Abstract
Afipia felis is a Gram-negative bacterium that causes some cases of human Cat Scratch Disease. A. felis can survive and multiply in several mammalian cell types, including macrophages, but the precise intracellular compartmentalization of A. felis-containing phagosomes is unknown. Here, we demonstrate that, in murine macrophages, most A. felis-containing phagosomes exclude lysosomal tracer loaded into macrophage lysosomes before, as well as endocytic tracer loaded after, establishment of an infection. Established Afipia-containing phagosomes possess neither early endosomal marker proteins [early endosome antigen 1 (EEA1), Rab5, transferrin receptor, trytophane aspartate containing coat protein (TACO)] nor late endosomal or lysosomal proteins [cathepsin D, β-glucuronidase, vacuolar proton-pumping ATPase, rab7, mannose-6-phosphate receptor, vesicle-associated membrane protein 8, lysosome-associated membrane proteins LAMP-1 and LAMP-2]. Those bacteria that will be found in a nonendosomal compartment enter the macrophage via an EEA1-negative compartment, which remains negative for LAMP-1. The smaller subpopulation of afipiae whose phagosomes will be part of the endocytic system enters into an EEA1-positive compartment, which also subsequently acquires LAMP-1. Killing of Afipia or opsonization with immune antibodies leads to a strong increase in the percentage of A. felis-containing phagosomes that interact with the endocytic system. We conclude that most phagosomes containing A. felis are disconnected from the endosome–lysosome continuum, that their unusual compartmentalization is decided at uptake, and that this compartmentalization requires bacterial viability.
Afipia felis (1) is a member of the α-2 subgroup of Gram-negative proteobacteria and was first described by English et al. (2) as the microorganism causing Cat-Scratch Disease, a usually focal, self-resolving lymphadenitis, typically occurring after a kitten scratch. Most cases of Cat-Scratch Disease are, however, caused by closely related Bartonella henselae (3, 4). A. felis is a facultative intracellular bacterium and can survive or multiply in amoebas (5), macrophages, and some epithelial and endothelial cells in vitro (3, 6). This fits nicely with the observation that Afipia was found both in macrophages and capillary walls when lymph node biopsies from Cat-Scratch Disease patients were studied (2).
Very little is known about the intracellular compartmentalization of A. felis-containing phagosomes (ACPs), except that only a minor percentage acquires preloaded lysosomal ferritin in macrophages when compared with phagosomes containing latex beads (3, 7). Maintenance of this unfused compartment depends on bacterial viability (7). Normal phagosomes, however, mature stepwise into phagolysosomes (8–13), and their maturation contributes to killing and degradation of ingested microorganisms and to presentation of their antigens to T cells (14).
Inhibition of phagolysosome formation is a feature that A. felis shares with several other intracellular pathogens, such as Mycobacterium tuberculosis, Legionella pneumophila, Chlamydia trachomatis, or the protozoan Toxoplasma gondii, with each of these occupying a different endocytic or, in some cases, nonendocytic niche (reviewed in refs. 10, 11, and 14).
In this study, we examined the molecular nature and composition of ACPs. We show that most afipiae enter macrophages into an ACP that does not belong to the endosome–lysosome continuum.
Materials and Methods
Basic Reagents.
All reagents were obtained from Sigma unless stated otherwise. Secondary antibodies to primary rabbit, mouse, and rat antibodies (labeled with either Alexa 488, fluorescein, or rhodamine) were purchased from Jackson ImmunoResearch and Molecular Probes.
Sources and Cultivation of Cell Lines and Bacteria.
J774E, a strongly mannose receptor-expressing murine macrophage-like cell line, was kindly donated by P. D. Stahl and M. Colombo (Washington University, St. Louis, MO) and grown at 37°C in 5% CO2 in RPMI 1640 medium/2 mM glutamine (GIBCO)/10% FCS (GIBCO). The type strain of A. felis (American Type Culture Collection 53690/DSM 7326) was purchased from the German Collection of Microorganisms. Further patient isolates of A. felis (B91-007360, B91-007391, B91-007334, all ref. 1; AfTA-1, ref. 4) were kindly provided by F. Quinn (Centers for Disease Control, Atlanta, GA) and M. Giladi (Tel Aviv Sourasky Medical Center, Tel Aviv) and Bordetella pertussis strains TIΔtox and Bp347 by R. Gross (Microbiology, University of Würzburg, Germany). Listeria innocua serovar 6a was from the Institute's strain collection.
Sources of Antibodies.
