Abstract
We have recently demonstrated that thrombin induces expression of the platelet-derived growth factor B-chain gene in endothelial cells (EC) through activation of the Y-box binding protein DNA-binding protein B (dbpB). We now present evidence that dbpB is activated by a novel mechanism: proteolytic cleavage leading to release from mRNA, nuclear translocation, and induction of thrombin-responsive genes. Cytosolic, full-length dbpB (50 kDa) was rapidly cleaved to a 30-kDa species upon thrombin stimulation of EC. This truncated, “active” dbpB exhibited nuclear localization and binding affinity for the thrombin response element sequence, which is distinct from the Y-box sequence. Oligo(dT) affinity chromatography revealed that cytosolic dbpB from control EC, but not active dbpB from thrombin-treated EC, was bound to mRNA. Latent dbpB immunoprecipitated from cytosolic extracts of control EC was activated by ribonuclease treatment. Furthermore, when EC cytosolic extracts were subjected to Nycodenz gradient centrifugation, latent dbpB fractionated with mRNA, whereas active dbpB fractionated with free proteins. The cytosolic retention domain of dbpB, which we localized to the region 247–267, was proteolytically cleaved during its activation. In contrast to full-length dbpB, truncated dbpB stimulated platelet-derived growth factor B-chain and tissue factor promoter activity by over 5-fold when transiently cotransfected with reporter constructs. These results suggest a novel mode of transcription factor activation in which an agonist causes release from mRNA of a latent transcription factor leading to its transport to the nucleus and its regulation of target gene expression.
Keywords: DNA-binding protein B, endothelium, platelet-derived growth factor, RNA-binding proteins
Multiple distinct mechanisms have been identified for the posttranslational activation of transcription factors in response to cellular agonists. In some cases [for example, signal transducers and activators of transcription (STAT) activation in response to the interferons], the transcription factor becomes phosphorylated upon association with an agonist-occupied cell-surface receptor, leading to dimerization and translocation to the nucleus (1). Other transcription factors are activated by phosphorylation or dephosphorylation reactions (2, 3) or by release from an inhibitory protein in the cytosol (e.g., NF-κB activation in response to cytokines) (4). Latent transcription factors also may be released from membranes in response to stimuli (5). In the current report, we describe a heretofore unrecognized mode of transcription factor activation in which a protein that is bound to mRNA, perhaps acting in this latent state as an RNA chaperone or regulator of translation (6), is released from mRNA in response to an agonist.
Thrombin is a coagulation system protease and a potent platelet-aggregating substance, which also induces multiple genes and functional changes in endothelial cells (EC) and other cell types (7–10). One thrombin-inducible gene is the platelet-derived growth factor (PDGF) B-chain (11–14). The underlying molecular mechanism for this induction remains poorly defined. We have previously identified both a 9-bp element in the PDGF B-chain promoter responsible for thrombin-induced transcription of this gene and a thrombin-inducible nuclear factor that bound this sequence (15). We recently purified and sequenced this nuclear factor and identified it as a truncated form of DNA binding protein B (dbpB), a member of the Y-box protein family (16).
Y-box proteins, which are highly conserved from Escherichia coli through humans, are DNA- and RNA-binding proteins that participate constitutively in the regulation of gene expression (reviewed in ref. 17). This class of transcription factor had not been previously shown to be activated in response to a cellular agonist. The activation of dbpB, defined as the acquisition of the ability to bind to the thrombin-response element in the PDGF B-chain promoter, occurred within minutes of thrombin treatment and was unaffected by preincubation of the cells with cycloheximide, suggesting that dbpB activation was a posttranslational event. In this report, we have tested the hypothesis that the activation of dbpB in response to thrombin occurs via release from mRNA and subsequent translocation to the nucleus.
Experimental Procedures
Materials.
Bovine α-thrombin was purchased from United States Biochemical. Endotoxin was removed from thrombin by using Acticlean ETOX prepacked columns from Sterogen (Arcadia, CA). DMEM/Ham's F-12 medium was from Irvine Scientific, and FBS was obtained from BioWhittaker. Tissue culture plastic was from Corning and Becton Dickinson. N2H-D-Phe-Pro-Arg-chloromethylketone was from Bachem, micrococcal nuclease (nuclease S7) and T4 kinase for end labeling of oligonucleotides were from Roche Molecular Biochemicals, [γ-32P]dATP and [α-32P]dCTP were from NEN, RNasin ribonuclease inhibitor was from Promega, Nonidet P-40 was from Pierce, and Trizol reagent, Lipofectin reagent, and oligo(dT)12–18 were from Life Technologies (Grand Island, NY). All other chemicals, unless otherwise noted, were purchased from Sigma.
Stimulation of EC with Thrombin and Preparation of Extracts for Electrophoretic Mobility-Shift Assay (EMSA).
