Abstract
The relationship between biological network architectures and evolution is unclear. Within the phylum nematoda olfaction represents a critical survival tool. For nematodes, olfaction contributes to multiple processes including the finding of food, hosts, and reproductive partners, making developmental decisions, and evading predators. Here we examine a dynamic nematode odor genetic network to investigate how divergence, diversity, and contribution are shaped by network topology. Our findings describe connectivity frameworks and characteristics that correlate with molecular evolution and contribution across the olfactory network. Our data helps guide the development of a robust evolutionary description of the nematode odor network that may eventually aid in the prediction of interactive and functional qualities of novel nodes.
1. Introduction
For nematodes, olfaction is a central mode of survival. Olfaction contributes to the finding of food, hosts, reproductive partners, in the making of developmental decisions, and to the evasion from predators. Studies into the olfactory system of the model nematode Caenorhabditis elegans have yielded detailed descriptions of the molecular and cellular pathways that subserve the olfactory system [1–4]. These signaling pathways appear highly conserved across very divergent nematode species, and the sensory neurons have clear anatomical orthologs in distantly related nematodes [5]. Within the olfactory system of C. elegans, the ability to detect dilute volatile odors is mostly conferred by three pairs of neurons termed the Amphid Wing cells type A (AWA), Amphid Wing cells type B (AWB), and the Amphid Wing cells type C (AWC) [3, 4]. These cells are primary sensory neurons located within the sensory amphid organ of the head that forms part of an anatomically distinct subclass of amphid neurons in that they do not extend processes through the amphid opening, but instead their distal ciliated endings terminate inside a glial sheath cell [6]. Here we describe a composite odor genetic network in C. elegans that encompasses all three pairs of volatile odor-detecting neurons. We used this network to identify orthologous genes of the odor network in the nematode Pristionchus pacificus. We chose the nematode P. pacificus based upon three criteria: (1) it exhibits a similar lifestyle to that of C. elegans, in that they are both self-fertilizing hermaphrodites that will feed on bacteria, (2) conservation of the olfactory signaling pathway of P. pacificus to C. elegans has been validated experimentally [7, 8], and (3) because it is sufficiently divergent to generate meaningful divergence data (estimated divergence between Caenorhabditis and Pristionchus is between 280 and 430 million years ago [9, 10]). One of our goals in this study was to compare selective pressures across the nodes of the odor network by comparing substitutions at silent sites to that of nonsilent sites; this analysis is best done using more divergent sequences [11]. Using this nematode odor network we searched for relationships between pathway position with divergence, diversity, and contribution across the network. Then, we designed an interaction map of the network and investigated relationships between various metrics of interaction with molecular evolution, and contribution across the network. Our data is not a definitive map of odor signaling in C. elegans but represents a snap shot of current data, and by uncovering robust associations between network topology, evolution, and function we may ultimately design a framework that facilitates a level of predictive power over novel nodes within the network.
2. Materials and Methods
2.1. Sequences
Pristionchus pacificus orthologs were located by cross-referencing matches using the orthology databases: InParanoid_7 [12], OrthoMCL database [13], and the OMA orthology matrix Browser [14]. For each node in our network WormBase (version WS231) has defined a curated ortholog using WormBase-Compara [15]; however, we corroborated these predictions using reciprocal Blast [16] searches, and, by inferring relationships by reconstructing phylogenetic trees (see Figures S1–S7 in Supplementary Materials available online at doi:10.1155/2012/548081), we outline all the evidence for orthology in Table 1. Orthologs were aligned using the multiple sequence alignment software MUSCLE v3.6 [17], and gaps were systematically stripped from all sequences after alignment, and phylogenies inferred using PhyML [18]. To determine orthologs, WormBase-Compara uses the databases TreeFam [http://www.treefam.org/], InParanoid_7, KOGs [http://www.ncbi.nlm.nih.gov/COG/], OMA. The genes used in our network (Table 1), and their corresponding ortholog in P. pacificus (WormBase identifier for P. pacificus is denoted Ppaxxx), were goa-1 (Go α subunit protein: Ppa-goa1); egl-30 (Gq α subunit protein: Ppa-egl30); dgk-1 (DGKθ, diacylglycerol kinase theta: Ppa-dgk1); eat-4 (vesicular glutamate transporter: Ppa-eat4); egl-4/pkg-1 (Protein Kinase G: Ppa-pkg1); tax-6 (calcineurin type A: Ppa-tax6); odr-1 (receptor guanylyl cyclase: Ppa17708); daf-11 (receptor guanylyl cyclase: Ppa-daf11); tax-2 (cyclic nucleotide-gated channel β subunit: Ppa-tax2); tax-4 (cyclic nucleotide-gated channel α subunit: Ppa-tax4); odr-3 (G-protein α subunit: Ppa-odr3); gpa-3 (G-protein α subunit: Ppa-gpa3); gpa-5 (G protein α subunit: Ppa10789); gpa-13 (G-protein α subunit: Ppa-gpa13); arr-1 (Arrestin: Ppa-arr1); rgs-3 (regulator of G-protein signaling: Ppa-rgs3). To detect C. elegans versus P. pacificus 1 : 1 candidate orthologs for the randomization study we selected orthologs from the orthology database, InParanoid_7 [12]. We only included orthologs that are represented by 100% bootstrap support, and from this approach we obtain 5,666 1 : 1 orthologs.
Table 1.