Polyclonal antibodies to Afipia were raised by injecting formaldehyde-killed bacteria subdermally into New Zealand White rabbits (special thanks to J. Thalhammer, University of Salzburg, Salzburg, Austria), those to L. innocua were a kind gift from H. Hof (University of Mannheim, Mannheim, Germany), and those to Bordetella were from Difco. Monoclonal murine anti-A. felis (Fig. 1A clone 1B6-4, subtype IgG3, titer 1:4,096) or clone 1B3-2, subtype IgG2b, titer 1:1,024 [used in experiments with lysosome-associated membrane protein (LAMP)-1 colocalization] were generously provided by D. Raoult and H. LePocher (Faculté de Médécine, Université de Marseille, Marseille, France; ref. 15). The generous gifts of polyclonal rabbit antibodies to (in alphabetical order) vacuolar ATPase (33-kDa “E” subunit) (D. Brown, Massachusetts General Hospital and Harvard University, Boston, MA, and Y. Moriyama, Osaka University, Osaka); to calnexin (A. Helenius, Eidgenössische Technische Hochschule, Zürich); to cathepsin D (E. Kominami, Juntendo University, Tokyo), to cation-dependent mannose-6-phosphate receptor = CD-M6PR (Y. Uchiyama, Osaka University, Osaka); to early endosome antigen (EEA1; G. Mills and M. Clague, University of Liverpool, Liverpool, U.K.); to β-glucuronidase (W. Sly, Washington University, St. Louis); to the Golgi protein GM130 (M. Lowe, University of Manchester, Manchester, U.K.); to murine lysosomal integral membrane protein 2 (H. Fujita, Kyushu University, Fukuoka, Japan); to rab5 (P. van der Sluijs, University of Utrecht, Utrecht, The Netherlands); to rab7 (I. Sandoval, Universidad Autonoma, Madrid); to tryptophane aspartate-containing coat protein (TACO; J. Pieters, Basel Institute for Immunology, Basel); to mitochondrial TOM-20 (K. Mihara, Kyushu University, Fukuoka, Japan); to vesicle-associated membrane protein (VAMP) 8 = endobrevin (W. Antonin and R. Jahn, Max Planck Institute for Biophysical Chemistry, Göttingen, Germany) were all highly appreciated. Monoclonal rat antibodies to murine LAMP-1 (clone 1D4B) and to murine LAMP-2 (clone ABL-93) were donated by U. Schaible (Max Planck Institute for Infection Biology, Berlin) or obtained from the Developmental Study Hybridoma Bank (maintained by the University of Iowa, Iowa City, IA). Monoclonal mouse antibody to the transferrin receptor (clone H68.4) was obtained from Zymed.
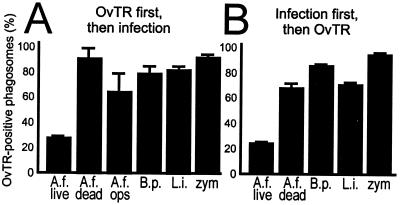
Figure 1.
Accessibility of phagosomes to OvTR. (A) OvTR was preloaded into J774E lysosomes and macrophages were infected for 1 h with live A. felis (A.f. live), heat-killed A. felis (A.f. dead), or live A. felis opsonized with monoclonal antibodies (A.f. ops), B. pertussis Bp347 (B.p.), L. innocua (L.i.), or zymosan (zym), and the infection was chased for 2 h. Bacteria colocalizing with red fluorophore were counted positive (three independent experiments with between 110 and 370 phagosomes counted per experiment per data set). Here and elsewhere, means and SD are shown. (B) Infection with the indicated bacteria was established for 16 h, followed by the addition of OvTR for 3 h. Analysis as in (A) with between 45 and 127 phagosomes counted for each data set in each of three independent experiments.
Immunofluorescence, Ovalbumin-Texas Red (OvTR) Colocalization, and Electron Microscopy Analyses.
Afipia were grown for 3 days on buffer-charcoal-yeast extract agar plates (1) at 30°C, Listeria overnight in brain–heart infusion (Difco) at 37°C, and Bordetella at 37°C in Stainer–Scholte liquid media (16). Infections were done in 24-well cell culture plates (Nunc) with 12-mm glass slides. Macrophages were grown for 2 days to 80% confluency and then infected at a multiplicity of infection of 10 (Afipia), 10 (Listeria), 10 (Bordetella), or 5 (zymosan) for 30 min at 4°C with centrifugation at 160 × g. After infection, media were removed and macrophages chased in fresh media at 37°C/5% CO2 for up to 120 min and fixed (17). Analysis of colocalization of bacteria with OvTR conjugate (Molecular Probes) was modified from ref. 18 as follows.
Mode 1. OvTR added before infection.
J774E cells were incubated in 1 ml of medium containing 15 μg of OvTR at 37°C/5% CO2 for 16 h and washed three times with PBS, and OvTR was chased into lysosomes for 2 h in label-free medium, followed by infection with particles for 1 h, another chase after removal of particles for 2 h, and further processing as described in ref. 18. In this and all other fluorescence experiments, zymosan or bacteria were prelabeled in 200 μg N-hydroxysuccinimide-fluorescein or N-hydroxysuccinimide-Texas Red (Molecular Probes) per milliliter and 100 mM sodium carbonate (pH 8.2) for 30 min at 4°C, then extensively washed. This labeling had no detectable effect on uptake efficiency or compartmentalization (data not shown). For opsonization, A. felis was preincubated in 1% monoclonal antibody 1B6-4 or 0.2% monoclonal antibody 1B3-2 in RPMI 1640 medium for 30 min at 37°C. Where indicated, afipiae were killed (>99% dead) by heating for 10 min at 70°C or by treating with 1% paraformaldehyde in PBS for 2 h at 4°C before infection.
Mode 2. OvTR added after infection.
Infection for 1 h was as described above, followed by a 16-h chase and addition of 1 ml of fresh medium containing 15 μg of OvTR. Next was an incubation for 3 h at 37°C/5% CO2. Samples were washed three times with PBS and immediately fixed and processed as described in ref. 18. As a control, permeabilized vs. nonpermeabilized infected macrophages were probed with polyclonal anti-Afipia to determine the percentage of Afipia inside vs. outside macrophages, which was determined to be 94% or more (data not shown). Electron microscopy of structural integrity of purified phagosomes was assessed as described in ref. 17.
Purification of Phagosomes.