The extracts for EMSA were prepared as described (16). EC were stimulated for 2 h with thrombin (10 units/ml). At the end of the incubation and before cell lysis, a 100× molar excess of the thrombin inhibitor N2H-D-Phe-Pro-Arg-chloromethylketone was added for 10 min at 37°C to inactivate thrombin bound to the cell surface. Cytosolic and nuclear extracts were prepared as described by Dignam et al. (18).
EMSA.
EMSA was performed as described (15, 16). Oligonucleotides used as probes for EMSA represented wild-type or mutated thrombin-response elements of the PDGF B-chain promoter (15). The thrombin-response element sequence CCACCCACC was changed as follows: ctCCACCCACC (wild type), ctAAATGCACC (4-base mutant), and ctAAGTTTGAAG (9-base mutant).
mRNA Isolation.
mRNA was isolated by oligo(dT) affinity chromatography by using an mRNA isolation kit from Roche Molecular Biochemicals. To copurify dbpB with mRNA, the procedure was changed. EC were lysed in ice-cold buffer (0.3 M KCl/20 mM Hepes/2 mM MgCl2/0.1% diethyl pyrocarbonate/0.5% Nonidet P-40/0.1 unit/μl RNasin) with 10 triturations by using a no. 26 needle. Nuclei and remains of the cell membrane were removed by centrifugation at 12,000 × g for 15 min at 4°C. EC cytosolic extract was dialyzed overnight in 0.2 M NaCl/10 mM Tris⋅HCl, pH 7.5/2 mM EDTA/0.2% Nonidet P-40/5 mM β-mercaptoethanol. Oligo(dT)–biotin complex was added to the extract, and the extract was applied to streptavidin agarose according to the mRNA isolation kit instructions. After rotation for 1 h at 4°C, the agarose beads were washed three times with buffer, then once with 25% formamide. The mRNA was eluted with 60% formamide, and all fractions were dialyzed overnight in EMSA buffer.
Nycodenz Gradient Fractionation.
Nonstimulated or thrombin-stimulated EC were washed three times with ice-cold PBS, scraped, and lysed in the buffer used by others (19) for purification of the Y-box protein MSY1 in association with mRNPs (0.3 M KCl/2 mM MgCl2/20 mM Hepes, pH 7.4/0.1% diethyl pyrocarbonate/0.5% Nonidet P-40). Lysis was achieved by 10 triturations through a no. 26 needle, and the lysate was centrifuged (12,000 × g, 15 min) at 4°C. RNasin (60 units/ml) was added to the extract, and 0.5–1 mg of protein (300 μl) was loaded onto 4 ml of Nycodenz gradient (20–60%). Centrifugation was performed at 150,000 × g for 20 h at 4°C. Twenty-five fractions (200 μl) were collected from each tube, dialyzed in buffer, and used in EMSA. RNA isolation from fractions was performed by using Trizol reagent, and an oligo(dT) probe was used to detect mRNA in Northern blotting.
Treatment with Ribonucleases.
Ribonuclease A (1 mg/ml) or micrococcal nuclease (5,000–10,000 units/ml) was added to the extract to digest RNA and release dbpB. The mixture was incubated 30 min at 37°C. To remove nucleotides and oligonucleotides generated during the incubation that could interfere with the detection of dbpB, the extract was dialyzed overnight in EMSA buffer and centrifuged at 12,000 × g for 15 min before being used in the EMSA.
Fusion Protein Constructs.
The pEGFP-C3 vector from CLONTECH was used to prepare fusion protein constructs of various species of dbpB with green fluorescent protein (GFP) at their N termini. cDNAs for five C-terminally truncated mutants of dbpB were obtained by introducing a stop codon by PCR after positions 621, 741, 801, 858, and 921. These dbpB cDNAs were cloned into the pEGFP-C3 vector by using the SacII and ApaI sites in the vector, and the sequence of the fusion protein was verified by DNA sequencing.
Transient Transfection of Bovine Aortic EC and NIH 3T3 Cells.
Bovine aortic EC were grown to 95% confluence on fibronectin-coated glass slides, washed with Optimem medium, and transfected with 0.5–1 μg of DNA/well by using Lipofectin reagent. After transfection for 6 h, the medium was replaced with full growth medium (DMEM/F12 with 10% FBS). The localization of GFP fusion proteins was determined after 36 h as described (16). NIH 3T3 cells were grown to 50–60% confluence in 6-well plates and transfected by using Lipofectin reagent with 1–2 μg of DNA/well for 4 h. After transfection, the medium was replaced with full growth medium. The extracts for luciferase activity assay were prepared 36 h after transfection. Luciferase activity was normalized to β-galactosidase activity from cotransfected pβGal-control vector (CLONTECH).
Results
Latent dbpB Is Associated with mRNA in EC.