Orthology support for each node within the odor-signaling network.
| C. elegans gene | P. pacificus ortholog | Evidence of orthology |
|---|---|---|
| goa-1 | PPA23648/Ppa-goa-1 | WormBase-Compara |
| odr-3 | PPA14189/Ppa-odr-3 | InParanoid; OMA; WormBase-Compara |
| egl-30 | PPA31690/Ppa-egl-30 | WormBase-Compara |
| gpa-3 | PPA09867/Ppa-gpa-3 | InParanoid; OMA; WormBase-Compara |
| gpa-5 | PPA10789 | OMA; WormBase-Compara |
| gpa-13 | PPA31402/Ppa-gpa-13 | WormBase-Compara |
| eat-4 | PPA15025/Ppa-eat-4 | InParanoid; WormBase-Compara |
| dgk-1 | PPA00312/Ppa-dgk-1 | InParanoid; OMA; WormBase-Compara |
| odr-1 | PPA17708 | InParanoid; OMA |
| daf-11 | PPA14907/Ppa-daf-11 | WormBase-Compara |
| tax-6 | PPA09320/Ppa-tax-6 | WormBase-Compara |
| pkg-1 | PPA27475/Ppa-pkg-1 | InParanoid; OMA; WormBase-Compara |
| tax-2 | PPA07436/Ppa-tax-2 | InParanoid; OMA; WormBase-Compara |
| tax-4 | PPA02388/Ppa-tax-4 | InParanoid; OMA; WormBase-Compara |
| 1 rgs-3 | PPA01970/Ppa-rgs-3 | InParanoid; OMA; WormBase-Compara |
| 1 arr-1 | PPA21763/Ppa-arr-1 | WormBase-Compara |
1Relationship support was confirmed by reconstructing phylogenetic trees for each gene except rgs-3 and arr-1 as they represent single-gene families, and therefore not possible to reconstruct phylogeny (see Figures S1–S7).
2.2. Analysis of Molecular Data
Synonymous (d S) and nonsynonymous (d N) substitution rates for orthologs were estimated using the methods of Yang and Nielsen [19] as implemented in yn00 in the PAML suite [20]. To test the null hypothesis that there is no above average selective pressure on these genes, we performed a randomization test where we determined the average d N/d S value for 50,000 randomly assembled networks and compared with the average d N/d S value for our odor network. Random networks were 16 genes in size and sampling permitted replacement. Measures of nucleotide diversity (π) were performed using DnaSP version 5 [21].
2.3. Network Analysis
Selection of vertices for the odor network was determined by mining literature databases. Network analysis was calculated using Cytoscape version 2.8.2 [22]. The network was treated as undirected and all network analyses available through Cytoscape version 2.8.2 were examined; these are average shortest path length, betweenness centrality, closeness centrality, clustering coefficient, degree, eccentricity, neighborhood connectivity, radiality, stress, and topological coefficient. The degree (k) of a node n is defined as the number of edges linked to n. The clustering coefficient (C n) reveals how connected the neighborhood of a node is by calculating the fraction of neighboring pairs, and for a node n it is defined as
| (1) |
where k n is the degree of n and e n is the number of connected pairs between all neighbors of n. Betweenness centrality (C b) is a measure of the fraction of shortest paths between node pairs (s, t) that pass through the node of interest, and for the node n it is calculated using the following formula:
| (2) |
where s and t are nodes different from n, σ st denotes the number of shortest paths from s to t, and σ st(n) is the number of shortest paths from s to t on which n lies.
All chemotaxis indices (for both wildtype and mutant animals) were mined from previous publications (see references below). The chemotaxis index difference (C.I.diff) for each mutant within each neuron (AWA, AWB, and AWC) was determined by calculating the difference between the wildtype chemotaxis index (C.I.wt) and mutant chemotaxis index (C.I.mut). For example, a mutant that presents an average AWA chemotaxis defect C.I. = 0.5, compared to the wildtype C.I. (C.I.wt-AWA) of 0.9, would have a C.I.diff-AWA = 0.4 (i.e., subtract 0.5 from 0.9). In the case of long-term adaptation (LTA) mutants the C.I.diff was calculated by subtracting the C.I. value for LTA from C.I.wt of unadapted animals. Wildtype odortaxis: 0.9 (C.I.wt AWA); −0.95 (C.I.wt AWB); 0.85 (C.I.wt AWC) [4, 23]. Wildtype long-term adaptation: 0.5 (C.I.diff AWA); 0.65 (C.I.diff AWC) [24, 25]. odr-3: 0.6 (C.I.diff AWA); −0.7 (C.I.diff AWB); 0.5 (C.I.diff AWC) [23, 26]. rgs-3: 0.85 (C.I.diff AWC) [27]. daf-11: 0.6 (C.I.diff AWB); 0.5 (C.I.diff AWC) [28]. dgk-1: 0 (C.I.diff AWC) [29]. eat-4: 0.55 (C.I.diff AWC) [30]. egl-30: 0.65 (C.I.diff AWC) [29].goa-1: 0.6 (C.I.diff AWA); 0.6 (C.I.diff AWC) [29]. odr-1: 0 (C.I.diff AWA); 0.85 (C.I.diff AWC) [31, 32]. tax-2: 0.2 (C.I.diff AWA); −0.8 (C.I.diff AWB), 0.75 (C.I.diff AWC) [23, 33]. tax-4: 0.5 (C.I.diff AWA); 0.75 (C.I.diff AWC) [23, 34]. egl-4: 0.8 (C.I.diff AWA); 0.65 (C.I.diff AWC) [25, 34]. The phenotype index (PI) was then calculated as follows:
| (3) |
The PI for LTA mutants was measured by calculating the relative contribution of the LTA mutant to the adaptation response. For example, if the LTA response of wildtype animals normally decreases the C.I. value to 0.3 from 0.9, then the C.I.diff = 0.6; if the mutant is fully required (i.e., there is no LTA response at all in the mutant) for the LTA response, then the PI is calculated as follows: 0.6/0.6 = 1; which reflects the total contribution of the gene product to LTA in wildtype animals but that which is defective in the LTA mutant. The weighted phenotype index (WPI) was calculated for each group of mutants as follows:
| (4) |
where P.I.mutx is the phenotype index (P.I.) for the mutant x (mutx), and wmutx represents the number of odors for which mutant x (mutx) exhibits a mutant phenotype.