The detailed protocol will be published elsewhere (17). Briefly, J774E macrophages (≈1–2 × 108 cells) were infected for 1 h, the noningested bacteria were washed away, and the infection was allowed to continue for 2 h at 37°C/5% CO2. Infected macrophages were washed in PBS, homogenization buffer (250 mM sucrose/0.5 mM EGTA/20 mM Hepes/KOH, pH 7.2), and homogenization buffer without EGTA, then lysed in a stainless steel homogenizer (Wheaton Scientific) in the presence of a protease inhibitor mix. The homogenate was incubated at 37°C for 5 min with Benzonase, and a postnuclear supernatant (PNS) was produced by centrifugation for 3 min at 440 × g and collection of the supernatant. The PNS was adjusted to 39% (wt/vol) sucrose/0.5 mM EGTA/20 mM Hepes/KOH (pH 7.2) and run for 1 h at 100,000 × g (SW40Ti Beckman centrifuge) in a discontinuous 65%/55%/39% (sample)/32.5%/10% sucrose gradient in homogenization buffer. The 55–65% fractions (some 2 ml) were removed, adjusted to ≈11% sucrose, placed on a 15% Ficoll (70, 000 Da) cushion, and centrifuged at 18,000 × g for 20 min (SW40Ti). The resulting pellet was resuspended in 11 ml of homogenization buffer and centrifuged at 18, 000 × g for 10 min (SW40Ti). The pellet was considered the product, contained ≈40% of the bacteria in the PNS (life cell count), was taken up in 0.1 ml of homogenization buffer, and the host proteins were collected from the supernatant after detergent treatment (0.2% Triton X-100) and centrifugation for 5 min at 20, 000 × g at 4°C in a minifuge. Protein concentration was determined with BSA as a standard (Bio-Rad DC Protein Assay). Western blot analysis was from samples boiled in 1× Laemmli sample buffer and run in a 10% SDS–polyacrylamide gel. Polyvinylidene difluoride membranes (Amersham Pharmacia Biotech) were used with chemiluminescence (Amersham Pharmacia Biotech) and horseradish peroxidase-coupled secondary antibodies (Dianova, Hamburg, Germany).
Results
Most A. felis-containing Phagosomes Do Not Acquire Luminal Endosomal or Lysosomal Tracers.
ACPs were reported to not mature into phagolysosomes compared with latex bead-containing phagosomes (3). We first tested this interpretation by using further assays of phagosome maturation with various microbial probes, i.e., B. pertussis, L. innocua, or a baker's yeast cell wall preparation (zymosan). These particles were chosen because the maturation kinetics of phagosomes containing latex beads can be different from that of phagosomes containing degradable material (19) and because maturation of phagosomes containing Gram-negative B. pertussis or Gram-positive L. innocua was reported to be normal (16, 20). Zymosan particles have long been an excellent tool to study undisturbed phagosome maturation (21, 22).
Macrophage (J774E) lysosomes were selectively prelabeled with OvTR (18), as determined by double immunofluorescence experiments with antibodies to late endosomal/lysosomal LAMP-1, which virtually completely colocalized with OvTR-preloaded organelles. Little colocalization was seen with antibodies to late endosomal rab7, as expected for a lysosomal compartment (data not shown). Only 27% of the phagosomes containing live A. felis were positive for Texas Red, i.e., matured to phagolysosomes, compared with 80–90% maturation of phagosomes containing either heat-killed or opsonized Afipia, or L. innocua, B. pertussis, or zymosan (Fig. 1A). Four other clinical isolates of A. felis (4, 6) that we have tested behaved identically, and maturation of ACPs was inhibited in primary murine bone marrow-derived macrophages as well (data not shown). Interestingly, ACPs colocalized much more frequently with lysosomal OvTR when afipiae were preincubated with monoclonal antibody raised to A. felis (15) (Fig. 1A). To further test whether ACPs would receive OvTR when the tracer was added to previously infected macrophages, macrophages with an established infection were exposed to OvTR for 3 h, followed by immediate fixation. Such treatment fills all levels of the endocytic system with OvTR. As fusion occurs largely between endocytic organelles and phagosomes of the same maturation stage (9, 23), any phagosome that fused with endocytic vesicles is expected to label positively. Only 24% of ACPs stained positive for OvTR, whereas ≈70–90% of the phagosomes containing either heat-killed Afipia or Listeria, Bordetella, or zymosan accumulated OvTR (Fig. 1B), suggesting that most ACPs are not accessible to the endocytic tracer. Formaldehyde fixation of bacteria or opsonization with monoclonal antibodies (15) before infection also leads to a much increased frequency of colocalization of Afipia-containing phagosomes with late endosomal/lysosomal LAMP-1 (62 ± 2% and 59 ± 5%, respectively, vs. 26 ± 2% with untreated bacteria).
Most A. felis-Containing Phagosomes Do Not Contain Endosomal or Lysosomal Marker Proteins.