We have previously shown that full-length, latent dbpB was constitutively present in the cytosol of EC and that its activation in response to thrombin stimulation did not require de novo protein synthesis. Because members of the Y-box protein family were known to bind RNA, we tested the hypothesis that latent dbpB was associated with RNA in the cytosol of unstimulated cells. Cytosolic extracts of control EC were subjected to RNA digestion by using ribonuclease A (1 mg/ml) or micrococcal nuclease (nuclease S7) (1,000–10,000 units/ml, 30 min at 37°C). An additional DNA-binding protein with the electrophoretic mobility of activated (thrombin-induced) dbpB was detected in the RNase-treated extracts (Fig. 1A). The protein activated by RNA digestion exhibited the same DNA-binding specificity as activated dbpB, suggesting that latent dbpB was associated with RNA in EC.
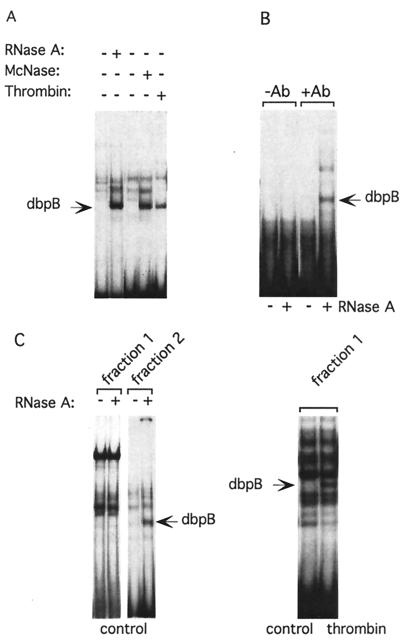
Figure 1.
(A) Latent dbpB is associated with RNA in nonstimulated EC. Cytosolic extracts were prepared from either control bovine aortic EC or EC treated with bovine α-thrombin (10 units/ml, 2 h), as described in Experimental Procedures. Extracts from control EC were incubated with RNase A (1 mg/ml) or micrococcal nuclease (10,000 units/ml) for 30 min at 37°C before EMSA with the thrombin-response element (CCACCCACC) oligonucleotide as a probe. Arrow shows the position of dbpB activated by thrombin treatment of intact cells or ribonuclease treatment of extracts from untreated EC; McNase, micrococcal nuclease. (B) Latent dbpB coimmunoprecipitates with RNA. Cytosolic extracts prepared from control EC were incubated with preimmune serum or anti-dbpB antiserum raised against a C-terminal peptide of dbpB, followed by precipitation with protein A/G Sepharose. Protein was eluted by heat treatment. Aliquots of the eluants were treated with RNase A to release dbpB coimmunoprecipitated in association with RNA, and EMSA was performed. −Ab, preimmune serum used as a control; +Ab, immune serum used for precipitation of dbpB; RNase A, sample treated with ribonuclease A. (C) Oligo(dT) affinity chromatography of dbpB. (Left) EC cytosolic extracts were prepared from control and thrombin-treated EC as described in Experimental Procedures and dialyzed overnight in 0.2 M NaCl/0.01 M Tris⋅HCl, pH 7.5/0.002 M EDTA/0.2% Nonidet P-40/0.005 M β-mercaptoethanol. Oligo(dT)–biotin complex was added to the extract, and the mixture was applied to streptavidin magnetic beads. After rotation for 1 h at 4°C, the unbound fraction (fraction 1) was removed, the beads were washed three times, then washed with 25% formamide, and RNA was eluted with 60% formamide (fraction 2). The fractions were dialyzed overnight in EMSA buffer, and aliquots were treated with RNase A and subjected to EMSA. (Right) Fraction 1 from thrombin-treated EC lysates was used for EMSA without RNase treatment to detect activated dbpB.
Immunoprecipitation experiments provided further evidence that latent dbpB was associated with RNA in untreated EC. When dbpB was immunoprecipitated from EC cytosolic extracts, no shifted band could be detected by EMSA. Only when the immunoprecipitate was treated with RNase did a gel-shift band appear corresponding to active dbpB (Fig. 1B). This suggested that dbpB was coprecipitated as a complex with RNA and that the binding to RNA prevented its detection by EMSA. To test this hypothesis, we subjected cytosolic extracts from nonstimulated human EC to oligo(dT) affinity chromatography. The mRNA fraction that was eluted from the affinity beads contained latent dbpB, which was detectable by EMSA only after digestion with RNase A (Fig. 1C). Treating the nonbound fraction with RNase A did not reveal dbpB, suggesting that all latent dbpB was associated with mRNA.
Active dbpB Is Released from mRNA.
To determine whether thrombin-activated dbpB showed similar binding to mRNA, cytosolic extracts of thrombin-stimulated EC were subjected to oligo(dT) affinity chromatography. Active dbpB, as measured by EMSA, was found only in the non-oligo(dT) bound fraction (Fig. 1C), suggesting that dbpB had been released from mRNA upon thrombin treatment of the cells. Activation of dbpB upon thrombin stimulation and its translocation to the nucleus occurred within minutes, suggesting a rapid release of dbpB from mRNA. When the oligo(dT) fraction was eluted and treated with RNase A, additional activated dbpB was revealed (data not shown), indicating that only a fraction of the dbpB bound to mRNA was released upon thrombin stimulation of EC. Active dbpB revealed by RNase treatment exhibited the same molecular size as thrombin-induced dbpB as determined by SDS/PAGE after crosslinking to labeled oligonucleotide. Because the RNase preparations that were used were free of proteolytic activity, we speculate that the protease that cleaves dbpB upon thrombin stimulation of EC is closely associated with dbpB or that the cleavage may be the result of autoproteolysis. The digestion of RNA itself, or possibly a change in dbpB conformation upon RNA digestion, may trigger this proteolysis.