3. Results
To compare divergence rate with pathway position in our network of dynamic odor transduction, we identified orthologous genes of the C. elegans odor network in P. pacificus, and divided the odor-signaling pathway members (Figure 1(a)) within this network into 3 categories: (I) regulators Class 1 (GPA-3 [35, 36], ODR-3 [26], GPA-5 [35, 36], GPA-13 [35, 36], ARR-1 [37] and RGS-3 [27]); (II) regulators Class 2 (EGL-4 [25], TAX-6 [38], ODR-1 [31], DAF-11 [28], TAX-2 [33], and TAX-4 [39]); (III) actuators (GOA-1 [40], EGL-30 [40], DGK-1 [40], and EAT-4 [41]). After binning pathway members into each category, we then calculated divergence within each group by comparing substitutions at silent sites to that of nonsilent sites and looked for patterns of correlation between topology and evolutionary rate. We did not observe a significant association between pathway position and divergence (pathway position: Spearman rank-order correlation coefficient r s = 0.37, P = 0.15, Kendall's τ = 0.31, P = 0.13). We also designed an orthologous intragenus network using Caenorhabditis japonica and compared substitutions at silent sites to that of non-silent sites and looked for a pattern of correlation with pathway position. Using this intragenus network we also did not observe significant association between pathway position and divergence (pathway position: r s = 0.28, P = 0.3, Kendall's τ = 0.17, P = 0.4), suggesting that the pathway position alone may not shape constraint in our odor network.
Figure 1.
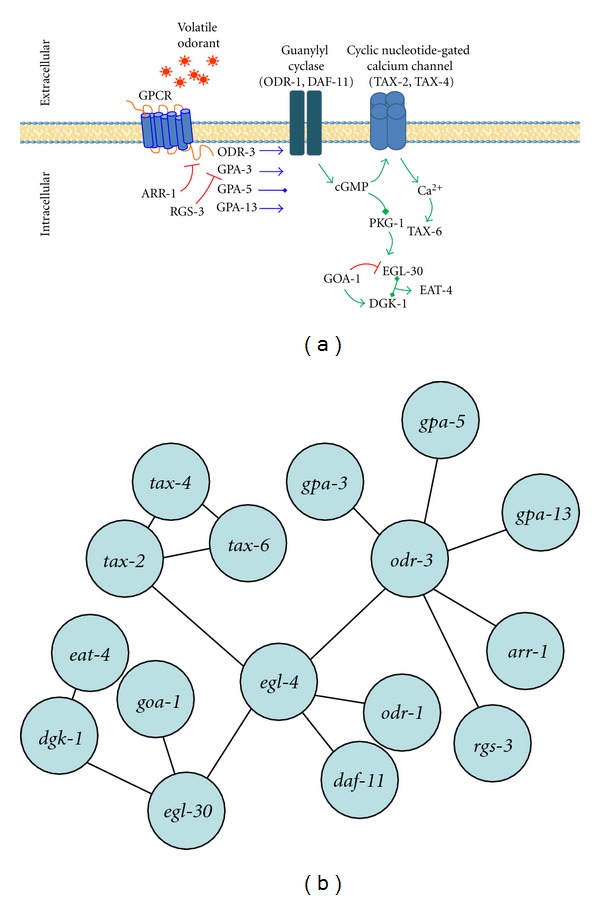
Odor-signaling pathway in C. elegans illustrated by pathway position and based on genetic interactions. (a) Summary of volatile odor signaling in C. elegans. (b) Schematic of an interaction-based odor network from C. elegans.
