe and the sample surface is followed by cantilever bending with AFM probe indentation during the trace (approach) cycle of the AFM experiment. On the retrace (retract) cycle, the sample surface is then stretched until the point of detachment.
B. An example of a typical experimental F–d curve for a trace and retrace cycle is presented; the stages are identified by the arrows. The meanings of Fmax, dmax and Wadh are also illustrated.
Fig. S2. The impact of carbon source on stress resistance is not dependent on the concentration of the carbon source. Resistance of wild-type (RM1000) C. albicans cells to osmotic stress (2 M NaCl) during exponential growth on different concentrations of glucose or lactate (0.1–4%). Means ± SEM for four independent experiments are presented. Relative to glucose-grown cells: *P < 0.05. Each concentration was also compared against its corresponding glucose or lactate 2% standard concentration: #P < 0.05.
Fig. S3. The impact of carbon source upon cell wall stress resistance is not dependent on Hog1, Mkc1 signalling or cellular ergosterol levels. Increased resistance to (B) Calcoflouor White (200 μg ml−1) and (C) Congo Red (300 μg ml−1) was observed in wild-type (RM1000), hog1Δ and mkc1Δ cells (Table S1) grown on lactate when compared to glucose-grown cells and relative to no-stress plates (A). Plates were incubated for 2–4 days at 30°C and then photographed. Pictures shown are representative of three independent experiments. (D) Resistance to amphotericin B (10 μg ml−1) and (E) resistance to miconazole (25 μg ml−1) of ergosterol depleted erg11 cells and corresponding wild-type strain. Relative to glucose-grown cells: *P < 0.05. The resistance of the erg11Δ strain was compared to the corresponding wild-type control, #P < 0.05.
Fig. S4. Stress resistance does not correlate inversely with growth rate.
A. Doubling times were calculated as the interval of time required by the cells to double from an OD600 = 0.5 to 1.0. The linear equation of the exponential phase slope was used for these calculations.
B–D. (B) Resistance to osmotic stress, (C) resistance to amphotericin B and (D) resistance to Congo Red of cells grown on different carbon sources were plotted against their relative doubling times under each growth condition. Resistance to Congo Red was assessed based on the number of dilutions (out of six plated) growing on Congo Red supplemented plates (Fig. 6C). No correlation was found between the rate of growth and level of resistance against the different stressors – Spearman correlation coefficient for (B) NaCl (2 M; P = 0.46), (C) amphotericin B (Ambisome; 10 μg ml−1; P = 0.062) and (D) Congo Red (300 μg ml−1; P = 0.65).
Table S1. List of fungal strains and clinical isolates used in this study.
Please note: Wiley-Blackwell are not responsible for the content or functionality of any supporting materials supplied by the authors. Any queries (other than missing material) should be directed to the corresponding author for the article.
References
- Adya AK, Canetta E. Atomic force microscopic investigation of commercial pressure sensitive adhesives for forensic analysis. Forensic Sci Int. 2011;210:16–25. doi: 10.1016/j.forsciint.2011.01.029. [DOI] [PubMed] [Google Scholar]
- Aguilar-Uscanga B, François JM. A study of the yeast cell wall composition and structure in response to growth conditions and mode of cultivation. Lett Appl Microbiol. 2003;37:268–274. doi: 10.1046/j.1472-765x.2003.01394.x. [DOI] [PubMed] [Google Scholar]