Gradient Fractionation of Cytosolic Extracts from Control and Thrombin-Stimulated EC.
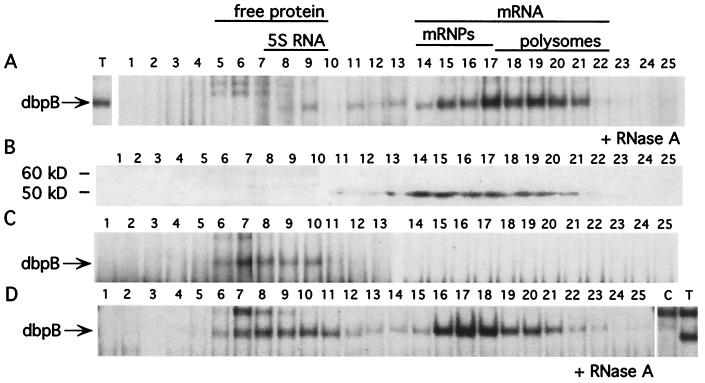
To confirm that latent dbpB was bound to mRNA and released after thrombin stimulation of EC, we performed Nycodenz gradient fractionation of cytosolic extracts from control and thrombin-treated EC. Gradient fractionation of extracts prepared from nonstimulated cells showed that latent dbpB copurified with mRNA (Fig. 2 A, B, and D). dbpB was detected in all fractions containing mRNA by Western blotting (Fig. 2B) and by EMSA, after ribonuclease treatment of fractions (Fig. 2 A and D). No latent dbpB was found in the fractions containing free proteins (fractions 5–9), as determined by Coomassie staining after SDS/PAGE of fractions, and fractionation of standard proteins (Fig. 2 A and B). In contrast, Nycodenz gradient fractionation of cytosolic extracts from thrombin-stimulated cells revealed that active dbpB cofractionated with free protein (Fig. 2C).
Figure 2.
Nycodenz gradient fractionation of cytosolic extracts from control and thrombin-stimulated EC. Lysates were prepared from nonstimulated or thrombin-stimulated EC and subjected to Nycodenz gradient fractionation as described in Experimental Procedures. Twenty-five fractions were collected from each tube, dialyzed, and used in EMSA after treatment with RNase to detect latent dbpB (A and D, fractions 15–21) or without any additional treatment (C) to detect thrombin-activated dbpB (C, fractions 6–10). Latent dbpB also was detected by Western blotting by using anti-dbpB antisera raised against the C-terminal peptide of dbpB (B). RNA isolation from fractions was performed by using Trizol reagent, and an oligo(dT) probe was used to detect mRNA by Northern blotting (not shown). Polysomal RNA (fractions 17–22) and 5S RNA (fractions 7–10) were detected by ethidium bromide staining of agarose gels (not shown). Free cytosolic protein (fractions 5–10) was detected by Coomassie staining of SDS/PAGE of individual fractions (not shown). C, cytosolic extract from unstimulated EC; T, cytosolic extract from thrombin-stimulated EC (arrow points to active dbpB in the extracts).
Identification of the Cytosolic Retention Domain of dbpB.
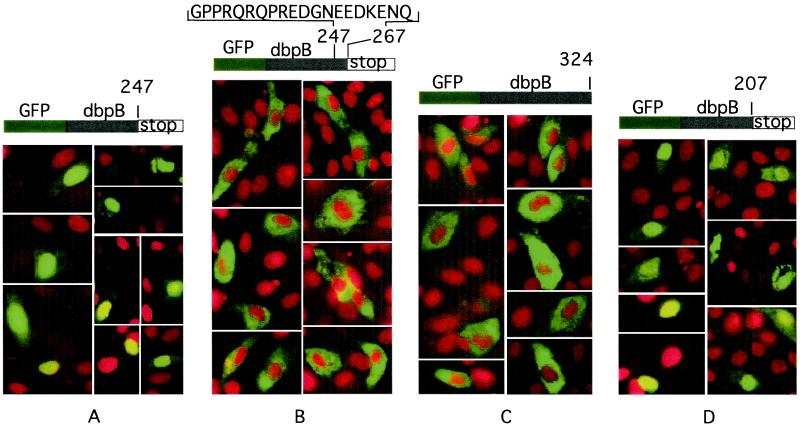
To identify the amino acids responsible for the binding of dbpB to mRNA, and therefore serving as a cytoplasmic retention domain for this protein, we prepared a series of expression vectors containing cDNA encoding truncated dbpB–GFP fusion proteins of varied length. Our previously published results indicated that the presence of GFP did not compromise the regulation of dbpB localization. When GFP–full-length dbpB was expressed in bovine EC, GFP was localized to the nucleus only after thrombin treatment (16). GFP–205-aa dbpB fusion protein expressed in COS-1 cells formed a complex with ThRE DNA in EMSA (data not shown). Truncations were made by introducing a stop codon after positions 621 (dbpB of 207 aa), 741 (247 aa), 810 (270 aa), and 921 (307 aa). cDNA constructs encoding GFP fusion proteins were made with all of the truncated mutants by using the EGFP-C3 vector. Bovine EC were transfected with these constructs, and fluorescent microscopy was performed 36 h later. The GFP fusion proteins of truncated dbpB of 207 aa and 247 aa were localized to the nucleus; however, the dbpB of 267 aa and all of the longer species were retained in the cytoplasm, as was full-length dbpB (Fig. 3 and not shown). These results suggested that the cytoplasmic retention domain of dbpB was located between positions 247 and 267 within the sequence RGPPRQRQPREDGNEEDKEN.