Next, we designed an interaction map of the network and tested for correlations between divergence and various topological metrics (Figure 1(b)). The interaction-based odor network is a composite map of the pathways within the three pairs of neurons that subserve volatile odor recognition in C. elegans (these cells are the AWA, AWB, and AWC). The interaction network was designed based upon genetic interactions and localization studies. Edges may represent interactions from only one of the three odor sensing pairs of neurons, but placed in a composite framework of all three pairs of odor-sensing neurons to obtain an informative number of vertices. The network was treated as undirected. The network comprised 16 nodes with a network diameter of 5, a characteristic path length of 2.825, and an average number of neighbors equal to 2. For the interaction network we observed that a strong correlation between number of neighbors and average neighborhood connectivity (correlation = 0.857; coefficient of determination R 2 = 0.648). Plotting the number of neighbors against average clustering coefficient yields a strong pattern of correlation using a linear function (correlation = 0.868; coefficient of determination R 2 = 0.753); we observed that a decrease in clustering coefficient with an increase in number of neighbors as is typical of many networks. Network analysis was performed and the output of the analyses was tested for correlations with divergence by calculating the evolutionary rate for each node by comparing substitutions at silent sites to that of non-silent sites with the orthologous genes of the odor network in P. pacificus. We observed a significant association in two cases: (1) measures of betweenness centrality and (2) measures of degree. By comparing measures of betweenness or measures of degree with divergence we observed significant negative correlations (betweenness: r s = −0.48, P = 0.02, Kendall's τ = −0.42, P = 0.03; degree: r s = −0.57, P = 0.01, Kendall's τ = −0.44, P = 0.03). Comparing measures of degree to measures of betweenness we observed a strong correlation (correlation = 0.936; R 2 = 0.877); this is typical of networks containing nodes of high influence that increase their influence with increases in their number of edges and is not specific for our odor network. Then, we calculated molecular diversity across the interaction network and searched for patterns of association with metrics from the following characteristics of the network: pathway position, betweenness centrality, and degree. In each case we did not observe a significant pattern of correlation (betweenness: r s = −0.35, P = 0.06, Kendall's τ = −0.3, P = 0.13; Pathway position: r s = 0.4, P = 0.08, Kendall's τ = 0.37, P = 0.06; degree: r s = −0.47, P = 0.06, Kendall's τ = −0.38, P = 0.06).
Next, we developed a metric of contribution within the network we termed the weighted phenotype index (WPI), and compared the total contribution of each group with divergence when organized by pathway position, betweenness, or degree. In the case of pathway position we observed a weak correlation between divergence and contribution (r = 0.44), and in the case of betweenness or degree we observed strong negative correlations with contribution and divergence (Figure 2(a) betweenness: r = −0.92; Figure 2(b) degree: r = −0.76). We also examined the correlation between quantities of betweenness or degree for each node in our network with contribution, and again we observed robust correlations (Figure 2(a) betweenness: r = 0.9; Figure 2(b) degree: r = 0.79). From this analysis we uncovered patterns of association between measures of betweenness, and degree with evolutionary rate and contribution.
Figure 2.
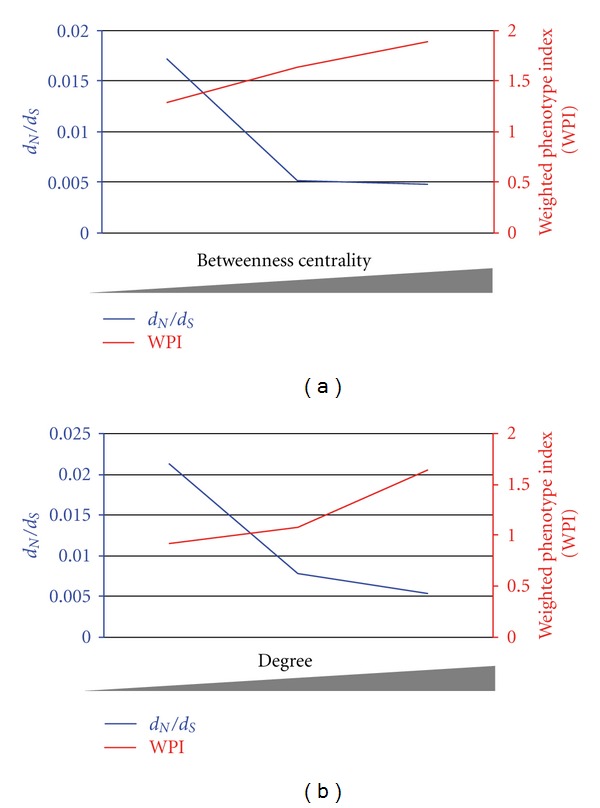
Plotting the relationship between divergence with measures of betweenness centrality or degree, and the relationship between contribution with measures of betweenness centrality or degree in a dynamic nematode odor network. (a) Divergence (left y-axis in blue) plotted against increasing measures of betweenness centrality (x-axis), and contribution (right y-axis in red) plotted against increasing measures of betweenness centrality. (b) Divergence (left y-axis in blue) plotted against increasing measures of degree (x-axis), and contribution (right y-axis in red) plotted against increasing measures of degree.
The most highly connected nodes within our network are that of odr-3 and egl-4 (also called pkg-1). ODR-3 is a Gα subunit protein that transduces multiple stimuli within the network [26]. EGL-4 is a protein kinase G that facilitates both primary signal transduction as well as desensitization responses within the network [25, 34]. To examine how molecular diversity varies across these hub vertices, we performed a sliding window examination of the coding region of each gene using 100-base-pair windows in increments of 20 base pairs. In the case of egl-4, we observed only a few polymorphic peaks, with each peak associating with areas between domains of functional importance (Figure 3(a)); these are the N-terminal low-complexity domain (LC), the coiled-coil domain (CC), two cGMP-binding domains (cNMP), and the serine/threonine kinase domain (S/T K). In the case of odr-3, we observed more variability overall compared with that of egl-4, but again with most variable peaks associating with areas between domains of functional significance (Figure 3(b)); in particular, the five alpha helices (G1–G5) that comprise the GTPase domain, the receptor interaction C-terminal (CT) as well as the fatty acid modification site (M) (Figure 3(b)). We observed similar scales for nucleotide diversity for both egl-4 (π = 0.27) and odr-3 (π = 0.25) with each gene exhibiting polymorphic peaks in areas between domains of functional importance. The similar pattern of diversity is in keeping with similar trends of divergence between egl-4 and odr-3: egl-4 d N/d S = 0.0041, d N = 0.1183, d S = 29.1777; odr-3 d N/d S = 0.0054, d N = 0.1132, and d S = 21.1204.