- Alonso-Monge R, Navarro-García F, Román E, Negredo AI, Eisman B, Nombela C, Pla J. The Hog1 mitogen-activated protein kinase is essential in the oxidative stress response and chlamydospore formation in Candida albicans. Eukaryot Cell. 2003;2:351–361. doi: 10.1128/EC.2.2.351-361.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amaral PF, Lehocky M, Barros-Timmons AM, Rocha-Leão MH, Coelho MA, Coutinho JA. Cell surface characterization of Yarrowia lipolytica IMUFRJ 50682. Yeast. 2006;23:867–877. doi: 10.1002/yea.1405. [DOI] [PubMed] [Google Scholar]
- Askew C, Sellam A, Epp E, Hogues H, Mullick A, Nantel A, Whiteway M. Transcriptional regulation of carbohydrate metabolism in the human pathogen Candida albicans. PLoS Pathog. 2009;5:e1000612. doi: 10.1371/journal.ppat.1000612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bain JM, Stubberfield C, Gow NA. Ura-status-dependent adhesion of Candida albicans mutants. FEMS Microbiol Lett. 2001;204:323–328. doi: 10.1111/j.1574-6968.2001.tb10905.x. [DOI] [PubMed] [Google Scholar]
- Barelle CJ, Priest CL, MacCallum DM, Gow NA, Odds FC, Brown AJ. Niche-specific regulation of central metabolic pathways in a fungal pathogen. Cell Microbiol. 2006;8:961–971. doi: 10.1111/j.1462-5822.2005.00676.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartizal C, Odds FC. Influences of methodological variables on susceptibility testing of caspofungin against Candida species and Aspergillus fumigatus. Antimicrob Agents Chemother. 2003;47:2100–2107. doi: 10.1128/AAC.47.7.2100-2107.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binning G, Quate CF, Gerber C. Atomic force microscopy. Phys Rev Lett. 1986;56:930–933. doi: 10.1103/PhysRevLett.56.930. [DOI] [PubMed] [Google Scholar]
- Blankenship JR, Fanning S, Hamaker JJ, Mitchell AP. An extensive circuitry for cell wall regulation in Candida albicans. PLoS Pathog. 2010;6:e1000752. doi: 10.1371/journal.ppat.1000752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown AJP, Odds FC, Gow NA. Infection-related gene expression in Candida albicans. Curr Opin Microbiol. 2007;10:307–313. doi: 10.1016/j.mib.2007.04.001. [DOI] [PubMed] [Google Scholar]
- Buchalter SE, Crain MR, Kreisberg R. Regulation of lactate metabolism in vivo. Diabetes Metab Rev. 1989;5:379–391. doi: 10.1002/dmr.5610050405. [DOI] [PubMed] [Google Scholar]
- Calderone RA. Candida and Candidiasis. 1st edn. Washington DC: ASM Press; 2002. [Google Scholar]
- Calderone RA, Clancy CJ. Candida and Candidiasis. 2nd edn. Washington DC: ASM Press; 2011. [Google Scholar]
- Cheetham J, Smith DA, Dantas A, Doris KS, Patterson MJ, Bruce CR, Quinn J. A single MAPKKK regulates the Hog1 MAPK pathway in the pathogenic fungus Candida albicans. Mol Biol Cell. 2007;18:4603–4614. doi: 10.1091/mbc.E07-06-0581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dague E, Bitar R, Ranchon H, Durand F, Yken HM, François JM. An atomic force microscopy analysis of yeast mutants defective in cell wall architecture. Yeast. 2010;27:673–684. doi: 10.1002/yea.1801. [DOI] [PubMed] [Google Scholar]
- Dalle F, Wächtler B, L'Ollivier C, Holland G, Bannert N, Wilson D, et al. Cellular interactions of Candida albicans with human oral epithelial cells and enterocytes. Cell Microbiol. 2009;12:248–271. doi: 10.1111/j.1462-5822.2009.01394.x. [DOI] [PubMed] [Google Scholar]
- De Nobel JG, Klis FM, Munnik T, Priem J, Van den Ende H. An assay of relative cell wall porosity in Saccharomyces cerevisiaeKluyveromyces lactis and Schizosaccharomyces pombe. Yeast. 1990;6:483–490. doi: 10.1002/yea.320060605. [DOI] [PubMed] [Google Scholar]