Figure 3.
Identification of the cytosolic retention domain of dbpB. The fusion protein GFP-1–247 aa dbpB was localized to the nucleus (A). The addition of 20 aa (1–267 aa dbpB) resulted in its retention in the cytoplasm (B), similar to full-length dbpB (C). GFP-1–207 aa dbpB was localized to the nucleus (D) as well as the 191-, 200-, and 205-aa fusion proteins (not shown). Truncated dbpB mutants were prepared by introducing stop codons after the designated amino acid. Transient transfection of bovine aortic EC and the preparation of slides for fluorescence microscopy were performed as described (16).
Activation of PDGF and Tissue Factor Promoters by a Truncated Form of dbpB.
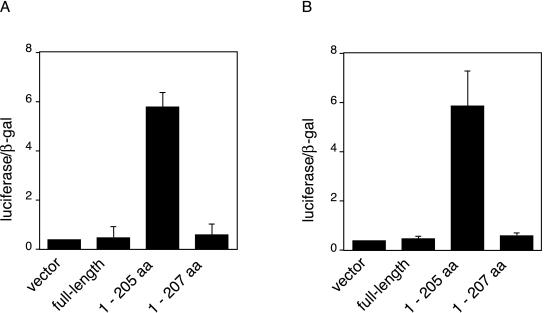
We wanted to verify that the proteolytically cleaved “active” dbpB that translocated to the nucleus exhibited functional activity in intact cells. We therefore tested the transcriptional activity of truncated dbpB on two thrombin-responsive promoters by using a coexpression system. We were uncertain of the exact length of naturally produced active dbpB because our microsequencing of tryptic peptides from the active dbpB that we purified did not yield any information in this region (16). As described above, truncated dbpB of ≈205 aa exhibited very similar size and DNA binding specificity to native, active dbpB. We therefore tested a series of expression vectors containing cDNA encoding truncated dbpB of about this size for the ability to stimulate transcription of reporter constructs containing either a fragment of the PDGF B-chain promoter (400 bp) or the tissue factor promoter (660 bp) driving expression of the luciferase gene. We and others have shown these two reporters to be thrombin-responsive. The coexpression of these reporter constructs with a truncated dbpB of 205 aa resulted in an over 5-fold increase in transcriptional activity compared with control expression vectors, including one encoding full-length dbpB (Fig. 4). Surprisingly, a slightly longer truncated dbpB (207 aa) had no activity, suggesting the functional importance of the new C terminus that is generated when cytosolic dbpB is cleaved and released from mRNA.
Figure 4.
Activation of PDGF and tissue-factor promoters by truncated dbpB. Truncated dbpB was prepared by introducing a stop codon into the cDNA after either 205 or 207 aa of the coding sequence. NIH 3T3 fibroblasts were transiently cotransfected with pcDNA3 containing the cDNAs for one of the dbpB species and either a 400-bp PDGF B-chain promoter–luciferase reporter construct (A) or a 660-bp tissue factor promoter–luciferase construct (B) as described in Experimental Procedures. Transfection (4 h) was followed by recovery in full growth medium for 36 h, at which time cells were lysed and assayed for luciferase activity. pβgal-control vector (CLONTECH) was cotransfected in all cases, and the activity of luciferase was normalized to β-galactosidase activity.
