Abstract
A compelling need exists for the development of technologies that facilitate and accelerate the discovery of novel protein biomarkers with therapeutic and diagnostic potential. Comparisons among shotgun proteome technologies, including capillary isotachophoresis (CITP)-based multidimensional separations and multidimensional liquid chromatography system, are therefore performed in this study regarding their abilities to address the challenges of protein complexity and relative abundance inherent in glioblastoma multiforme derived cancer stem cells. Comparisons are conducted using a single processed protein digest with equal sample loading, identical second dimension separation (reversed phase liquid chromatography) and mass spectrometry conditions, and consistent search parameters and cutoff established by the target-decoy determined false discovery rate.
Besides achieving superior overall proteome performance in total peptide, distinct peptide, and distinct protein identifications, analytical reproducibility of the CITP proteome platform coupled with the spectral counting approach is determined by a Pearson R2 value of 0.98 and a coefficient of variation of 15% across all proteins quantified. In contrast, extensive fraction overlapping in strong cation exchange greatly limits the ability of multidimensional liquid chromatography separations for mining deeper into the tissue proteome as evidenced by the poor coverage in various protein functional categories and key protein pathways. The CITP proteomic technology, equipped with selective analyte enrichment and ultrahigh resolving power, is expected to serve as a critical component in the overall toolset required for biomarker discovery via shotgun proteomic analysis of tissue specimens.
Keywords: Biomarker, Capillary Electrophoresis, Mass Spectrometry, Strong Cation Exchange Chromatography, Tissue Proteomics
INTRODUCTION
Although blood is the preferred assay specimen for a final diagnostic test, the biological materials analyzed for biomarker discovery need not be plasma. In addition to the analytical challenges posed by the complexity and depth of the plasma proteome, many biomarkers elaborated from diseased tissues as the result of leakage and secretion are massively diluted into circulating blood with ultimate concentrations which are five to seven orders of magnitude lower in abundance than the most highly concentrated plasma proteins. If biomarkers specific for a particular disease arise locally from the affected tissue, it is highly plausible that a gradient of concentration diminishes with increasing distance from the site of disease. Thus, it has been proposed that a better way to discover biomarkers might be to study tissues involved in disease or the fluids bathing theses tissues [1, 2].
Still, biologically relevant proteomics data can only be generated if the tissue samples investigated consist of homogeneous cell populations, in which no unwanted cells of different types and/or development stages obscure the results. For example, SM22, a dominant protein in smooth muscle cells, has been widely reported to be abnormally expressed in many solid tumors. However, a recent proteomic study conducted by Li and co-workers has revealed that the highly expressed SM22 was mainly found in smooth muscle layers, blood vessels, and myofibroblasts instead of gastric cancer cells[3]. Thus, laser capture microdissection (LCM) technology[4, 5] has been developed to provide a rapid and straightforward method for procuring homogeneous subpopulations of cells for biochemical and molecular biological analyses.
However, in the absence of protein amplification, proteomic analysis of microdissected-procured specimens is severely constrained by sample amounts ranging from 103–105 cells, corresponding to a total protein content of 0.1–10 µg [5]. While two-dimensional polyacrylamide gel electrophoresis (2-D PAGE) analyses of tissue samples have been attempted [6–14], including the analysis of microdissected tumor tissues [15–19], these studies required significant manual effort and time to extract sufficient levels of protein for analysis, while providing little information on protein expression beyond a relatively small number of high abundance proteins. On the other hand, the reported tissue proteomic studies employing multidimensional liquid chromatography separations [20–23] have been mainly based on the analysis of entire tissue sections instead of targeted cell populations. The minimal quantity of available protein samples from microdissected cell populations has restricted recent analyses [24–32] to the use of only a single chromatography separation prior to tandem MS analysis and greatly limited the ability for mining deeper into the tissue proteome.
Combined capillary isoelectric focusing (CIEF)/nano-reversed phase liquid chromatography (nano-RPLC) separations coupled with electrospray ionization-mass spectrometry (ESI-MS) have been demonstrated to enable ultrasensitive analysis of minute proteins extracted from microdissected fresh-frozen [33–35] and formalin-fixed and paraffin-embedded (FFPE) tissues [36–38]. As evaluated in our recent analyses of human saliva and mouse brain mitochondrial samples [39–41], a capillary isotachophoresis (CITP)-based proteomic platform not only enables selective enrichment of low abundance proteins, but also provides superior resolving power than that achieved using CIEF/nano-RPLC separations. In this work, comparisons among shotgun proteome technologies, including multidimensional CITP and liquid chromatography separations [42–44], are performed regarding their abilities to address the challenges of protein complexity and relative abundance inherent in cancer stem cells as a model tissue proteome system.
MATERIALS AND METHODS
Materials
Fused-silica capillaries (50 µm i.d./375 µm o.d. and 100 µm i.d./375 µm o.d.) were acquired from Polymicro Technologies (Phoenix, AZ). Acetic acid, dithiothreitol (DTT), iodoacetamide (IAM), and trifluoroacetic acid (TFA) were purchased from Sigma (St. Louis, MO). Acetonitrile, ammonium acetate, ammonium formate, hydroxypropyl cellulose (average MW 100,000), sodium dodecyl sulfate (SDS), tris(hydroxymethyl)aminomethane (Tris), and urea were obtained from Fisher Scientific (Pittsburgh, PA). Pharmalyte 3–10 was purchased from Amersham Pharmacia Biotech (Uppsala, Sweden). Sequencing grade trypsin was acquired from Promega (Madison, WI). All solutions were prepared using water purified by a Neu-Ion system (Baltimore, MD) equipped with a UV sterilizing lamp and a 0.05 µm membrane final filter.
Preparation of Glioblastoma Derived Cancer Stem Cells
Neural stem cells were isolated from glioblastoma multiforme (GBM) tissues and cultured [45] in Dr. Deric M. Park’s laboratory at the University of Pittsburgh Cancer Institute. Pelleted cells were suspended in a buffer that consisted of 50 mM Tris (pH 8.0), 65 mM DTT, and 10% glycerol. The cells were disrupted by homogenization with glass beads in a Mini-BeadBeater (BioSpec Products, Bartlesville, OK). The soluble proteins were collected in the supernatant by centrifugation at 20,000 g for 30 min.