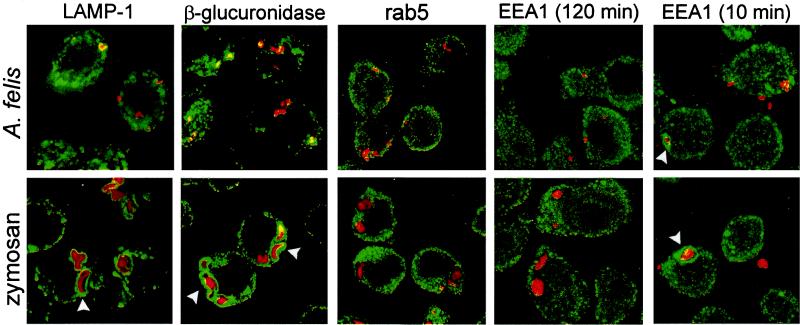
The above experiments suggested that most ACPs were not part of the endosome–lysosome continuum. To test this hypothesis, we performed confocal laser scanning immunofluorescence microscopy of J774E macrophages infected for 2 h using a number of antibodies to typical resident proteins of early and late endosomes or lysosomes (8, 10) and to other subcellular compartments. We found that only some 25–30% of ACPs were positive for any of the late endosomal/lysosomal marker proteins LAMP-1, LAMP-2, β-glucuronidase, the GTPase rab7, or the generally endocytic VAMP-8 (endobrevin) (24, 25) (Table 1 and Fig. 2), whereas some 90% of the zymosan-containing phagosomes were positive. Neither of the early endosomal markers EEA1, rab5, TACO, or transferrin receptor were present on more than ≈5% of any phagosome type after a 2-h chase (Table 1 and Fig. 2), suggesting that established ACPs are not early endosomal compartments either. None of the phagosomes colocalized frequently with antibodies to diagnostic proteins for endoplasmic reticulum (ER; calnexin) or the trans-Golgi (CD-M6PR) (Table 1).
Table 1.
Organelle marker content of phagosomes containing A. felis or zymosan analyzed by confocal immunofluorescence microscopy
| Marker protein | Organelle | Phagosomes containing
|
|
|---|---|---|---|
| A. felis (%) | Zymosan (%) | ||
| rab5 | EE | 4 ± 4 (444) | 7 ± 7 (322) |
| EEA1 | EE | 2 ± 3 (447) | 3 ± 2 (169) |
| Transferrin receptor | EE | 4 ± 2 (321) | 9 ± 4 (281) |
| TACO | EE/PM* | 4 ± 2 (242) | ND |
| VAMP-8 | EE/LE | 36 ± 11 (672) | 72 ± 12 (352) |
| rab7 | LE | 4 ± 3 (292) | ND |
| CD-M6PR | Golgi/LE | 1 ± 1 (482) | 5 ± 6 (220) |
| LAMP-1 | LE/lys | 26 ± 2 (255) | 84 ± 9 (332) |
| LAMP-2 | LE/lys | 29 ± 4 (158) | 89 ± 12 (254) |
| β-Glucuronidase | lys | 33 ± 12 (614) | 77 ± 11 (242) |
| Calnexin | ER | 4 ± 2 (400) | 9 ± 4 (236) |
J774E cells were infected for 1 h, chased for 2 h, and processed with the respective antibodies as described in Materials and Methods. Analysis was by confocal immunofluorescence microscopy. Data indicate percentage of phagosomes positive for the respective marker protein and are from 3 to 10 independent experiments. Mean and SDs and number of phagosomes counted (in parentheses) are indicated.
EE, early endosome; LE, late endosome; lys, lysosome; PM, plasma membrane; ND, not determined.
Predominantly PM and early phagosomes (27).
Figure 2.
Most ACPs lack endocytic marker proteins. J774E macrophages containing Texas Red-labeled A. felis or zymosan internalized for 30 min at 4°C and 160 × g, chased for 10 or 120 min, formaldehyde fixed, decorated with the indicated antibodies to compartmental organelle markers (LAMP-1 and β-glucuronidase for late endosomal/lysosomal organelles, rab5 and EEA1 for early endocytic structures), detected by a secondary green fluorescent Alexa 488-labeled antibody, and colocalization with bacteria-bound Texas Red was investigated by confocal laser scanning microscopy on a Leica microscope. Photographs are shown as overlays. Note strong accumulation of LAMP-1 and β-glucuronidase around zymosan phagosomes at 120 min (arrowheads) and of EEA1 around some phagosomes (arrowheads) early after entry.
Afipia Does Not Enter Through a Compartment with Characteristics of an Early Endosome.
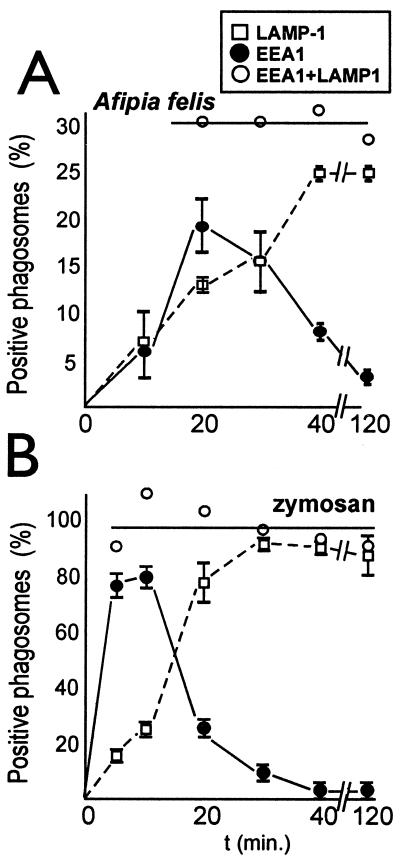
Because established ACPs did not show any feature of the endocytic pathway, we determined whether this unusual compartmentalization is already established as bacteria are taken up or whether they would first be found in an endocytic compartment, which they would later force to develop in an unusual direction. ACPs or zymosan-containing phagosomes were chased for various times at 37°C (Fig. 3). Most adherent A. felis enter the macrophages between 10 and 20 min after starting of the 37°C chase, whereas most zymosan particles are taken up within the first 5 min (data not shown). Early endosomal EEA1 can be found very frequently on early zymosan-containing phagosomes and, a little delayed because of a retarded uptake of the bacteria, on a subpopulation of ACPs (Fig. 3). Once all bound particles have been taken up (after 5 min for zymosan, ≈20 min for Afipia), the added percentages for EEA1 and LAMP-1 colocalization (Fig. 3, ○) on either phagosome type was roughly constant, as would be expected if the presence of these proteins would be mutually exclusive and if they quickly succeeded each other, as has, in fact, been shown previously (26). These added percentages represent the portion of phagosomes that are first EEA1 positive and then become positive for LAMP-1, being 30% for ACPs and almost 100% for zymosan-containing phagosomes. These data are in excellent agreement with the results of the above OvTR chase experiments (Fig. 1A). To conclude, this experiment demonstrates that only 30% of ACPs, but 100% of zymosan-containing phagosomes, belong to the endosomal–lysosomal continuum, as defined by the consecutive presence of EEA1 and LAMP-1.