- Eisman B, Alonso-Monge R, Román E, Arana D, Nombela C, Pla J. The Cek1 and Hog1 mitogen-activated protein kinases play complementary roles in cell wall biogenesis and chlamydospore formation in the fungal pathogen Candida albicans. Eukaryot Cell. 2006;5:347–358. doi: 10.1128/EC.5.2.347-358.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enjalbert B, Nantel A, Whiteway M. Stress-induced gene expression in Candida albicans: absence of a general stress response. Mol Biol Cell. 2003;14:1460–1467. doi: 10.1091/mbc.E02-08-0546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enjalbert B, Smith DA, Cornell MJ, Alam I, Nicholls S, Brown AJ, Quinn J. Role of the Hog1 stress-activated protein kinase in the global transcriptional response to stress in the fungal pathogen Candida albicans. Mol Biol Cell. 2006;17:1018–1032. doi: 10.1091/mbc.E05-06-0501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fidel PL, Sobel JD. Murine models of Candida vaginal infections. In: Zak O, editor. Handbook of Animal Models of Infection. New York, NY, USA: Academic Press; 1999. pp. 741–748. [Google Scholar]
- Gancedo JM. The early steps of glucose signalling in yeast. FEMS Microbiol Rev. 2008;32:673–704. doi: 10.1111/j.1574-6976.2008.00117.x. [DOI] [PubMed] [Google Scholar]
- Görner W, Durchschlag E, Wolf J, Brown EL, Ammerer G, Ruis H, Schüller C. Acute glucose starvation activates the nuclear localization signal of a stress-specific yeast transcription factor. EMBO J. 2002;21:135–144. doi: 10.1093/emboj/21.1.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gounalaki N, Thireos G. Yap1p, a yeast transcriptional activator that mediates multidrug resistance, regulates the metabolic stress response. EMBO J. 1994;13:4036–4041. doi: 10.1002/j.1460-2075.1994.tb06720.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedbacker K, Carlson M. SNF1/AMPK pathways in yeast. Front Biosci. 2008;13:2408–2420. doi: 10.2741/2854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henriques M, Azeredo J, Oliveira R. The involvement of physico-chemical interactions in the adhesion of Candida albicans and Candida dubliniensis to epithelial cells. Mycoses. 2007;50:391–396. doi: 10.1111/j.1439-0507.2007.01387.x. [DOI] [PubMed] [Google Scholar]
- Hertz H. In Miscellaneous Papers. London: MacMillian; 1986. [Google Scholar]
- Ihmels J, Bergmann S, Gerami-Nejad M, Yanai I, McClellan M, Berman J, Barkai N. Rewiring of the yeast transcriptional network through the evolution of motif usage. Science. 2005;309:938–940. doi: 10.1126/science.1113833. [DOI] [PubMed] [Google Scholar]
- Johnston M. Feasting, fasting and fermenting. Glucose sensing in yeast and other cells. Trends Genet. 1999;15:29–33. doi: 10.1016/s0168-9525(98)01637-0. [DOI] [PubMed] [Google Scholar]
- Kawahata M, Masaki K, Fujii T, Iefuji H. Yeast genes involved in response to lactic acid and acetic acid: acidic conditions caused by the organic acids in Saccharomyces cerevisiae cultures induce expression of intracellular metal metabolism genes regulated by Aft1p. FEMS Yeast Res. 2006;6:924–936. doi: 10.1111/j.1567-1364.2006.00089.x. [DOI] [PubMed] [Google Scholar]
- Kim JH, Johnston M. Two glucose-sensing pathways converge on Rgt1 to regulate expression of glucose transporter genes in Saccharomyces cerevisiae. J Biol Chem. 2006;281:26144–26149. doi: 10.1074/jbc.M603636200. [DOI] [PubMed] [Google Scholar]
- Koyama T, Makita M, Shibata N, Okawa Y. Influence of oxidative and osmotic stresseson the structure of the cell wall mannan of Candida albicans serotype A. Carbohydr Res. 2009;44:2195–2200. doi: 10.1016/j.carres.2009.08.002. [DOI] [PubMed] [Google Scholar]