Proteins collected in the supernatant were denatured, reduced, and alkylated by sequentially adding urea, DTT, and IAM with final concentrations of 8 M, 10 mg/mL, and 20 mg/mL, respectively. The solution was incubated at 37 °C for 1 hr in the dark and then diluted 8-fold with 100 mM ammonium acetate at pH 8.0. Trypsin was added at a 1:40 (w/w) enzyme to substrate ratio and the solution was incubated at 37 °C overnight. Tryptic digests were desalted using a Peptide MacroTrap column (Michrom Bioresources, Auburn, CA), lyophilized to dryness using a SpeedVac (Thermo, San Jose, CA), and then stored at −80 °C.
Transient CITP/Capillary Zone Electrophoresis (CZE)-Based Multidimensional Separations
Similar to the procedures described in our previous studies [38, 40, 41], a 80-cm long CITP capillary (100 µm i.d./365 µm o.d.) coated with hydroxypropyl cellulose was initially filled with a background electrophoresis buffer of 0.1 M acetic acid at pH 2.8. The samples containing protein digests processed from neural stem cells were prepared in a 2% pharmalyte solution. A 50-cm long sample plug, corresponding to 4.0 µL sample volume, was hydrodynamically injected into the capillary. A positive electric voltage of 24 kV was then applied to the inlet reservoir filled with a 0.1 M acetic acid solution.
The cathodic end of the capillary was housed inside a stainless steel needle using a coaxial liquid sheath flow configuration [46, 47]. A sheath liquid composed of 0.1 M acetic acid was delivered at a flow rate of 1 µL/min using a Harvard Apparatus 22 syringe pump (South Natick, MA). The stacked and resolved peptides in the CITP/CZE capillary were sequentially fractionated and loaded into individual wells on a moving microtiter plate.
Peptides collected in individual wells were sequentially injected into dual trap columns (3 cm × 200 µm i.d. × 365 µm o.d.) packed with 5 µm porous C18 reversed-phase particles. Each peptide fraction was subsequently analyzed by nano-RPLC equipped with an Ultimate dual-quaternary pump (Dionex, Sunnyvale, CA) and a dual nano-flow splitter connected to two pulled-tip fused-silica capillaries (50 µm i.d. × 365 µm o.d.). These two 15-cm long capillaries were packed with 3-µm Zorbax Stable Bond (Agilent, Palo Alto, CA) C18 particles.
Nano-RPLC separations were performed in parallel in which a dual-quaternary pump delivered two identical 2-hr organic solvent gradients with an offset of 1 hr. Peptides were eluted at a flow rate of 200 nL/min using a 5–45% linear acetonitrile gradient over 100 min with the remaining 20 min for column regeneration and equilibration. The peptide eluants were monitored using a linear ion-trap mass spectrometer (LTQ, ThermoFinnigan, San Jose, CA) equipped with an electrospray ionization interface and operated in a data dependent mode. Full scans were collected from 400 – 1400 m/z and 5 data dependent MS/MS scans were collected with dynamic exclusion set to 30 sec.
Strong Cation-Exchange (SCX) Coupled with Nano-RPLC-ESI-MS
A SCX column (5 cm length × 300 µm i.d.) packed with 5-µm poly(2-sulfoethyl aspartamide) particles were purchased from PolyLC (Columbia, MD). The salt gradient for SCX was the same as that employed by Liu and co-workers for the characterization of the human plasma proteome [48]. Briefly, peptides were eluted at a flow rate of 10 µL/min using a 10–255 mM ammonium formate gradient (containing 25% acetonitrile) over 50 min with the remaining 20 min for column regeneration and equilibration. Peptides collected in individual wells of a microtiter plate were lyophilized to dryness and reconstituted in a 0.1% TFA solution containing 2% acetonitrile. Peptide fractions resolved from SCX were subsequently and individually analyzed using nano-RPLC-ESI-MS by following the same procedures described previously.
MS Data Analysis
Raw LTQ data were converted to peak list files by msn_extract.exe (ThermoFinnigan). Open Mass Spectrometry Search Algorithm (OMSSA) [49] developed at the National Center for Biotechnology Information was used to search the peak list files against the UniProt sequence library (April 20, 2006) with decoyed sequences appended. This decoyed database was constructed by reversing all sequences and appending them to the end of the sequence library. Searches were performed using the following parameters: fully tryptic, 1.5 Da precursor ion mass tolerance, 0.4 Da fragment ion mass tolerance, 1 missed cleavage, alkylated Cys as a fixed modification and variable modification of Met oxidation. Searches were run in parallel on a 14 node, 28 CPU Linux cluster (Linux Networx, Bluffdale, UT).
False discovery rates (FDRs) were determined using a target-decoy search strategy introduced by Elias and co-workers [50]. An E-value threshold corresponding to a 1% FDR in total peptide identifications was used as a cutoff in this analysis as this correlates with the maximum sensitivity versus specificity in our previous work [51]. No other cutoffs, such as requiring a minimum number of distinct peptides per protein identification, were utilized.
Many proteome research laboratories routinely discard single peptide identifications to significantly reduce the FDR of distinct protein identifications. However, as previously demonstrated by Dr. Gygi’s [52] and our laboratories [51], these single-protein identifications, after filtering, are mostly correct and often represent 30–50% of a proteome dataset. For example, several low abundance proteins, including CD74, CD117, CD45, and S-100, were identified by single peptide hits during the proteome analysis of FFPE liver tissues and were successfully validated by subsequent immunohistochemistry measurements [38]. Thus, as advocated by Elias and Gygi [52] and also implemented in our proteome studies, the target-decoy search strategy is employed as the routine practice to design more stringent criteria for single-peptide identifications.
The UniProt sequence library consists of entries from both SwissProt and TrEMBL. Due to the minimally redundant nature of SwissProt, only peptides mapping to the SwissProt subset were reported here. After generation of search data, the OMSSA XML result files were parsed using a Java parser and loaded into an Oracle 10g database for analysis and reporting using in-house software.