Figure 3.
Kinetics of acquisition of early endosomal EEA1 or late endosomal LAMP-1 by phagosomes containing either Afipia or zymosan. APCs (A) and zymosan-containing phagosomes (B) (please note different scales in A and B). Experiments were done as in Fig. 2 with a spin at 160 × g for 30 min in a tabletop ELISA plate centrifuge with swingout rotor at 4°C, followed by removal of media and addition of fresh 37°C media (being set as 0 min), plus chase for up to 120 min. Glass slide preparations were fixed at the indicated times and rings of antibody staining by using anti-LAMP-1 (□) or anti-EEA1 (●) were counted positive in confocal microscopy. For every time tested, added colocalization frequencies are indicated as well (○), equaling the number of phagosomes that are part of the endocytic system. Results are from three independent experiments with a minimum of 50 phagosomes counted per time and sample type.
Biochemical Analysis of A. felis-Containing Phagosomes.
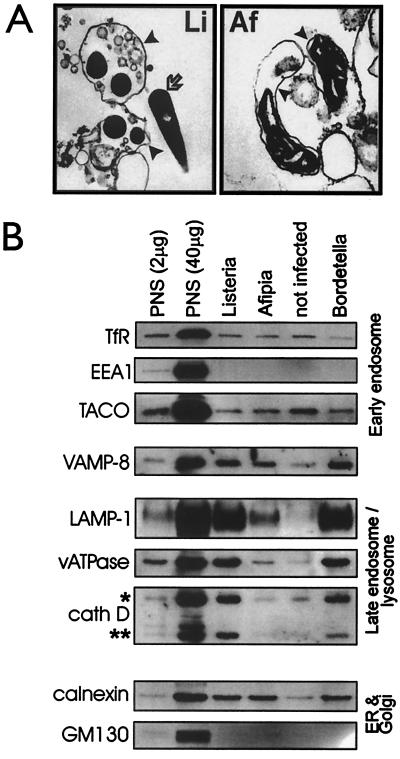
To analyze further the compartmentalization of ACPs, we established a phagosome purification protocol and purified phagosomes containing either A. felis, noninterfering B. pertussis, or L. innocua. Macrophages were infected for 1 h, followed by a 2-h chase and phagosome isolation. Isolated phagosomes were analyzed for their content in various subcellular marker proteins, which was compared with the accumulation of markers in “pseudophagosome preparations” (from uninfected macrophages) to analyze the level of organelle contamination of our preparations. Also, enrichment of marker proteins relative to the starting homogenate (PNS) was followed by using the same protein concentration in the lanes containing PNS or phagosome preparations (2 μg) or 20-fold concentrated PNS (40 μg) in one lane, respectively (Fig. 4B). Although phagosomes with Listeria or Bordetella contained all of the late endosomal/lysosomal marker proteins LAMP-1, proton-pumping vacuolar ATPase (33-kDa E subunit), mature cathepsin D (all Fig. 4B), β-glucuronidase, acid β-galactosidase, and lysosomal integral membrane protein 2 (all not shown), they accumulated none of the early endosomal marker proteins EEA1, transferrin receptor (all Fig. 4B) or rab5 (data not shown), as is expected after a 2-h chase. These phagosomes were also not enriched in the TACO protein previously reported to be an important retained factor during biogenesis of Mycobacterium-containing phagosomes (27); in fact, all phagosome preparations contained less TACO than the pseudophagosome preparation (Fig. 4B). All of the phagosome preparations had increased concentrations of ER marker compared with the pseudophagosome preparation (Fig. 4B). Equal small contaminations from endosomes, lysosomes (Fig. 4B), or mitochondria (marker TOM20; ref. 17) were determined, and no accumulation of a Golgi marker protein (GM130) was seen (Fig. 4B). In additional experiments, the macrophage plasma membrane was labeled by binding horseradish peroxidase, and only 0.1–0.2% of the horseradish peroxidase activity in the PNS was recovered in our phagosome preparations, independent of the phagocytic probe used (17). In summary, these data suggest a high degree of purity of our phagosome preparations.
Figure 4.
Analysis of isolated phagosomes. Phagosomes containing either A. felis, B. pertussis TIΔtox, or L. innocua, or a pseudophagosome fraction (not infected) from uninfected macrophages were isolated as described in Materials and Methods. (A) Samples were processed for electron microscopy (L. innocua, magnification ×10,000; A. felis, ×24,000). Phagosome membranes are marked by arrowheads and a free longitudinally cut Listeria by an arrow. (B) Phagosomal protein was analyzed by Western blot analysis by using antibodies to organelle marker proteins (see Materials and Methods). *, immature and intermediate 48- to 51-kDa precursor forms of cathepsin D (cath D); **, mature form (31 kDa). Either 2 or 40 μg protein of the PNS fractions or 2 μg protein of each phagosomal fraction were analyzed. Acronyms and sources of antibodies are listed in Materials and Materials. Each result is representative for at least three independent experiments.