- Kruppa M, Greene RR, Noss I, Lowman DW, Williams DL. C. albicans increases cell wall mannoprotein, but not mannan, in response to blood, serum and cultivation at physiological temperature. Glycobiology. 2011;21:1173–1180. doi: 10.1093/glycob/cwr051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulkarni RK, Hollingsworth PJ, Volz PA. Variation in cell surface features of Candida albicans with respect to carbon sources. Sabouraudia. 1980;18:255–260. [PubMed] [Google Scholar]
- Lavoie H, Hogues H, Whiteway M. Rearrangements of the transcriptional regulatory networks of metabolic pathways in fungi. Curr Opin Microbiol. 2009;12:655–663. doi: 10.1016/j.mib.2009.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee KK, MacCallum DM, Jacobsen MD, Walker LA, Odds FC, Gow NA, Munro CA. Elevated cell wall chitin in Candida albicans confers echinocandin resistance in vivo. Antimicrob Agents Chemother. 2011;56:208–217. doi: 10.1128/AAC.00683-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenz MC, Fink GR. The glyoxylate cycle is required for fungal virulence. Nature. 2001;412:83–86. doi: 10.1038/35083594. [DOI] [PubMed] [Google Scholar]
- Lorenz MC, Bender JA, Fink GR. Transcriptional response of Candida albicans upon internalization by macrophages. Eukaryot Cell. 2004;3:1076–1087. doi: 10.1128/EC.3.5.1076-1087.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacCallum DM, Castillo L, Nather K, Munro CA, Brown AJ, Gow NA, Odds FC. Property differences among the four major Candida albicans strain clades. Eukaryot Cell. 2009;8:373–387. doi: 10.1128/EC.00387-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacCallum DM, Coste A, Ischer F, Jacobsen MD, Odds FC, Sanglard D. Genetic dissection of azole resistance mechanisms in Candida albicans and their validation in a mouse model of disseminated infection. Antimicrob Agents Chemother. 2010;54:1476–1483. doi: 10.1128/AAC.01645-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martchenko M, Levitin A, Hogues H, Nantel A, Whiteway M. Transcriptional rewiring of fungal galactose-metabolism circuitry. Curr Biol. 2007;17:1007–1013. doi: 10.1016/j.cub.2007.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller DJ, Dufrêne YF. Force nanoscopy of living cells. Curr Biol. 2011;21:R212–R216. doi: 10.1016/j.cub.2011.01.046. [DOI] [PubMed] [Google Scholar]
- Munro CA, Selvaggini S, de Bruijn I, Walker L, Lenardon MD, Gerssen B, et al. The PKC, HOG and Ca2+ signalling pathways co-ordinately regulate chitin synthesis in Candida albicans. Mol Microbiol. 2007;63:1399–1413. doi: 10.1111/j.1365-2958.2007.05588.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navarro-Garcia F, Alonso-Monge R, Rico H, Pla J, Sentandreu R, Nombela C. A role for the MAP kinase gene MKC1 in cell wall construction and morphological transitions in Candida albicans. Microbiology. 1998;144:411–424. doi: 10.1099/00221287-144-2-411. [DOI] [PubMed] [Google Scholar]
- Navarro-Garcia F, Eisman B, Fiuza SM, Nombela C, Pla J. The MAP kinase Mkc1p is activated under different stress conditions in Candida albicans. Microbiology. 2005;151:2737–2749. doi: 10.1099/mic.0.28038-0. [DOI] [PubMed] [Google Scholar]
- Negredo A, Monteoliva L, Gil C, Pla J, Nombela C. Cloning, analysis and one-step disruption of the ARG56 gene of Candida albicans. Microbiology. 1997;143:297–302. doi: 10.1099/00221287-143-2-297. [DOI] [PubMed] [Google Scholar]
- Netea MG, Gow NA, Munro CA, Bates S, Collins C, Ferwerda G, et al. Immune sensing of Candida albicans requires cooperative recognition of mannans and glucans by lectin and Toll-like receptors. J Clin Invest. 2006;166:1642–1650. doi: 10.1172/JCI27114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niimi M, Kamiyama A, Tokunaga M. Respiration of medically important Candida species and Saccharomyces cerevisiae in relation to glucose effect. J Med Vet Mycol. 1988;26:195–197. [PubMed] [Google Scholar]