RESULTS AND DISCUSSION
Increasing evidence support the hypothesis that a variety of cancers, including GBM, the most common and malignant brain tumor, arise from malignant transformation of undifferentiated stem-progenitor cells. Similar to normal neural stem cell (multipotent cells in the central nervous system capable of differentiating into neurons, astrocytes, and oligodendrocytes), GBM derived stem cells express primitive markers, self-renew, and are able to differentiate to multiple lineages upon activation of appropriate pathways [53, 54]. The presence of malignant stem-like cells within the heterogeneous cell populations in GBM is thought to be responsible for recurrence, infiltration into surrounding white matter, and radiation-chemoresistance. Therefore, treatment strategies directed against the malignant stem cell compartment of GBM has the potential to limit proliferation and invasion to allow a greater tumor resection, and more durable response to radiation and chemotherapy.
Neural stem cells were isolated from GBM tissues and cultured [45] in Dr. Park’s laboratory as part of ongoing research collaboration toward the identification of stem and differentiation markers in determining cellular behavior, including growth, motility, proliferative index, and survival. Similar to primary cell cultures, glioblastoma derived cancer stem cells in culture have a very limited lifespan before they senesce. Thus, these small cancer stem cell populations isolated from tumor tissues and enriched in culture represent an excellent opportunity for conducting comparisons among shotgun proteome techniques targeting tissue proteomics, including transient CITP/CZE-based multidimensional separations and multidimensional protein identification technology (MuDPIT) [42–44].
Evaluation of overall proteome performance
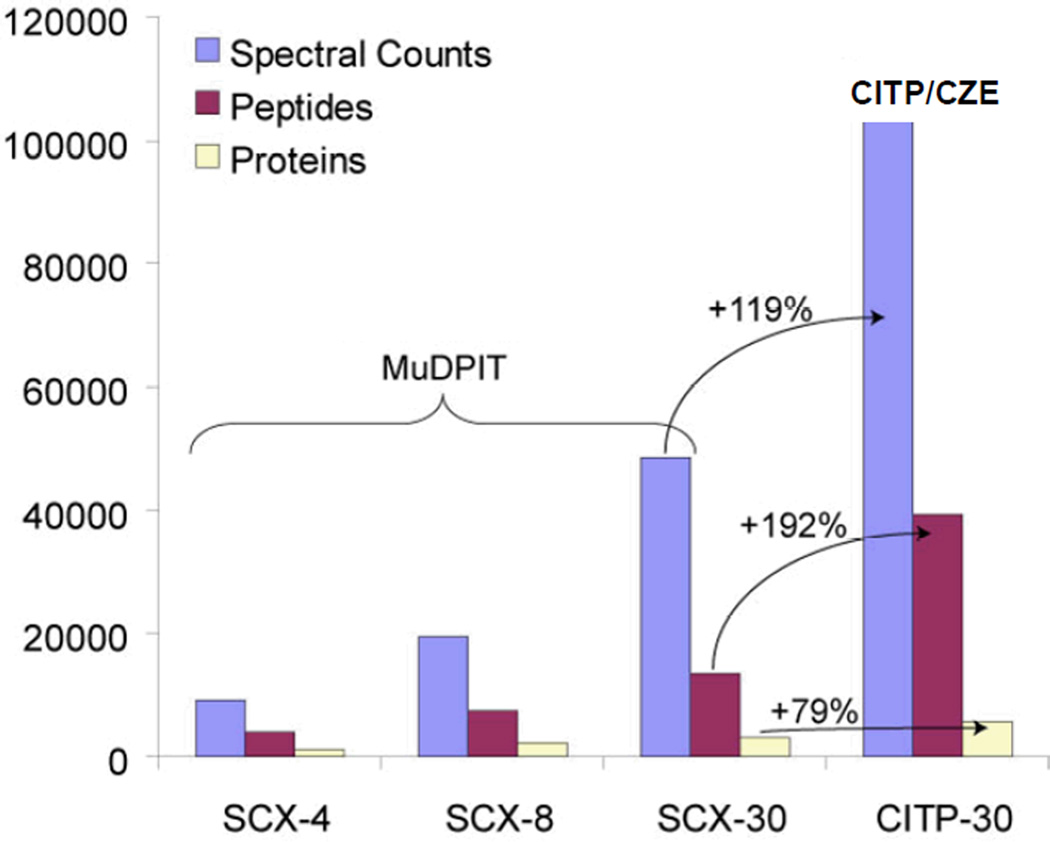
Comparisons were achieved using a single processed tryptic digest of cancer stem cells, equal loading of 15 µg digest into the CITP capillary and the SCX column, same nano-RPLC-ESI-MS conditions, and identical search parameters together with a consistent cutoff as established by the target-decoy determined FDR [50, 51]. By enhancing the overall separation peak capacity and the MS duty cycle, the numbers of total spectral counts (or total peptide identifications), distinct peptides, and distinct proteins summarized in Fig. 1 all increased with increasing the number of SCX fractions from 4, 8, and 30. A 30 CITP/CZE-fraction measurement, however, resulted in further enhancements in total peptide, distinct peptide, and distinct protein identifications over a 30-fraction SCX run by 119%, 192%, and 79%, respectively.
Figure 1.
Summary of proteomic results of GBM derived neural stem cells using MuDPIT and CITP/CZE-based multidimensional separations.