Figure 3.
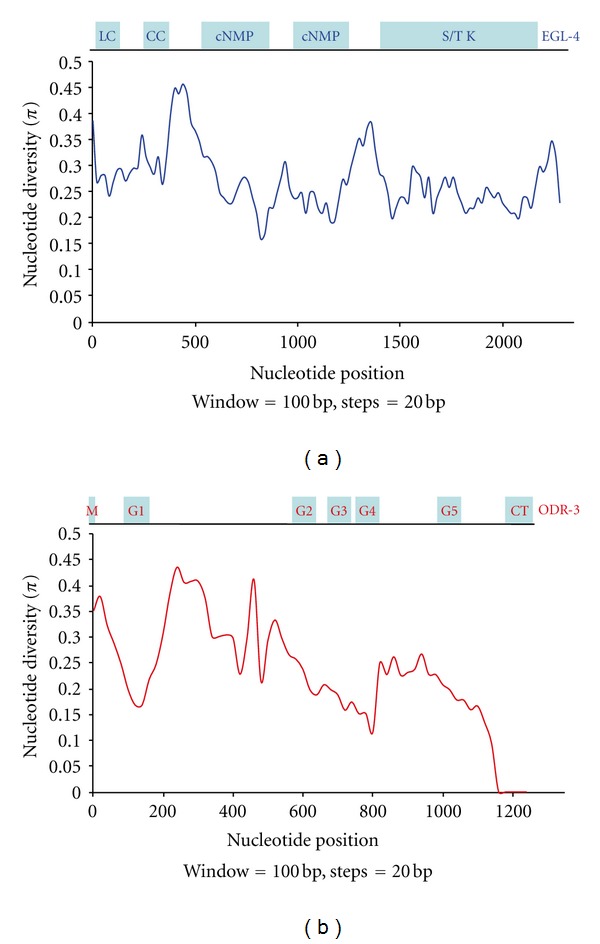
Sliding windows of nucleotide diversity within the coding regions of the genes egl-4 and odr-3. Nucleotide position in the alignments (x-axis) is plotted against nucleotide diversity (π) on the y-axis. (a) Nucleotide diversity in egl-4. The shaded boxes on top of the graph illustrate the various functional domains within the EGL-4 protein. LC: low-complexity domain; CC: coiled-coil domain; cNMP: cyclic nucleotide-binding domain, S/TK: serine/threonine kinase domain. (b) Nucleotide diversity in odr-3. The shaded boxes on top of the graph illustrate the various functional domains within the ODR-3 protein. M: fatty-acid modification site; G1–G5: five alpha helices that comprise the GTPase domain; CT: C-terminal receptor interaction domain. In each case a 100-base-pair window was analyzed in 20-base-pair increments.
Overall, nodes within the odor network are undergoing purifying selection, with an average d N/d S value across the network of 0.012 for P. pacificus versus C. elegans. To place this value in a context of global divergence rates between C. elegans and P. pacificus we examined divergence across 5,666 pairs of 1 : 1 orthologs, then generated randomized data sets (50,000 in total) comprising an equal number of nodes as our odor network (i.e., 16), and examined the frequency of mean d N/d S values in each case (Figure 4, blue bars). From this analysis we found an average d N/d S value = 0.14 which is more than an order of magnitude higher (~11.6X) than what we observed for our dynamic odor network. To control for the number of Gα subunit proteins within our odor network we also conducted a randomization trial whereby d N/d S averages from 50,000 random data sets of 16 nodes that contain 6 Gα subunit genes in each case were generated (Figure 4, red bars). In this control experiment we still observed a significantly higher average d N/d S value (d N/d S average = 0.106) than for our odor network. Taken together, this suggests that the dynamic odor-signaling network is shaped by a more pronounced purifying selection than what guides global constraint across the genome.
Figure 4.
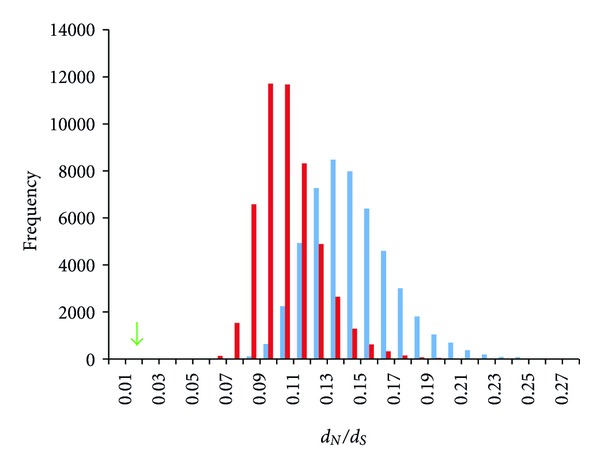
Frequency distribution of average d N/d S values from 50,000 randomization sets comprising 16 node groupings from 5,666 1 : 1 orthologs between C. elegans and P. pacificus (blue bars). Red bars indicate frequency distribution of average d N/d S values from 50,000 randomization sets of 16 node groupings that include 6 Gα subunit proteins in each; this control group was examined to investigate potential bias in our network by having 6 Gα subunit proteins within our odor genetic network. The x-axis represents binned divergence categories plotted against the frequency on the y-axis. The green arrow indicates the actual average d N/d S value of the nematode odor network.