Discussion
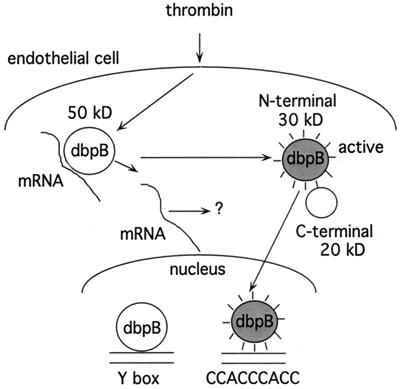
Y-box binding proteins have been postulated to represent a link between translational and transcriptional regulation of gene expression due to their DNA and RNA binding capacities, but the nature of such a link had not been elucidated experimentally. Several Y-box proteins have been found in cells in association with RNA (17, 20–22); however, stimuli, which cause a switch in Y-box protein function from translational to transcriptional regulation or translocation of Y-box proteins from cytosol to nucleus, have not been previously identified. Our study of the mechanism of action of the first identified extracellular stimulator of a Y-box protein thrombin has led us to uncover a novel pathway of transcriptional activation. We have found that dbpB translocation to the nucleus and its transcriptional activity are the result of its cleavage and release from an “inhibitory factor” in the cytosol—i.e., mRNA (Fig. 5). This is an analogous process to release of NF-κB from IκB (4), which is triggered by phosphorylation events, or by SREBP release from membranes, which is triggered by sterol content (5). Under basal conditions, latent dbpB is associated with mRNA, which is consistent with a role in translational regulation (6, 17, 23–26) or mRNA chaperoning (27). Only upon cleavage and release from mRNA does active truncated dbpB, which exhibits a distinct DNA binding specificity from full-length dbpB, regulate transcription of thrombin-dependent genes.
Figure 5.
Model for agonist-induced dbpB activation. Latent dbpB bound to mRNA under basal conditions may act as a regulator of translation or as an RNA chaperone. Thrombin stimulation leads to dbpB cleavage, release from mRNA, translocation to the nucleus, and the regulation of thrombin-responsive gene expression (e.g., the PDGF B-chain gene). Truncated, activated dbpB has a distinct consensus binding site from that of full-length dbpB.
To elucidate further dbpB mechanism of activation, we identified the region of the protein responsible for the cytosolic retention of dbpB. Using fusion proteins of truncated dbpB and GFP, we localized the cytosolic retention (mRNA binding) domain to the region encompassing amino acids 247–267. Y-box proteins have been reported to form direct contacts with RNA, and the highly conserved cold-shock domain, as well as some C-terminal regions, have been shown to be required for this binding (28–31). Matsumoto et al. (31) reported that the C-terminal tail domain is essential both for translational regulation and stable incorporation of the Y-box protein FRGY2 into ribonucleoprotein particles. The cold-shock domain alone was shown to bind mRNA both in vitro and when injected into oocytes (31) and was retained in the cytoplasm. However, the short form of dbpB created upon thrombin stimulation contains additional sequence beyond the cold-shock domain, including a putative nuclear localization signal (basic sequence between amino acids 186 and 205). Our results show that further truncation of the protein (186 aa) results in its inability to translocate to the nucleus and its retention in the cytoplasm.
The specificity of Y-box protein–RNA binding is controversial; Y-box proteins have been reported to bind any RNA longer than 70–80 nt (28), as well as to recognize specific sequences in RNA (22, 32–34) or to bind only transcripts derived from intronless genes (26). DbpB has some similarities to the viral RNA-binding protein Tat (32); these two proteins are homologous in their RNA-binding domains (35). The large amount of dbpB in EC (≈0.1% of total cellular protein) suggests that it is bound to multiple messages. However, only a small fraction of it is activated by thrombin and released from mRNA, possibly from specific transcripts leading to a change in their rate of translation. The release of dbpB from mRNA in response to cell stimulation by thrombin is not the only way to dissociate Y-box protein from mRNA. During oocyte maturation, the chaperone nucleoplasmin regulates FRGY2 binding to mRNA, thus relieving the transcriptional repression of histone H4 mRNA (36).
We recently reported that thrombin caused the formation of a truncated dbpB that bound specifically to an oligonucleotide corresponding to the thrombin-response element and that its nuclear appearance or inhibition of appearance with signaling inhibitors correlated with expression of the thrombin-responsive gene PDGF B-chain. However, we had difficulty directly demonstrating functional activity of truncated dbpB (16). Transient coexpression of expression vectors containing truncated dbpB cDNA with a thrombin-responsive PDGF promoter–reporter construct revealed no stimulation of transcriptional activity. We postulated that this was due to either the requirement of a second thrombin-induced factor for induction or that our cDNAs were not expressing a protein of the exact size of naturally activated dbpB. (We had identified dbpB by microsequencing of tryptic peptides derived from the protein we purified; however, we did not obtain sequence corresponding to the C terminus of the protein.) We have shown in this report that truncated dbpB of 205 aa, but not full-length dbpB, stimulated transcription driven by two thrombin-responsive promoters, PDGF and tissue factor. The addition of only 2 aa to the 205-aa protein resulted in a lack of activity, indicating the functional importance of the newly formed C terminus of dbpB in its transcriptional activity.
We have examined to date two thrombin-responsive promoters; however, we believe the dbpB mechanism of regulation of gene expression by thrombin may be more general. It has been recently reported that the induction of protein C receptor expression by thrombin depends on a very similar thrombin-response element in its promoter (37) and a thrombin-inducible nuclear factor that may be activated dbpB. In addition, the promoters of other thrombin-responsive genes have potential binding sites for activated dbpB, including thrombomodulin, PAR-1, and vascular endothelial growth factor.