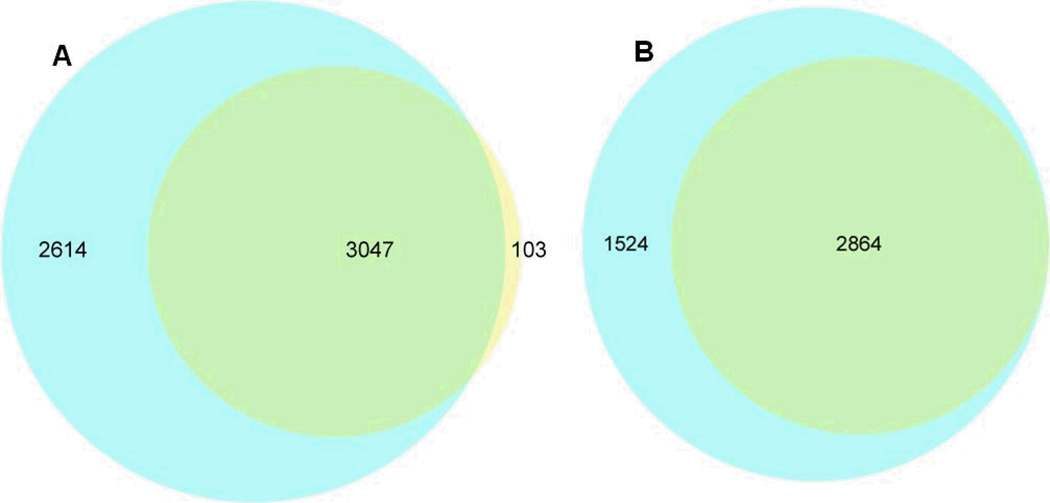
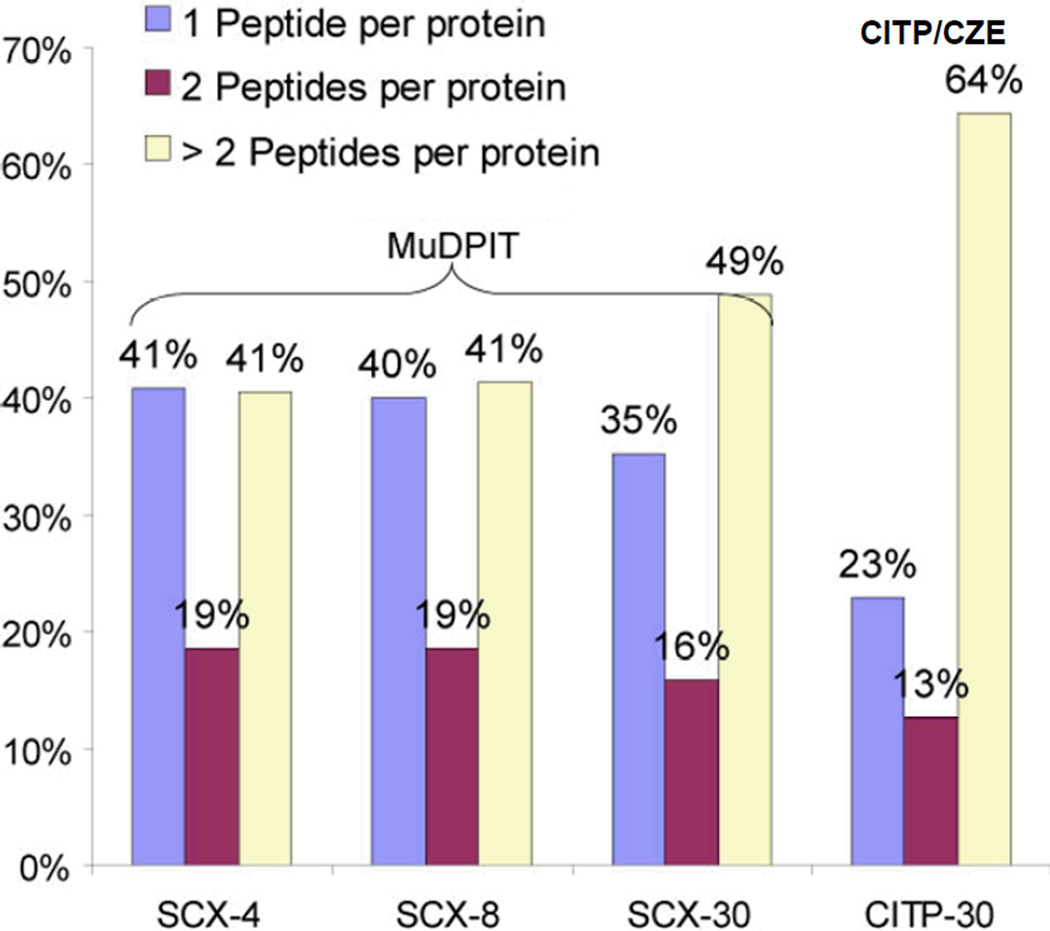
There were very good agreements between proteomic results obtained from 30 fractions SCX- and CITP-based shotgun techniques on a core set of proteins identified by at least one (Fig. 2A) or two (Fig. 2B) distinct peptides. The Venn diagrams illustrated that the CITP-based proteomic platform uniquely detected 2,614 proteins by one or more distinct peptides or 1,524 proteins by two or more distinct peptides. Proteins identified by two or more distinct peptides using MuDPIT were completely included in the CITP measurements. In addition to increasing the number of protein identifications, the percentage of proteins identified by two or more distinct peptides also increased from 60% (19% + 41%) in 4- and 8-fraction runs to 65% (16% + 49%) in a 30-fraction MuDPIT experiment (Fig. 3). This percentage further increased to 77% using transient CITP/CZE-based multidimensional separations.
Figure 2.
The Venn diagrams comparing proteins detected using MuDPIT (small circle) and CITP/CZE-based multidimensional separations (large circle). Proteins are identified by at least one (A) or two (B) distinct peptides.
Figure 3.
Summary of the percentages of proteins identified by one, two, or more distinct peptides using MuDPIT and CITP/CZE-based shotgun proteome techniques.
Other measures of proteome data quality are the average numbers of spectral counts and distinct peptide identifications per protein. Increase in the number of distinct peptide identifications per protein is interpreted as an improvement in the confidence and the sequence coverage of a protein identification [55]. The number of spectral counts becomes very important when implementing spectral counting-based label-free quantification [56–58], as the expression levels of each protein are determined by the number of tandem MS events. Although CITP delivered 79% more protein identifications (Fig. 1), CITP still outperformed MuDPIT with respect to both measures: an average of 18.8 spectral counts and 6.9 distinct peptides per protein in CITP versus an average 15.4 spectral counts and 4.3 distinct peptides of each protein in MuDPIT.