In contrast to phagosomes containing Bordetella or Listeria, ACPs contained only minute amounts of all above endosomal or lysosomal marker proteins with some enrichment in vacuolar ATPase, LAMP-1, VAMP-8/endobrevin, and calnexin over the pseudophagosome preparation (Fig. 4B). Enrichment in late endosomal markers was probably because of the LAMP-1-positive phagolysosomal subpopulation of ACPs present in the infected cells. We have not as yet identified any host protein that is distinctively present on ACPs during the preparation. Selective disruption of ACPs was excluded as a cause for this phenomenon, as ACPs are intact to at least the same degree as are phagosomes containing noncytolytic Listeria (as judged by electron microscopy; Fig. 4A).
Discussion
In this study, we present evidence that ≈70% of ACPs do not have any endocytic characteristic, suggesting that these phagosomes represent a novel phagosome compartment. This hypothesis is supported by the facts that neither did ovalbumin preloaded into lysosomes enter most ACPs, nor did ovalbumin loaded into the endocytic system after infection, and the percentages of ACPs positive for late endosomal/lysosomal resident proteins (immunofluorescence) were as low as those for OvTR colocalization. Furthermore, these marker proteins were present only at low concentrations, as suggested by the biochemical evidence; early endosomal proteins were rarely detectable in any of the samples tested at 2-h postinfection and, even early during infection, only some 30% of ACPs showed endocytic characteristics. In addition, our preliminary data show a redirection of biogenesis of ACPs in primary murine bone marrow-derived macrophages and human peripheral blood monocytes as well (A.S., A.L., and A.H., unpublished data).
Other compartments that share some features with ACPs are phagosomes containing either facultative intracellular L. pneumophila, the obligate intracellular pathogen C. trachomatis, or the parasite T. gondii. Very recent evidence suggests that legionellae also enter macrophages directly into a nonendocytic compartment (28) that is closely associated with mitochondria at early, and ribosomes at later, stages of the infection (30). Failure to form such a compartment is correlated with killing of bacteria (31). In fact, the proportion of ACPs positive for LAMP-1 (25–30%) compares very well with the 20–35% LAMP-1-positive L. pneumophila-containing phagosomes in macrophages (28, 32). Interestingly, nonendocytic Legionella-containing compartments will, by an unknown mechanism, turn into endocytic ones after some 18 h (33). We have, by electron microscopy, not yet seen a close association of mitochondria or ribosomes with ACPs; therefore, it is unlikely that ACPs and Legionella-containing phagosomes are the same compartment.
C. trachomatis-containing phagosomes also do not contain detectable endosomal markers (34, 35) and are spacious compartments that intersect with the secretory system of the host cell (36). Chlamydiae may enter macrophages via a nonendocytic compartment too, although this question has not as yet been directly addressed. Finally, Toxoplasma-containing phagosomes are different from all others in that they are formed when the parasite enters the host cell through a narrow collar-like structure in the host cell plasma membrane (37). This collar strips off most of the plasma membrane's integral proteins and leads to formation of a phagosome that contains very few host proteins (37, 38). We have seen neither very spacious phagosomes nor collar-like structures during or after infection with A. felis.
Whereas all of the above phagosomes do not show endocytic characteristics, macrophage phagosomes containing Ehrlichia chaffensis (39) or pathogenic mycobacteria (40, 41) do, although they do not fuse with preexisting lysosomes. Mycobacterium-containing phagosomes are accessible to both externally added horseradish peroxidase (41) and to cholera toxin B subunit (40), and they receive immature cathepsin D from a biosynthetic pool (42). Therefore, Afipia-, Legionella-, Toxoplasma-, or Chlamydia-containing phagosomes can be classified into the “nonendocytic” group of phagosomes with a restricted or missing interaction with the endocytic system, whereas phagosomes containing pathogenic mycobacteriae, Brucella (43, 44), or Ehrlichia (39) continuously interact with the endosomal system. Phagosomes containing Salmonella are a mosaic case of trafficking in that they show a lack of fusion with lysosomes yet they acidify and acquire lysosomal glycoproteins (45–47).
Unusual trafficking of ACPs requires that the bacteria are alive, because heat-killing or formaldehyde fixation of afipiae promotes phagolysosome formation. This is in line with previous observations that killing of intracellular A. felis by antibiotics during infection promotes phagolysosome formation (7). Similarly, opsonization of live bacteria with immune antibodies leads to a strongly increased level of phagolysosome formation of ACPs (and an increased uptake of afipiae; data not shown). These data strongly suggest that A. felis must enter macrophages via an as-yet-unspecified macrophage receptor (14) different from the Ig receptors to end up in an unusual compartment, and that continued bacterial metabolism is required for the maintenance of this compartment. Bound immune antibodies normalize biogenesis not only of ACPs but also of phagosomes containing some other relevant pathogens, such as Legionella, Chlamydia, Toxoplasma, or Mycobacterium (reviewed in ref. 10) but not of phagosomes containing Brucella abortus (43).