One might speculate that the dbpB mechanism of activation is a paradigm followed by other RNA-binding proteins (e.g., heterogeneous ribonucleoprotein particle protein K, CBTF122, and TFIIIA) that have been reported to act as regulators of transcription by unknown mechanisms (38–40). The stimuli activating these RNA-binding proteins have not been identified to date. Heterogeneous ribonucleoprotein particle protein K, a regulator of 15-lipoxygenase translation, has been reported to activate c-myc gene transcription by an undefined mechanism (38). CBTF122, present in association with mRNA in germ cells, was found to regulate the transcription of the GATA-2 gene during the gastrula stage of development (39). TFIIIA bound to 5S rRNA in cytosol has been reported to play a role in the regulation of 5S rRNA gene transcription (40). One or more of these RNA binding proteins may be activated upon extracellular stimulation in a similar mechanism to dbpB activation by thrombin.
Acknowledgments
We thank Amy Bunting and Paula Hoang for the cell culture assistance and Drs. Guy M. Chisolm and Mei-Zhen Cui for the tissue factor promoter reporter construct. We also thank Drs. Paul Copeland, Donna Driscoll, Paul Fox, and Nicholas Tripoulas for helpful discussions. This work was supported by National Institutes of Health Grant HL 29582 (to P.E.D.) and by a fellowship award (to O.I.S.) from the American Heart Association, Northeast Ohio Affiliate. Human umbilical vein EC were provided by the cords collected through the Birthing Services Department at the Cleveland Clinic Foundation and the Perinatal Clinical Research Center (National Institutes of Health General Clinical Research Center award RR-00080) at the Cleveland Metrohealth Hospital.
Abbreviations
- dbpB
DNA-binding protein B
- EC
endothelial cell
- EMSA
electrophoretic mobility-shift assay
- GFP
green fluorescent protein
- PDGF
platelet-derived growth factor
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Darnell J E., Jr J Interferon Cytokine Res. 1998;18:549–554. doi: 10.1089/jir.1998.18.549. [DOI] [PubMed] [Google Scholar]
- 2.Boulikas T. Crit Rev Eukaryotic Gene Expression. 1995;5:1–77. [PubMed] [Google Scholar]
- 3.Meek D W. Cell Signalling. 1998;10:159–166. doi: 10.1016/s0898-6568(97)00119-8. [DOI] [PubMed] [Google Scholar]
- 4.Ghosh S, Baltimore D. Nature (London) 1990;344:678–682. doi: 10.1038/344678a0. [DOI] [PubMed] [Google Scholar]
- 5.Wang X, Sato R, Brown M S, Hua X, J L, Goldstein J L. Cell. 1994;77:53–62. doi: 10.1016/0092-8674(94)90234-8. [DOI] [PubMed] [Google Scholar]
- 6.Chen C Y, Gherzi R, Andersen J S, Gaietta G, Jurchott K, Royer H-D, Mann M, Karin M. Genes Dev. 2000;14:1236–1248. [PMC free article] [PubMed] [Google Scholar]
- 7.Carveth H J, Shaddy R E, Whatley R E, McIntyre T M, Prescott S M, Zimmerman G A. Semin Thromb Hemostasis. 1992;18:126–134. doi: 10.1055/s-2007-1002417. [DOI] [PubMed] [Google Scholar]
- 8.Brass L F, Molino M. Thromb Haemostasis. 1997;78:234–241. [PubMed] [Google Scholar]
- 9.Garcia J G, Verin A D, Schaphorst K L. Semin Thromb Hemostasis. 1996;22:309–315. doi: 10.1055/s-2007-999025. [DOI] [PubMed] [Google Scholar]
- 10.Coughlin S R. Proc Natl Acad Sci USA. 1999;96:11023–11027. doi: 10.1073/pnas.96.20.11023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Daniel T O, Gibbs V C, Milfay D F, Garovoy M R, Williams L T. J Biol Chem. 1986;261:9579–9582. [PubMed] [Google Scholar]
- 12.Daniel T O, Gibbs V C, Milfay D F, Williams L T. J Biol Chem. 1987;262:11893–11896. [PubMed] [Google Scholar]
- 13.Shankar R, de la Motte C A, DiCorleto P E. J Biol Chem. 1992;267:9376–9382. [PubMed] [Google Scholar]
- 14.Grandaliano G, Choudhury G G, Poptic E, Woodruff K, Barnes J L, Abboud H E. J Am Soc Nephrol. 1998;9:583–589. doi: 10.1681/ASN.V94583. [DOI] [PubMed] [Google Scholar]