Comparison of separation performance
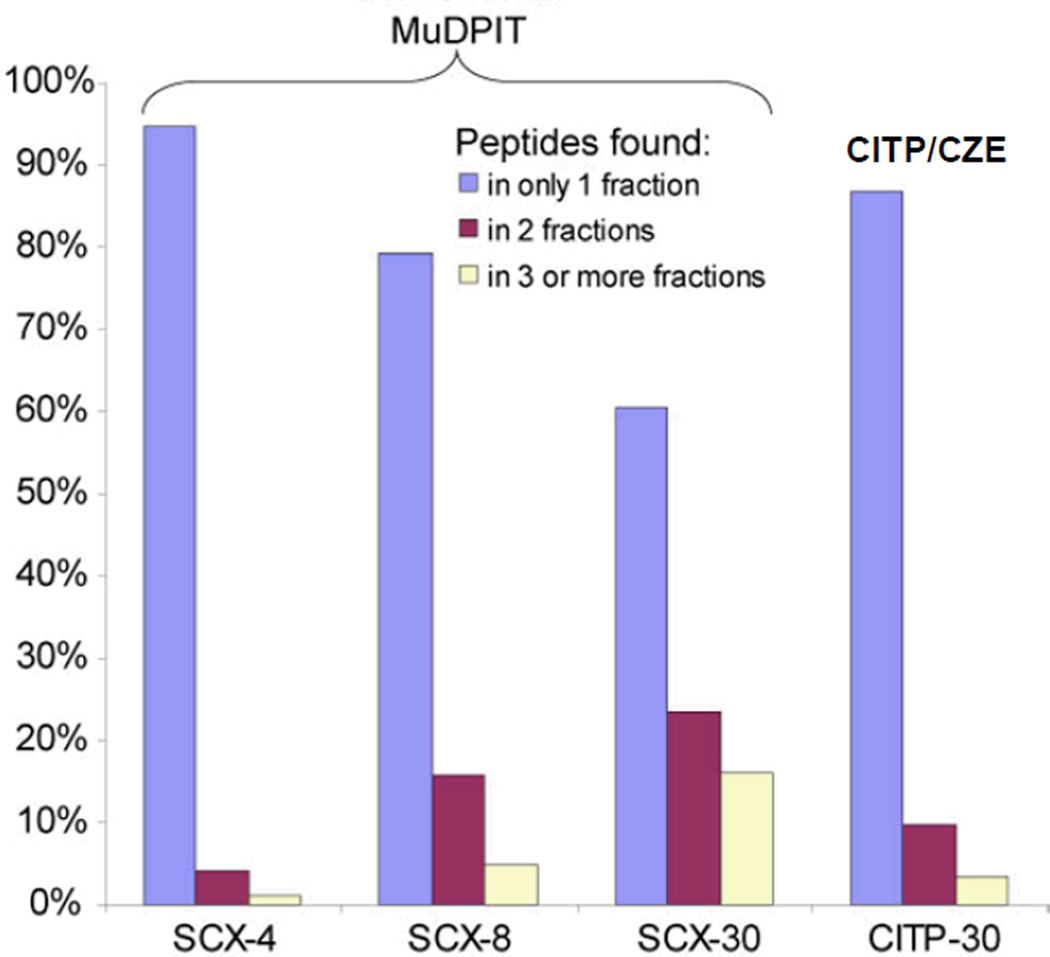
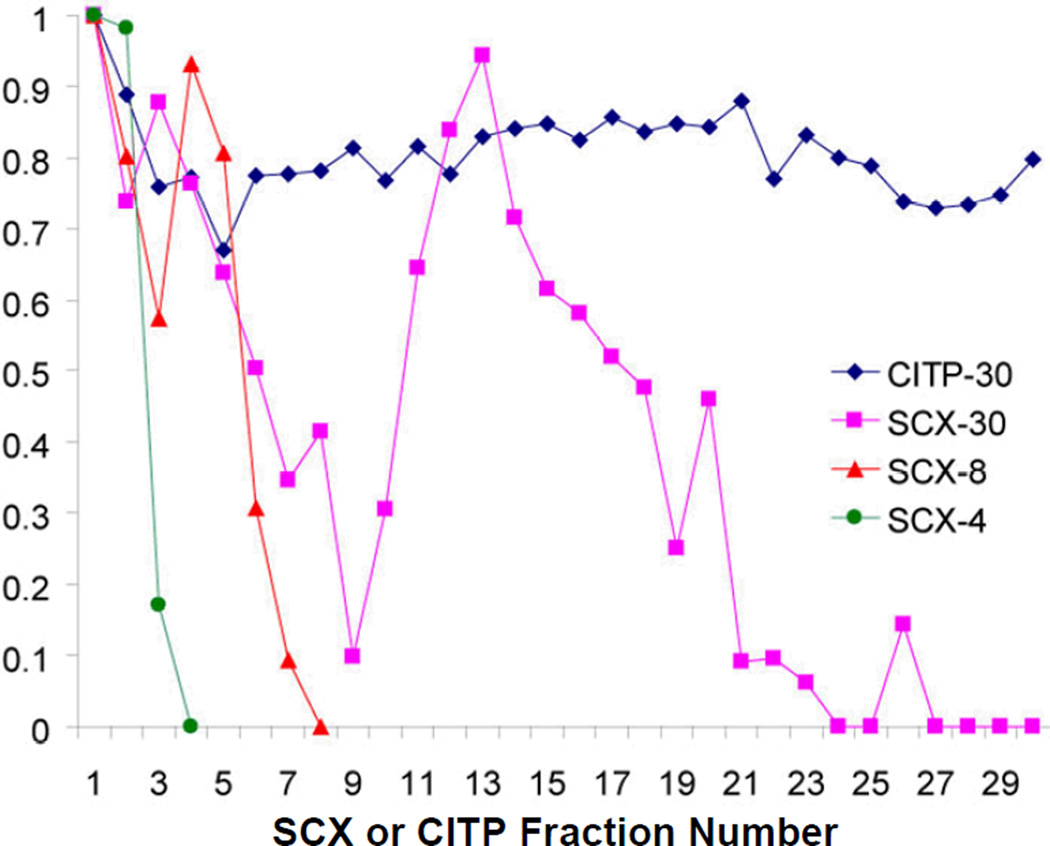
The high resolving power of transient CITP/CZE as the first separation dimension is clearly demonstrated by significantly low peptide fraction overlapping shown in Fig. 4. Approximately 89% of distinct peptides were identified in only a single CITP fraction. Of the remaining peptides, 8% were identified in two fractions and 3% were measured in three and more fractions. Only a 4-fraction MuDPIT run exhibited lower fraction overlapping than that observed in CITP, however, at the expense of the overall peak capacity critically needed for resolving complex proteome mixtures.
Figure 4.
Summary of the percentages of peptides identified from single or multiple CITP and SCX fractions using MuDPIT and CITP/CZE-based multidimensional separations, respectively.
The percentage of distinct peptides identified in only a single SCX fraction decreased from 94% in 4 fractions to 80% in 8 fractions and 61% in 30 fractions. In contrast, this percentage determined from the CITP measurements was only reduced from 95% in 15 fractions to 89% in 30 fractions. Thus, the intrinsically high resolution of transient CITP/CZE allows the number of fractions sampled in the first separation dimension to be increased for enhancing the overall peak capacity of multidimensional separations and the resulting proteome coverage, particularly for the characterization of small cell populations and limited tissue samples.