All of our phagosome preparations had increased concentrations of ER marker compared with the pseudophagosome preparation. This finding is particularly interesting, as ER proteins are clearly enriched in preparations of highly purified latex bead-containing phagosomes as well (8). Whether such acquisition of ER by bacteria-containing phagosomes occurs via the fusion of autophagic organelles (potentially containing ER) with phagosomes or phagolysosomes or whether it occurs via a particular “stickiness” of phagosomes for ER fragments remains to be determined. That only a few percent of ACPs are positive for calnexin, as judged by immunofluorescence analysis, and that all phagosome types, unusual or not, seem to have similar levels of ER contents (Fig. 4B) speaks for an “unspecific” attachment. ER vesicles could be a major contributor to vesicular nonphagosomal impurities in our phagosome preparations (Fig. 4A). This would not exclude a potentially interesting and biologically significant role for ER elements in a subpopulation of phagosomes.
The increased concentration of TACO in the pseudophagosome fraction can be explained by the decrease of the absolute amount of organelle contamination protein when phagosomes are contained in a sample and a constant protein concentration (in our experiments, 2 μg/lane) is applied to the gel: If phagosomes have no TACO at all but there is some on a contaminating organelle, then its concentration is exacerbated in pseudophagosome samples.
It will be interesting to determine the precise macrophage surface receptor for Afipia and also to identify representative proteins of the ACPs to determine the exact compartmentalization of these phagosomes.
Acknowledgments
We are very grateful to many colleagues (see Materials and Methods) for generous gifts of antibodies and cells. We cordially thank W. Goebel and our other colleagues at the Lehrstuhl für Mikrobiologie for their hospitality and support, and we thank C. Gehrig, G. Griffiths, and G. Krohne for help with electron microscopy. This study was supported by a grant (Ha 1929/3-3) and a Heisenberg Fellowship (Ha 1929/3-4) from the Deutsche Forschungsgemeinschaft and by a grant from the Fonds der Chemischen Industrie (to A.H.).
Abbreviations
- ACPs
Afipia-containing phagosomes
- EEA1
early endosome antigen 1
- LAMP
lysosome-associated membrane protein
- OvTR
Ovalbumin-Texas Red conjugate
- TACO
tryptophane aspartate containing coat protein
- VAMP
vesicle-associated membrane protein
- PNS
postnuclear supernatant
- ER
endoplasmic reticulum
References
- 1.Brenner D J, Hollis D G, Moss C W, English C K, Hall G S, Vincent J, Radosevic J, Birkness K A, Bibb W F, Quinn F D, et al. J Clin Microbiol. 1991;29:2450–2460. doi: 10.1128/jcm.29.11.2450-2460.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.English C K, Wear D J, Margileth A M, Lissner C R, Walsh G P. J Am Med Assoc. 1988;259:1347–1352. doi: 10.1001/jama.259.9.1347. [DOI] [PubMed] [Google Scholar]
- 3.Brouqui P, Raoult D. Microb Pathog. 1993;15:187–195. doi: 10.1006/mpat.1993.1069. [DOI] [PubMed] [Google Scholar]
- 4.Giladi M, Avidor B, Kletter Y, Abulafia S, Slater L N, Welch D F, Brenner D J, Steigerwalt A G, Whitney A M, Ephros M. J Clin Microbiol. 1998;36:2499–2502. doi: 10.1128/jcm.36.9.2499-2502.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.La Scola B, Raoult D. Lancet. 1999;353:1330. doi: 10.1016/s0140-6736(99)00906-x. [DOI] [PubMed] [Google Scholar]
- 6.Birkness K A, George V G, White E H, Stephens D S, Quinn F D. Infect Immun. 1992;60:2281–2287. doi: 10.1128/iai.60.6.2281-2287.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Le Pocher H, Brouqui P, Raoult D. J Antimicrob Chemother. 1998;42:825–829. doi: 10.1093/jac/42.6.825. [DOI] [PubMed] [Google Scholar]
- 8.Garin J, Diez R, Kieffer S, Dermine J F, Duclos S, Gagnon E, Sadoul R, Rondeau C, Desjardins M. J Cell Biol. 2000;152:165–180. doi: 10.1083/jcb.152.1.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Desjardins M, Nzala N N, Corsini R, Rondeau C. J Cell Sci. 1997;110:2303–2314. doi: 10.1242/jcs.110.18.2303. [DOI] [PubMed] [Google Scholar]
- 10.Haas A. Mol Membr Biol. 1998;15:103–121. doi: 10.3109/09687689809074522. [DOI] [PubMed] [Google Scholar]
- 11.Méresse S, Steele-Mortimer O, Moreno E, Desjardins M, Finlay B, Gorvel J P. Nat Cell Biol. 1999;1:E183–E188. doi: 10.1038/15620. [DOI] [PubMed] [Google Scholar]
- 12.Pitt A, Mayorga L S, Stahl P D, Schwartz A L. J Clin Invest. 1992;90:1978–8193. doi: 10.1172/JCI116077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Duclos S, Desjardins M. Cell Microbiol. 2000;2:365–377. doi: 10.1046/j.1462-5822.2000.00066.x. [DOI] [PubMed] [Google Scholar]