- 15.Scarpati E, DiCorleto P. J Biol Chem. 1996;271:3025–3032. doi: 10.1074/jbc.271.6.3025. [DOI] [PubMed] [Google Scholar]
- 16.Stenina O I, Poptic E J, DiCorleto P E. J Clin Invest. 2000;106:579–587. doi: 10.1172/JCI9075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Matsumoto K, Wolffe A P. Trends Cell Biol. 1998;8:318–323. doi: 10.1016/s0962-8924(98)01300-2. [DOI] [PubMed] [Google Scholar]
- 18.Dignam J, Lebovitz R, Roeder R. Nucleic Acids Res. 1983;11:1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tafuri S, Familari M, Wolffe A. J Biol Chem. 1993;268:12213–12220. [PubMed] [Google Scholar]
- 20.Tafuri S, Wolffe A. J Biol Chem. 1993;268:24255–24261. [PubMed] [Google Scholar]
- 21.Bouvet P, Matsumoto K, Wolffe A. J Biol Chem. 1995;270:28297–28303. doi: 10.1074/jbc.270.47.28297. [DOI] [PubMed] [Google Scholar]
- 22.Bouvet P, Wolffe A. Cell. 1994;77:931–941. doi: 10.1016/0092-8674(94)90141-4. [DOI] [PubMed] [Google Scholar]
- 23.Evdokimova V M, Wei C L, Sitikov A S, Simonenko P N, Lazarev O A, Vasilenko K S, Ustinov V A, Hershey J W, Ovchinnikov L P. J Biol Chem. 1995;270:3186–3192. doi: 10.1074/jbc.270.7.3186. [DOI] [PubMed] [Google Scholar]
- 24.Matsumoto K, Meric F, Wolffe A P. J Biol Chem. 1996;271:22706–22712. doi: 10.1074/jbc.271.37.22706. [DOI] [PubMed] [Google Scholar]
- 25.Davydova E K, Evdokimova V M, Ovchinnikov L P, Hershey J W. Nucleic Acids Res. 1997;25:2911–2916. doi: 10.1093/nar/25.14.2911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Braddock M, Muckenthaler M, White M R H, Thorburn A M, Sommerville J, Kingsman A J, Kingsman S M. Nucleic Acids Res. 1994;22:5255–5264. doi: 10.1093/nar/22.24.5255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Evdokimova V M, Kovrigina E A, Nashchekin D V, Davydova E K, Hershey J W, Ovchinnikov L P. J Biol Chem. 1998;273:3574–3581. doi: 10.1074/jbc.273.6.3574. [DOI] [PubMed] [Google Scholar]
- 28.Marello K, LaRovere J, Sommerville J. Nucleic Acids Res. 1992;20:5593–5600. doi: 10.1093/nar/20.21.5593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ladomery M, Sommerville J. Nucleic Acids Res. 1994;22:5582–5589. doi: 10.1093/nar/22.25.5582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ranjan M, Tafuri S R, Wolffe A P. Genes Dev. 1993;7:1725–1736. doi: 10.1101/gad.7.9.1725. [DOI] [PubMed] [Google Scholar]
- 31.Matsumoto K, Meric F, Wolffe A P. J Biol Chem. 1996;271:22706–22712. doi: 10.1074/jbc.271.37.22706. [DOI] [PubMed] [Google Scholar]
- 32.Shen Q, Wu R, Leonard J L, Newburger P E. J Biol Chem. 1998;273:5443–5446. doi: 10.1074/jbc.273.10.5443. [DOI] [PubMed] [Google Scholar]
- 33.Triqueneaux G, Velten M, Franzon P, Dautry F, Jacquemin-Sablon H. Nucleic Acids Res. 1999;27:1926–1934. doi: 10.1093/nar/27.8.1926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Swamynathan S K, Nambiar A, Guntaka R V. Biochem J. 2000;348:297–305. [PMC free article] [PubMed] [Google Scholar]
- 35.Churcher M J, Lamont C, Hamy F, Dingwall C, Green S M, Lowe A D, Butler J G, Gait M J, Karn J. J Mol Biol. 1993;230:90–110. doi: 10.1006/jmbi.1993.1128. [DOI] [PubMed] [Google Scholar]
- 36.Meric F, Matsumoto K, Wolffe A P. J Biol Chem. 1997;272:12840–12846. doi: 10.1074/jbc.272.19.12840. [DOI] [PubMed] [Google Scholar]
- 37.Gu J-M, Fukudome K, Esmon C T. J Biol Chem. 2000;275:12481–12488. doi: 10.1074/jbc.275.17.12481. [DOI] [PubMed] [Google Scholar]
- 38.Ladomery M. BioEssays. 1997;19:903–909. doi: 10.1002/bies.950191010. [DOI] [PubMed] [Google Scholar]
- 39.Michelotti E F, Michelotti G A, Aronsohn A I, Levens D. Mol Cell Biol. 1996;16:2350–2360. doi: 10.1128/mcb.16.5.2350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Brzostowski J, Robinson C, Orford R, Elgar S, Scarlett G, Peterkin T, Malartre M, Kneale G, Wormington M, Guille M. EMBO J. 2000;19:3683–3692. doi: 10.1093/emboj/19.14.3683. [DOI] [PMC free article] [PubMed] [Google Scholar]