While increasing the number of CITP fractions, the amount of individual (not total) peptides collected in each of CITP fractions and subsequently loaded into each of nano-RPLC columns still remained the same as long as there was no over-sampling. By maintaining the same amount of individual peptides introduced and resolved in nano-RPLC-MS, the enhancement in the overall peak capacity further reduced matrix effects by simplifying the complexity of co-eluting peptides prior to MS detection and sequencing. Matrix effects negatively affect the dynamic range and detection sensitivity of MS measurements in three critical ways. First, high abundance peptides compete for ionization and leave less current available for low abundance peptides. Second, the presence of highly abundant ions limits the opportunity of low abundance ions for entering the ion trap and being selected for tandem MS sequencing. Last, the MS detector itself has a finite dynamic range and can be saturated by the presence of higher abundance ions.
In addition to analyzing fraction overlapping, the ratio of new distinct peptides (comparing with distinct peptides collectively determined in all prior fractions) to total peptides identified in each SCX or CITP fraction was also evaluated and summarized in Fig. 5. Extensive fraction overlapping in SCX clearly limits its ability to identify new peptides. Comparisons among 4-, 8-, and 30-fraction SCX runs indicated that the new peptide ratio reached a maximum around the salt concentration of 110–130 mM ammonium formate, corresponding to the elution of mainly triply charged peptides. The association of this maximum new peptide ratio with 3+ charged peptides was also in good agreement with the highest number of peptide identifications achieved from triply charged peptides in the work of Peng and co-workers [44].
Figure 5.
The ratio of new distinct peptides to total peptides found in each SCX or CITP fraction.
In contrast, the CITP-based multidimensional separations delivered high new peptide ratios evenly across all 30 fractions. Transient CITP/CZE separations resolve peptides due to their differences in electrophoretic mobility which is influenced by the charge and size of the peptide ions, and provides better selectivity over charge-based SCX. It is known that capillary electrophoresis, in general, offers superior separation efficiency than that typically achieved in LC, further contributing to enhancement in the overall resolving power.
Assessment of proteome reproducibility
Detection of high confidence proteins was achieved with the reproducibility of greater than 85% within triplicate technical replicates of the same cancer stem cell sample using the CITP-based proteomic platform. Besides protein identification, the CITP technology was coupled with the spectral counting quantification approach [56–58] for determining protein expression profile within GBM derived cancer stem cells. The spectral counts of individual proteins were normalized against the average number of spectral counts across all runs and termed expression values. Several measures, including the coefficient of variation (CV) and the Pearson correlation coefficient, were employed to assess quantitative proteomic data quality and reproducibility.
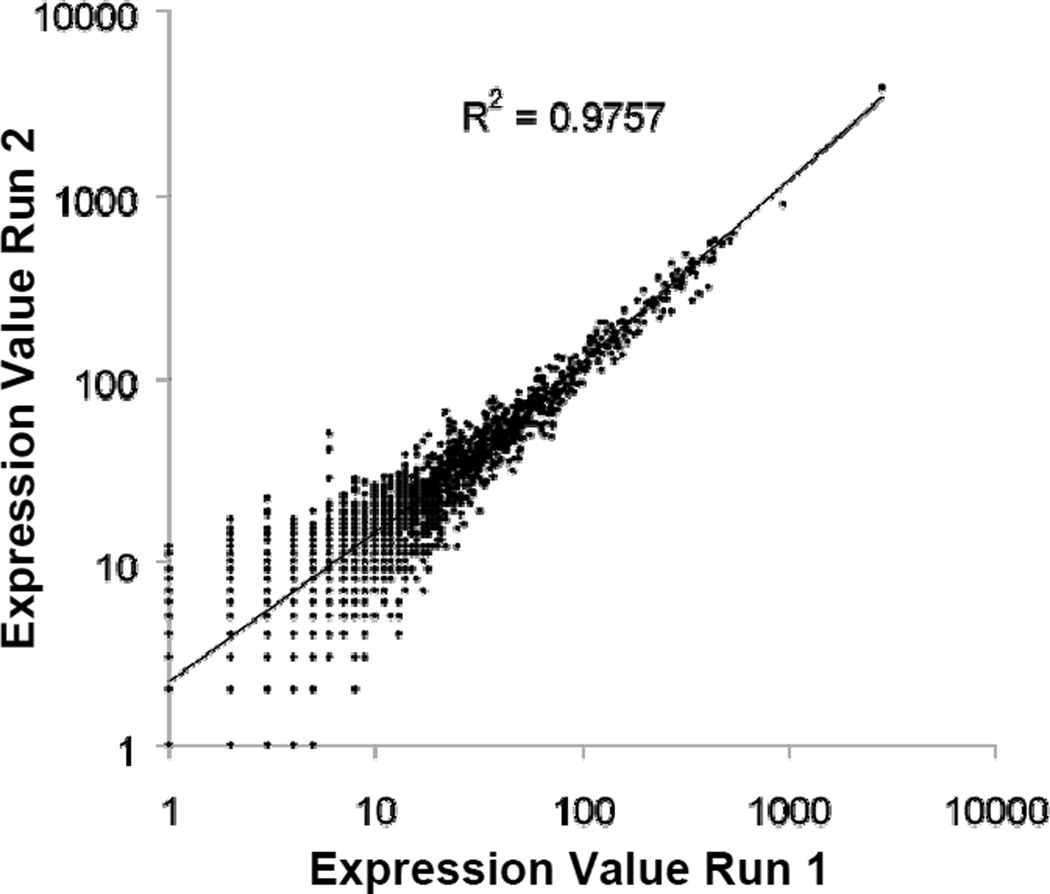
Correlation of two runs of the same tryptic digest of neural stem cells is shown in Fig. 6. All separations were performed on different CITP capillaries and nano-RPLC columns. All proteins with one or more spectral counts were used to calculate the correlation yielding a Pearson R2 value of 0.98 over a dynamic range of greater than 103. This value compares favorably with a recent report of a Pearson R2 correlation of 0.88 among technical replicates using MuDPIT [59]. Furthermore, the CV among CITP measurements was less than 10% for over half of proteins quantified and averaged 15% across all proteins.