4. Discussion
Positional rate variation (PRV) has been tested within numerous biosynthetic pathways where it has been demonstrated that upstream genes (perhaps on account of greater pleiotropy) undergo more selective constraint than downstream genes [42–44]. However, it is unlikely that this effect holds true for all signaling pathways as input and network architecture will vary widely. In the case of sensory signaling pathways that subserve an environment versus perception “arms race,” the idea of upstream genes exhibiting greater diversity may be more applicable. This trend was observed in a study of the Drosophila innate immune system where the authors demonstrated that recognition genes undergo higher levels of positive selection than immune effector genes [45]. In our network we did not observe a significant correlation between pathway position and divergence, which was in keeping with previous observations from data on intragenus networks of odor signaling in Caenorhabditis [46]. However, by comparing measures of degree or centrality with divergence, we did observe significant negative correlations. This may suggest that pathway position is not the only factor in shaping constraint or change in our network, but rather network characteristics such as connectivity may play major roles in guiding molecular evolution. Negative correlations between divergence with node relevance, and positive correlations between essentiality with node relevance have been reported previously and may represent a general trend of many biological networks [47, 48].
By comparing global levels of divergence between C. elegans and P. pacificus we found a pervasive theme of constraint across the genome; however, through our randomization study (Figure 4) we found that global levels of constraint are an order of magnitude lower than the level of constraint across the odor network. This suggests that the odor-signaling pathway represents a fixed circuit whose network properties are preserved by strong purifying selection. Low levels of divergence have been reported within Caenorhabditis for members of the olfactory pathway [46], and previously we have found that a large component in Regulators Class 1 and 2 has undergone extensive nematode-specific gene duplication events, namely, the Gα subunit proteins and guanylyl cyclase proteins [49, 50]. Many of these genes are either exclusively or highly expressed in primary sensory neurons [26, 28, 31, 35, 36, 51]. This genetic expansion facilitates multiple capacities that shape developmental and survival strategies through intercellular and intracellular processing of polymodal sensory input. Many of these duplicates are present in all four major clades of the phylum nematoda [49] suggesting that the odor-signaling network arose early in nematode evolution and has undergone neofunctionalization events that are preserved by strong functional constraint.
Supplementary Material
The Supplementary information file contains Figures of relationships inferred by reconstructing phylogenetic trees for seven families of genes within our odor signaling network. These are: 1) guanylyl cyclases; 2) diacylglycerol kinases; 3) vesicular glutamate transporters; 4) G protein alpha subunits; 5) protein kinase G; 6) calcineurin; and 7) cyclic nucleotide gated channel subunits.
Acknowledgments
The authors are very grateful to Rolf Sommer and Christian Rodelsperger for providing them with Pristionchus pacificus coding sequence files and to Scott Hamilton for critical reading of this manuscript. They also wish to thank their two anonymous reviewers for their excellent suggestions.
References
- 1.Ward S. Chemotaxis by the nematode Caenorhabditis elegans: identification of attractants and analysis of the response by use of mutants. Proceedings of the National Academy of Sciences of the United States of America. 1973;70(3):817–821. doi: 10.1073/pnas.70.3.817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dusenbery DB. Countercurrent separation: a new method for studying behavior of small aquatic organisms. Proceedings of the National Academy of Sciences of the United States of America. 1973;70(5):1349–1352. doi: 10.1073/pnas.70.5.1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bargmann CI, Horvitz HR. Chemosensory neurons with overlapping functions direct chemotaxis to multiple chemicals in C. elegans . Neuron. 1991;7(5):729–742. doi: 10.1016/0896-6273(91)90276-6. [DOI] [PubMed] [Google Scholar]
- 4.Bargmann CI, Hartwieg E, Horvitz HR. Odorant-selective genes and neurons mediate olfaction in C. elegans . Cell. 1993;74(3):515–527. doi: 10.1016/0092-8674(93)80053-h. [DOI] [PubMed] [Google Scholar]
- 5.Stretton AOW, Davis RE, Angstadt JD. Neural control of behaviour in Ascaris . Trends in Neurosciences. 1985;8(6):294–300. [Google Scholar]
- 6.Perkins LA, Hedgecock EM, Thomson JN, Culotti JG. Mutant sensory cilia in the nematode Caenorhabditis elegans . Developmental Biology. 1986;117(2):456–487. doi: 10.1016/0012-1606(86)90314-3. [DOI] [PubMed] [Google Scholar]