- O'Hanlon DE, Moench TR, Cone RA. In vaginal fluid, bacteria associated with bacterial vaginosis can be suppressed with lactic acid but not hydrogen peroxide. BMC Infect Dis. 2011;11:200. doi: 10.1186/1471-2334-11-200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odds FC, Van Nuffel L, Gow NA. Survival in experimental Candida albicans infections depends on inoculum growth conditions as well as animal host. Microbiology. 2000;146:1881–1889. doi: 10.1099/00221287-146-8-1881. [DOI] [PubMed] [Google Scholar]
- Perlroth J, Choi B, Spellberg B. Nosocomial fungal infections: epidemiology, diagnosis, and treatment. Med Mycol. 2007;45:321–346. doi: 10.1080/13693780701218689. [DOI] [PubMed] [Google Scholar]
- Pfaller MA, Diekema DJ. Epidemiology of invasive mycoses in North America. Crit Rev Microbiol. 2010;36:1–53. doi: 10.3109/10408410903241444. [DOI] [PubMed] [Google Scholar]
- Piekarska K, Mol E, van den Berg M, Hardy G, van den Burg J, van Roermund C, et al. Peroxisomal fatty acid beta-oxidation is not essential for virulence of Candida albicans. Eukaryot Cell. 2006;5:1847–1856. doi: 10.1128/EC.00093-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plaine A, Walker L, Da Costa G, Mora-Montes HM, McKinnon A, Gow NA, et al. Functional analysis of Candida albicans GPI-anchored proteins: roles in cell wall integrity and caspofungin sensitivity. Fungal Genet Biol. 2008;45:1404–1414. doi: 10.1016/j.fgb.2008.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price MS, Betancourt-Quiroz M, Price JL, Toffaletti DL, Vora H, Hu G, et al. Cryptococcus neoformans requires a functional glycolytic pathway for disease but not persistence in the host. mBio. 2011;2:e00103–e00111. doi: 10.1128/mBio.00103-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramsdale M, Selway L, Stead D, Walker J, Yin Z, Nicholls SM, et al. MNL1 regulates weak acid induced stress responses of the fungal pathogen Candida albicans. Mol Biol Cell. 2008;19:4393–4403. doi: 10.1091/mbc.E07-09-0946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodaki A, Bohovych IM, Enjalbert B, Young T, Odds FC, Gow NA, Brown AJP. Glucose promotes stress resistance in the fungal pathogen Candida albicans. Mol Biol Cell. 2009;20:4845–4855. doi: 10.1091/mbc.E09-01-0002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabina J, Brown V. Glucose sensing network in Candida albicans: a sweet spot for fungal morphogenesis. Eukaryot Cell. 2009;8:1314–1320. doi: 10.1128/EC.00138-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- San José C, Monge RA, Pérez-Díaz R, Pla J, Nombela C. The mitogen-activated protein kinase homolog HOG1 gene controls glycerol accumulation in the pathogenic fungus Candida albicans. J Bacteriol. 1996;178:5850–5852. doi: 10.1128/jb.178.19.5850-5852.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanglard D, Ischer F, Parkinson T, Falconer D, Bille J. Candida albicans mutations in the ergosterol biosynthetic pathway and resistance to several antifungal agents. Antimicrob Agents Chemother. 2003;47:2404–2412. doi: 10.1128/AAC.47.8.2404-2412.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sardi JC, Duque C, Mariano FS, Marques MR, Höfling JF, Gonçalves RB. Adhesion and invasion of Candida albicans from periodontal pockets of patients with chronic periodontitis and diabetes to gingival human fibroblasts. Med Mycol. 2011;50:43–49. doi: 10.3109/13693786.2011.586133. [DOI] [PubMed] [Google Scholar]