Figure 6.
Pearson correlation plot of all proteins quantified in two runs of the same tryptic digest obtained from GBM derived cancer stem cells.
Protein functional subclass and pathway coverage
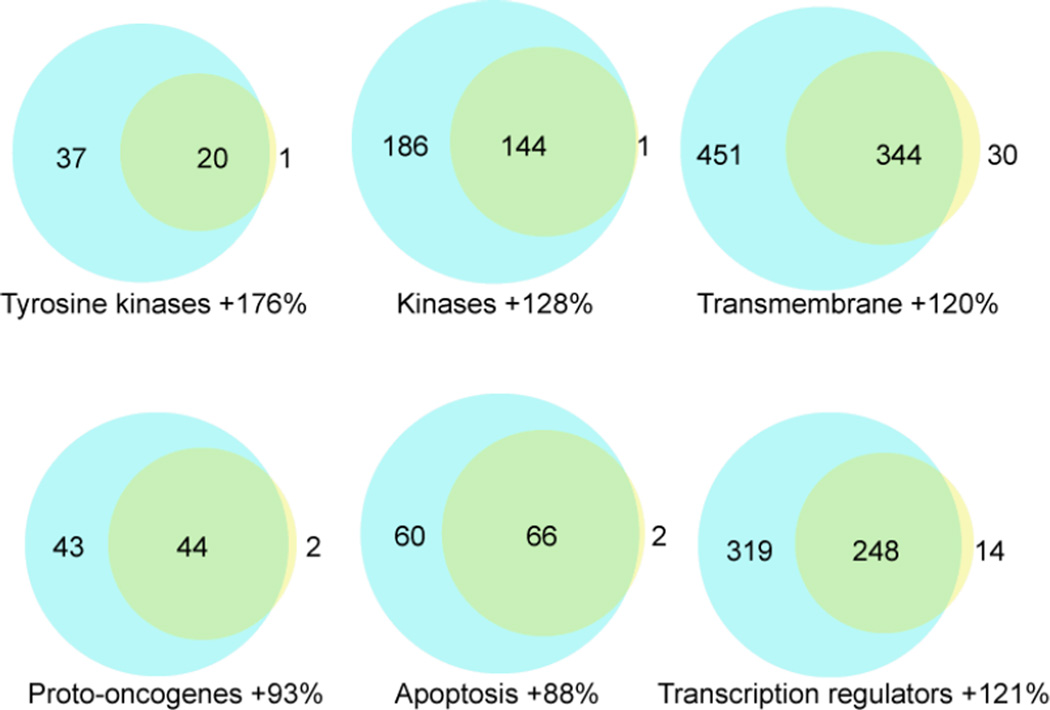
The Venn diagrams shown in Fig. 7 specifically compare the proteome coverage among various functional categories, including tyrosine kinases, kinases, transmembrane, proto-oncogenes, apoptosis, and transcription regulators, achieved using the CITP-based proteomic platform and MuDPIT. The CITP measurements routinely detected about two fold more proteins in any given subclass, even for very low abundance proteins such as tyrosine kinases and proto-oncogenes. Similar to comparative proteome results shown in Fig. 2, very few proteins in these subclasses were uniquely identified by MuDPIT.
Figure 7.
The Venn diagrams comparing proteins identified within different functional categories using MuDPIT (small circle) and CITP/CZE-based multidimensional separations (large circle).
The approach of grouping individual expression changes into pathways of actions is not only useful in trying to interpret genomic and proteomic datasets, but also allows common biological themes to emerge from the comparative proteomic analyses [60]. Comparative proteomics involving measurements in changes of biological pathways or functions are expected to provide relevant therapy-associated biomarkers and networks, and the initial steps and control points in signaling cascades and compensatory processes that drive the progression of therapeutic response and/or drug resistance. From a practical perspective, the evaluation of a comparative proteomic dataset within a biological context is essential for high-throughput data validation, prioritization of follow-on biomarker selection and validation experiments.
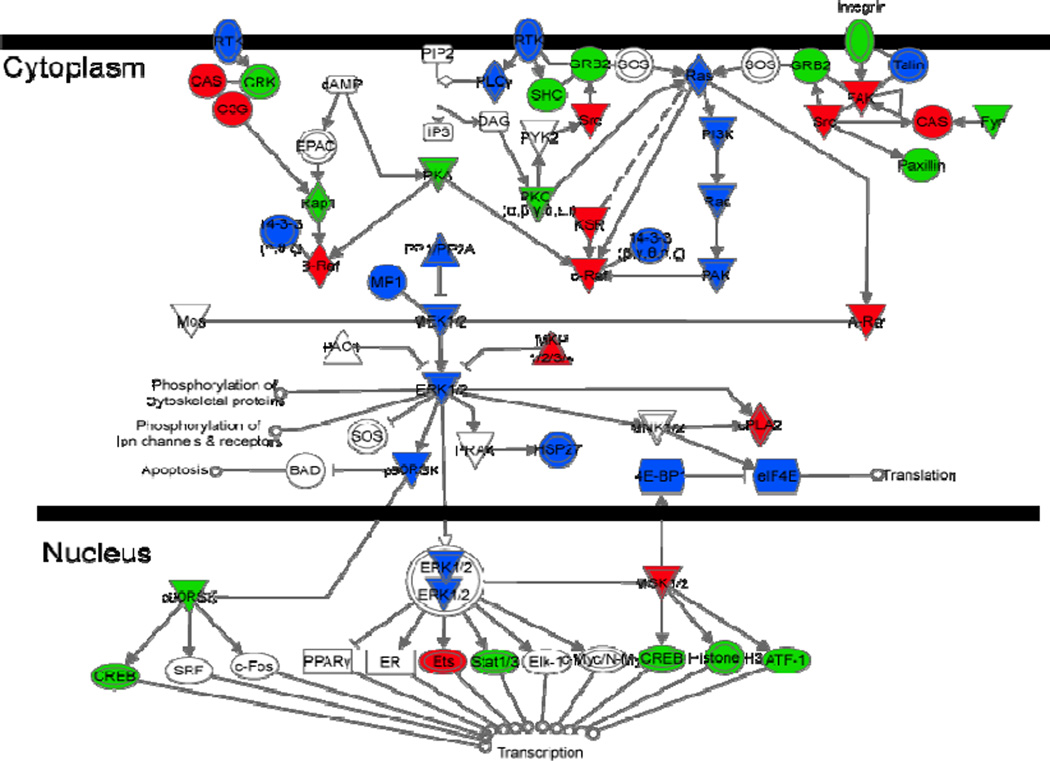
The coverage of key protein networks related to neural cancer stem cells such as the ERK/MAPK pathway (Fig. 8) was analyzed using the Ingenuity System™ and compared among the results achieved by the CITP proteomic platform and MuDPIT. Clearly, combined CITP/nano-RPLC separations, due to its excellent resolving power, accomplished significantly greater coverage in this key pathway than that obtained from the multidimensional liquid chromatography system. Many biologically relevant proteins, including MKP, the Raf family, and Src in the ERK/MAPK pathway, were only identified by the CITP-based shotgun proteomic measurements.