- 7.Kroetz SM, Srinivasan J, Yaghoobian J, Sternberg PW, Hong RL. The cGMP signaling pathway affects feeding behavior in the necromenic nematode Pristionchus pacificus . PLoS ONE. 2012;7(4) doi: 10.1371/journal.pone.0034464.e34464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hong RL, Witte H, Sommer RJ. Natural variation in Pristionchus pacificus insect pheromone attraction involves the protein kinase EGL-4. Proceedings of the National Academy of Sciences of the United States of America. 2008;105(22):7779–7784. doi: 10.1073/pnas.0708406105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dieterich C, Clifton SW, Schuster LN, et al. The Pristionchus pacificus genome provides a unique perspective on nematode lifestyle and parasitism. Nature Genetics. 2008;40(10):1193–1198. doi: 10.1038/ng.227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sommer RJ. Pristionchus pacificus . WormBook. 2006;14:1–8. doi: 10.1895/wormbook.1.102.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kryazhimskiy S, Plotkin JB. The population genetics of dN/dS. PLoS Genetics. 2008;4(12) doi: 10.1371/journal.pgen.1000304.e1000304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Östlund G, Schmitt T, Forslund K, et al. Inparanoid 7: new algorithms and tools for eukaryotic orthology analysis. Nucleic Acids Research. 2009;38(1):D196–D203. doi: 10.1093/nar/gkp931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li L, Stoeckert CJ, Jr., Roos DS. OrthoMCL: identification of ortholog groups for eukaryotic genomes. Genome Research. 2003;13(9):2178–2189. doi: 10.1101/gr.1224503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schneider A, Dessimoz C, Gonnet GH. OMA Browser—exploring orthologous relations across 352 complete genomes. Bioinformatics. 2007;23(16):2180–2182. doi: 10.1093/bioinformatics/btm295. [DOI] [PubMed] [Google Scholar]
- 15.Yook K, Harris TW, Bieri T, et al. WormBase 2012: more genomes, more data, new website. Nucleic Acids Research. 2012;40(D1):D735–D741. doi: 10.1093/nar/gkr954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Altschul SF, Madden TL, Schäffer AA, et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Research. 1997;25(17):3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Research. 2004;32(5):1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Guindon S, Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Systematic Biology. 2003;52(5):696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- 19.Yang Z, Nielsen R. Estimating synonymous and nonsynonymous substitution rates under realistic evolutionary models. Molecular Biology and Evolution. 2000;17(1):32–43. doi: 10.1093/oxfordjournals.molbev.a026236. [DOI] [PubMed] [Google Scholar]
- 20.Yang Z. PAML: a program package for phylogenetic analysis by maximum likelihood. Computer Applications in the Biosciences. 1997;13(5):555–556. doi: 10.1093/bioinformatics/13.5.555. [DOI] [PubMed] [Google Scholar]
- 21.Librado P, Rozas J. DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics. 2009;25(11):1451–1452. doi: 10.1093/bioinformatics/btp187. [DOI] [PubMed] [Google Scholar]
- 22.Shannon P, Markiel A, Ozier O, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Research. 2003;13(11):2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Troemel ER, Kimmel BE, Bargmann CI. Reprogramming chemotaxis responses: sensory neurons define olfactory preferences in C. elegans . Cell. 1997;91(2):161–169. doi: 10.1016/s0092-8674(00)80399-2. [DOI] [PubMed] [Google Scholar]
- 24.Colbert HA, Bargmann CI. Odorant-specific adaptation pathways generate olfactory plasticity in C. elegans . Neuron. 1995;14(4):803–812. doi: 10.1016/0896-6273(95)90224-4. [DOI] [PubMed] [Google Scholar]
- 25.L’Etoile ND, Coburn CM, Eastham J, Kistler A, Gallegos G, Bargmann CI. The cyclic GMP-dependent protein kinase EGL-4 regulates olfactory adaptation in C. elegans . Neuron. 2002;36(6):1079–1089. doi: 10.1016/s0896-6273(02)01066-8. [DOI] [PubMed] [Google Scholar]
- 26.Roayaie K, Crump JG, Sagasti A, Bargmann CI. The Gα protein ODR-3 mediates olfactory and nociceptive function and controls cilium morphogenesis in C. elegans olfactory neurons. Neuron. 1998;20(1):55–67. doi: 10.1016/s0896-6273(00)80434-1. [DOI] [PubMed] [Google Scholar]
- 27.Ferkey DM, Hyde R, Haspel G, et al. C. elegans G protein regulator RGS-3 controls sensitivity to sensory stimuli. Neuron. 2007;53(1):39–52. doi: 10.1016/j.neuron.2006.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Birnby DA, Link EM, Vowels JJ, Tian H, Colacurcio PL, Thomas JH. A transmembrane guanylyl cyclase (DAF-11) and Hsp90 (DAF-21) regulate a common set of chemosensory behaviors in Caenorhabditis elegans . Genetics. 2000;155(1):85–104. doi: 10.1093/genetics/155.1.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Matsuki M, Kunitomo H, Iino Y. Goα regulates olfactory adaptation by antagonizing G qα-DAG signaling in Caenorhabditis elegans . Proceedings of the National Academy of Sciences of the United States of America. 2006;103(4):1112–1117. doi: 10.1073/pnas.0506954103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nuttley WM, Harbinder S, van der Kooy D. Regulation of distinct attractive and aversive mechanisms mediating benzaldehyde chemotaxis in Caenorhabditis elegans . Learning and Memory. 2001;8(3):170–181. doi: 10.1101/lm.36501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.L’Etoile ND, Bargmann CI. Olfaction and odor discrimination are mediated by the C. elegans guanylyl cyclase ODR-1. Neuron. 2000;25(3):575–586. doi: 10.1016/s0896-6273(00)81061-2. [DOI] [PubMed] [Google Scholar]