- Schaber J, Adrover MA, Eriksson E, Pelet S, Petelenz-Kurdziel E, Klein D, et al. Biophysical properties of Saccharomyces cerevisiae and their relationship with HOG pathway activation. Eur Biophys J. 2010;39:1547–1546. doi: 10.1007/s00249-010-0612-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sexton JA, Brown V, Johnston M. Regulation of sugar transport and metabolism by the Candida albicans Rgt1 transcriptional repressor. Yeast. 2007;24:847–860. doi: 10.1002/yea.1514. [DOI] [PubMed] [Google Scholar]
- Smith DA, Nicholls S, Morgan BA, Brown AJ, Quinn J. A conserved stress-activated protein kinase regulates a core stress response in the human pathogen Candida albicans. Mol Biol Cell. 2004;15:4179–4190. doi: 10.1091/mbc.E04-03-0181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soumya A, Mohamed M, Fatimazahra B, Hassan L, Abdellah H, Fatima H, Saad IK. Study of microbial adhesion on some wood species: theoretical prediction. Mikrobiologiia. 2011;80:47–52. [PubMed] [Google Scholar]
- Stanhill A, Schick N, Engelberg D. The yeast ras/cyclic AMP pathway induces invasive growth by suppressing the cellular stress response. Mol Biol Cell. 1999;19:7529–7538. doi: 10.1128/mcb.19.11.7529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueno K, Matsumoto Y, Uno J, Sasamoto K, Sekimizu K, Kinjo Y, Chibaba H. Intestinal resident yeast Candida glabrata requires Cyb2p-mediated lactate assimilation to adapt in mouse intestine. PLoS ONE. 2011;6:e24759. doi: 10.1371/journal.pone.0024759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vieira N, Casal M, Johansson B, MacCallum DM, Brown AJ, Paiva S. Functional specialization and differential regulation of short-chain carboxylic acid transporters in the pathogen Candida albicans. Mol Microbiol. 2010;75:1337–1354. doi: 10.1111/j.1365-2958.2009.07003.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vylkova S, Carman AJ, Danhof HA, Collette JR, Zhou H, Lorenz MC. The fungal pathogen Candida albicans autoinduces hyphal morphogenesis by raising extracellular pH. mBio. 2011;2:e00055–e00011. doi: 10.1128/mBio.00055-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker LA, Munro CA, de Bruijn I, Lenardon MD, McKinnon A, Gow NA. Stimulation of chitin synthesis rescues Candida albicans from echinocandins. PLoS Pathog. 2008;4:e1000040. doi: 10.1371/journal.ppat.1000040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willis AM, Coulter WA, Hayes JR, Bell P, Lamey PJ. Factors affecting the adhesion of Candida albicans to epithelial cells of insulin-using diabetes mellitus patients. J Med Microbiol. 2000;49:291–293. doi: 10.1099/0022-1317-49-3-291. [DOI] [PubMed] [Google Scholar]
- Wilson D, Tutulan-Cunita A, Jung W, Hauser NC, Hernandez R, Williamson T, et al. Deletion of the high-affinity cAMP phosphodiesterase encoded by PDE2 affects stress responses and virulence in Candida albicans. Mol Microbiol. 2007;65:841–856. doi: 10.1111/j.1365-2958.2007.05788.x. [DOI] [PubMed] [Google Scholar]
- Yin Z, Smith RJ, Brown AJP. Multiple signalling pathways trigger the exquisite sensitivity of yeast gluconeogenic mRNAs to glucose. Mol Microbiol. 1996;20:751–761. doi: 10.1111/j.1365-2958.1996.tb02514.x. [DOI] [PubMed] [Google Scholar]
- Yoshijima Y, Murakami K, Kayama S, Liu D, Hirota K, Ichikawa T, Miyake Y. Effect of substrate surface hydrophobicity on the adherence of yeast and hyphal Candida. Mycoses. 2010;53:221–226. doi: 10.1111/j.1439-0507.2009.01694.x. [DOI] [PubMed] [Google Scholar]
- Zaman S, Lippman SI, Zhao X, Broach JR. How Saccharomyces responds to nutrients. Annu Rev Genet. 2008;42:27–81. doi: 10.1146/annurev.genet.41.110306.130206. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