- 14.Schaible UE, Collins H L, Kaufmann S H. Adv Immunol. 1999;71:267–377. doi: 10.1016/s0065-2776(08)60405-8. [DOI] [PubMed] [Google Scholar]
- 15.Yu X, Raoult D. Am J Clin Pathol. 1994;101:603–606. doi: 10.1093/ajcp/101.5.603. [DOI] [PubMed] [Google Scholar]
- 16.Schneider B, Gross R, Haas A. Infect Immun. 2000;68:7039–7048. doi: 10.1128/iai.68.12.7039-7048.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lührmann, A. & Haas, A. (2001) Methods Cell Sci., in press. [DOI] [PubMed]
- 18.Swanson M S, Isberg R R. Infect Immun. 1996;64:2585–2594. doi: 10.1128/iai.64.7.2585-2594.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Griffiths G. Protoplasma. 1996;195:37–58. [Google Scholar]
- 20.Schwan W R, Demuth A, Kuhn M, Goebel W. Infect Immun. 1994;62:4795–4803. doi: 10.1128/iai.62.11.4795-4803.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang Y L, Goren M B. J Cell Biol. 1987;104:1749–1754. doi: 10.1083/jcb.104.6.1749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Düzgünes N, Majumdar S, Goren M B. Methods Enzymol. 1993;221:234–238. doi: 10.1016/0076-6879(93)21020-9. [DOI] [PubMed] [Google Scholar]
- 23.Ward D M, Pevsner J, Scullion M A, Vaughn M, Kaplan J. Mol Biol Cell. 2000;11:2327–2333. doi: 10.1091/mbc.11.7.2327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Antonin W, Holroyd C, Tikkanen R, Höning S, Jahn R. Mol Biol Cell. 2000;11:3289–3298. doi: 10.1091/mbc.11.10.3289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mullock B M, Smith C W, Ihrke G, Bright N A, Lindsay M, Parkinson E J, Brooks D A, Parton R G, James D E, Luzio J P, Piper R C. Mol Biol Cell. 2000;11:3137–3153. doi: 10.1091/mbc.11.9.3137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Steele-Mortimer O, Méresse S, Gorvel J-P, Toh B-H, Finlay B B. Cell Microbiol. 1999;1:33–49. doi: 10.1046/j.1462-5822.1999.00003.x. [DOI] [PubMed] [Google Scholar]
- 27.Ferrari G, Langen H, Naito M, Pieters J. Cell. 1999;97:435–447. doi: 10.1016/s0092-8674(00)80754-0. [DOI] [PubMed] [Google Scholar]
- 28.Joshi A D, Sturgill-Koszycki S, Swanson M S. Cell Microbiol. 2001;3:99–114. doi: 10.1046/j.1462-5822.2001.00093.x. [DOI] [PubMed] [Google Scholar]
- 30.Horwitz M A. J Exp Med. 1983;158:1319–1331. doi: 10.1084/jem.158.4.1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Brand B, Sadosky A B, Shuman H A. Mol Microbiol. 1994;14:797–808. doi: 10.1111/j.1365-2958.1994.tb01316.x. [DOI] [PubMed] [Google Scholar]
- 32.Roy C R, Berger K H, Isberg R R. Mol Microbiol. 1998;28:663–674. doi: 10.1046/j.1365-2958.1998.00841.x. [DOI] [PubMed] [Google Scholar]
- 33.Sturgill-Koszycki S, Swanson M S. J Exp Med. 2000;192:1261–1272. doi: 10.1084/jem.192.9.1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Taraska T, Ward D M, Ajioka R S, Wyrick P B, Davis-Kaplan S R, Davis C H, Kaplan J. Infect Immun. 1996;64:3713–3727. doi: 10.1128/iai.64.9.3713-3727.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Heinzen R A, Scidmore M A, Rockey D D, Hackstadt T. Infect Immun. 1996;64:796–809. doi: 10.1128/iai.64.3.796-809.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hackstadt T, Rockey D D, Heinzen R A, Scidmore M A. EMBO J. 1996;15:964–977. [PMC free article] [PubMed] [Google Scholar]
- 37.Mordue D G, Desai N, Dustin M, Sibley L D. J Exp Med. 1999;190:1783–1792. doi: 10.1084/jem.190.12.1783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mordue D G, Hakansson S, Niesman I, Sibley L D. Exp Parasitol. 1999;92:87–99. doi: 10.1006/expr.1999.4412. [DOI] [PubMed] [Google Scholar]
- 39.Barnewall R E, Ohashi N, Rikihisa Y. Infect Immun. 1999;67:2258–2265. doi: 10.1128/iai.67.5.2258-2265.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Russell D G, Dant J, Sturgill-Koszycki S. J Immunol. 1996;156:4764–4773. [PubMed] [Google Scholar]
- 41.de Chastellier C, Lang T, Thilo L. Eur J Cell Biol. 1995;68:167–182. [PubMed] [Google Scholar]
- 42.Ullrich H J, Beatty W L, Russell D G. Eur J Cell Biol. 1999;78:739–748. doi: 10.1016/S0171-9335(99)80042-9. [DOI] [PubMed] [Google Scholar]
- 43.Arenas G N, Staskevich A S, Aballay A, Mayorga L S. Infect Immun. 2000;68:4255–4263. doi: 10.1128/iai.68.7.4255-4263.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pizarro-Cerda J, Moreno E, Gorvel J P. Microbes Infect. 1998;2:829–835. doi: 10.1016/s1286-4579(00)90368-x. [DOI] [PubMed] [Google Scholar]
- 45.Garcia-del Portillo F, Finlay B B. J Cell Biol. 1995;129:81–97. doi: 10.1083/jcb.129.1.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ishibashi Y, Arai T. FEMS Microbiol Immunol. 1990;2:35–44. doi: 10.1111/j.1574-6968.1990.tb03476.x. [DOI] [PubMed] [Google Scholar]
- 47.Beuzón C R, Méresse S, Unsworth K E, Ruiz-Albert J, Garvis S, Waterman S R, Ryder T A, Boucrot E, Holden D W. EMBO J. 2000;19:3235–3249. doi: 10.1093/emboj/19.13.3235. [DOI] [PMC free article] [PubMed] [Google Scholar]