- 32.Sze JY, Ruvkun G. Activity of the Caenorhabditis elegans UNC-86 POU transcription factor modulates olfactory sensitivity. Proceedings of the National Academy of Sciences of the United States of America. 2003;100(16):9560–9565. doi: 10.1073/pnas.1530752100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Coburn CM, Bargmann CI. A putative cyclic nucleotide-gated channel is required for sensory development and function in C. elegans . Neuron. 1996;17(4):695–706. doi: 10.1016/s0896-6273(00)80201-9. [DOI] [PubMed] [Google Scholar]
- 34.Daniels SA, Ailion M, Thomas JH, Sengupta P. egl-4 Acts through a transforming growth factor-β/SMAD pathway in Caenorhabditis elegans to regulate multiple neuronal circuits in response to sensory cues. Genetics. 2000;156(1):123–141. doi: 10.1093/genetics/156.1.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jansen G, Thijssen KL, Werner P, van der Horst M, Hazendonk E, Plasterk RHA. The complete family of genes encoding G proteins of Caenorhabditis elegans . Nature Genetics. 1999;21(4):414–419. doi: 10.1038/7753. [DOI] [PubMed] [Google Scholar]
- 36.Lans H, Rademakers S, Jansen G. A network of stimulatory and inhibitory Gα-subunits regulates olfaction in Caenorhabditis elegans . Genetics. 2004;167(4):1677–1687. doi: 10.1534/genetics.103.024786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Palmitessa A, Hess HA, Bany IA, Kim YM, Koelle MR, Benovic JL. Caenorhabditus elegans arrestin regulates neural G protein signaling and olfactory adaptation and recovery. The Journal of Biological Chemistry. 2005;280(26):24649–24662. doi: 10.1074/jbc.M502637200. [DOI] [PubMed] [Google Scholar]
- 38.Kuhara A, Inada H, Katsura I, Mori I. Negative regulation and gain control of sensory neurons by the C. elegans calcineurin TAX-6. Neuron. 2002;33(5):751–763. doi: 10.1016/s0896-6273(02)00607-4. [DOI] [PubMed] [Google Scholar]
- 39.Komatsu H, Mori I, Rhee JS, Akaike N, Ohshima Y. Mutations in a cyclic nucleotide-gated channel lead to abnormal thermosensation and chemosensation in C. elegans . Neuron. 1996;17(4):707–718. doi: 10.1016/s0896-6273(00)80202-0. [DOI] [PubMed] [Google Scholar]
- 40.Miller KG, Emerson MD, Rand JB. G(o)α and diacylglycerol kinase negatively regulate the G(q)α pathway in C. elegans . Neuron. 1999;24(2):323–333. doi: 10.1016/s0896-6273(00)80847-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lee RYN, Sawin ER, Chalfie M, Horvitz HR, Avery L. EAT-4, a homolog of a mammalian sodium-dependent inorganic phosphate cotransporter, is necessary for glutamatergic neurotransmission in Caenorhabditis elegans . Journal of Neuroscience. 1999;19(1):159–167. doi: 10.1523/JNEUROSCI.19-01-00159.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rausher MD, Miller RE, Tiffin P. Patterns of evolutionary rate variation among genes of the anthocyanin biosynthetic pathway. Molecular Biology and Evolution. 1999;16(2):266–274. doi: 10.1093/oxfordjournals.molbev.a026108. [DOI] [PubMed] [Google Scholar]
- 43.Ramsay H, Rieseberg LH, Ritland K. The correlation of evolutionary rate with pathway position in plant terpenoid biosynthesis. Molecular Biology and Evolution. 2009;26(5):1045–1053. doi: 10.1093/molbev/msp021. [DOI] [PubMed] [Google Scholar]
- 44.Waxman D, Peck JR. Pleiotropy and the preservation of perfection. Science. 1998;279(5354):1210–1213. [PubMed] [Google Scholar]
- 45.Sackton TB, Lazzaro BP, Schlenke TA, Evans JD, Hultmark D, Clark AG. Dynamic evolution of the innate immune system in Drosophila . Nature Genetics. 2007;39(12):1461–1468. doi: 10.1038/ng.2007.60. [DOI] [PubMed] [Google Scholar]
- 46.Jovelin R, Dunham JP, Sung FS, Phillips PC. High nucleotide divergence in developmental regulatory genes contrasts with the structural elements of olfactory pathways in Caenorhabditis. Genetics. 2009;181(4):1387–1397. doi: 10.1534/genetics.107.082651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wuchty S. Evolution and topology in the yeast protein interaction network. Genome Research. 2004;14(7):1310–1314. doi: 10.1101/gr.2300204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fraser HB, Hirsh AE, Steinmetz LM, Scharfe C, Feldman MW. Evolutionary rate in the protein interaction network. Science. 2002;296(5568):750–752. doi: 10.1126/science.1068696. [DOI] [PubMed] [Google Scholar]
- 49.O’Halloran DM, Fitzpatrick DA, McCormack GP, McInerney JO, Burnell AM. The molecular phylogeny of a nematode-specific clade of heterotrimeric G-protein α-subunit genes. Journal of Molecular Evolution. 2006;63(1):87–94. doi: 10.1007/s00239-005-0215-z. [DOI] [PubMed] [Google Scholar]
- 50.Fitzpatrick DA, O’Halloran DM, Burnell AM. Multiple lineage specific expansions within the guanylyl cyclase gene family. BMC Evolutionary Biology. 2006;6, article 26 doi: 10.1186/1471-2148-6-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yu S, Avery L, Baude E, Garbers DL. Guanylyl cyclase expression in specific sensory neurons: a new family of chemosensory receptors. Proceedings of the National Academy of Sciences of the United States of America. 1997;94(7):3384–3387. doi: 10.1073/pnas.94.7.3384. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The Supplementary information file contains Figures of relationships inferred by reconstructing phylogenetic trees for seven families of genes within our odor signaling network. These are: 1) guanylyl cyclases; 2) diacylglycerol kinases; 3) vesicular glutamate transporters; 4) G protein alpha subunits; 5) protein kinase G; 6) calcineurin; and 7) cyclic nucleotide gated channel subunits.