Figure 8.
Comparison of the coverage in the ERK/MAPK pathway achieved by MuDPIT and CITP/CZE-based shotgun proteome techniques. Red: proteins only identified by CITP; blue: proteins identified by CITP with higher confidence (larger numbers of spectral counts and distinct peptide identifications per protein) than that achieved by MuDPIT; green: proteins identified by both CITP and MuDPIT with approximately equal confidence.
CONCLUSION
Since the sizes of human tissue biopsies are becoming significantly smaller due to the advent of minimally-invasive methods and early detection and treatment of lesions, a more effective discovery-based proteomic technology is critically needed to enable comprehensive and sensitive studies of protein profiles that will have diagnostic and therapeutic relevance. By employing small cancer stem cell populations isolated from GBM tissues and enriched in culture, comparisons among shotgun proteome techniques, including transient CITP/CZE-based multidimensional separations and MuDPIT, were performed and reported in this work. Comparisons were conducted using a single processed protein digest with equal sample loading, identical second dimension separation and MS conditions, and consistent search parameters and cutoff established by the target-decoy determined FDR.
For the evaluation of overall proteome performance, a CITP/CZE measurement demonstrated significant enhancements in total peptide, distinct peptide, and distinct protein identifications over a corresponding SCX run by 119%, 192%, and 79%, respectively. On the basis of at least two or more distinct peptides leading to each protein identification, the Venn diagram indicated the complete inclusion of proteins identified by MuDPIT within the CITP-based proteomic results. CITP further outperformed MuDPIT with respect to the average numbers of distinct peptide identifications and spectral counts per protein which lead to improvements in the sequence coverage and the confidence of protein quantification.
As evaluated by the measurements of CV and the Pearson correlation coefficient, the CITP-based proteome platform coupled with the spectral counting approach was demonstrated to be highly sensitive and reproducible in determining protein relative abundance in complex tissue proteome. In contrast, extensive fraction overlapping in SCX greatly limited the ability of MuDPIT for mining deeper into the tissue proteome. As evidenced by the coverage among various protein functional categories, CITP routinely detected about two fold more proteins in any given subclass, even for very low abundance proteins such as tyrosine kinases and proto-oncogenes. Combined CITP/nano-RPLC separations, due to its excellent resolving power, further accomplished superior coverage in key pathways than that of MuDPIT. Many biologically relevant proteins, including MKP, the Raf family, and Src in the ERK/MAPK pathway, were only identified by the CITP-based shotgun proteomic technique.
ACKNOWLEDGEMENT
This work was supported by NIH grants RR022667 to WW, GM073723 to CSL, and NS055419 to BMB.
REFERENCES
- 1.Rifai N, Gillette MA, Carr SA. Nat Biotechnol. 2006;24:971–983. doi: 10.1038/nbt1235. [DOI] [PubMed] [Google Scholar]
- 2.Cottingham K. J Proteome Res. 2007;6:2052–2052. doi: 10.1021/pr070753l. [DOI] [PubMed] [Google Scholar]
- 3.Li N, Zhang J, Liang Y, Shao J, Peng F, Sun M, Xu N, Li X, Wang R, Liu S, Lu Y. J Proteome Res. 2007;6:3304–3312. doi: 10.1021/pr0702363. [DOI] [PubMed] [Google Scholar]
- 4.Emmert-Buck MR, Bonner RF, Smith PD, Chuaqui RF, Zhuang Z, Goldstein SR, Weiss RA, Liotta LA. Science. 1996;274:998–991001. doi: 10.1126/science.274.5289.998. [DOI] [PubMed] [Google Scholar]
- 5.Bonner RF, Emmert-Buck M, Cole K, Pohida T, Chuaqui R, Goldstein S, Liotta LA. Science. 1997;278:1481–1481. doi: 10.1126/science.278.5342.1481. [DOI] [PubMed] [Google Scholar]
- 6.Ruepp SU, Tonge RP, Shaw J, Wallis N, Pognan F. Toxicol Sci. 2002;65:135–150. doi: 10.1093/toxsci/65.1.135. [DOI] [PubMed] [Google Scholar]
- 7.De Souza AI, McGregor E, Dunn MJ, Rose ML. Proteomics. 2004;4:578–586. doi: 10.1002/pmic.200300660. [DOI] [PubMed] [Google Scholar]
- 8.Banks RE, Dunn MJ, Forbes MA, Stanley A, Pappin D, Naven T, Gough M, Harnden P, Selby PJ. Electrophoresis. 1999;20:689–700. doi: 10.1002/(SICI)1522-2683(19990101)20:4/5<689::AID-ELPS689>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- 9.Ornstein DK, Gillespie JW, Paweletz CP, Duray PH, Herring J, Vocke CD, Topalian SL, Bostwick DG, Linehan WM, Petricoin EF, Emmert-Buck MR. Electrophoresis. 2000;21:2235–2242. doi: 10.1002/1522-2683(20000601)21:11<2235::AID-ELPS2235>3.0.CO;2-A. [DOI] [PubMed] [Google Scholar]
- 10.Lawrie LC, Curran S, McLeod HL, Fothergill JE, Murray GI. Mol Pathol. 2001;54:253–258. doi: 10.1136/mp.54.4.253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Craven RA, Totty N, Harnden P, Selby PJ, Banks RE. Am J Pathol. 2002;160:815–822. doi: 10.1016/S0002-9440(10)64904-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Somiari RI, Sullivan A, Russell S, Somiari S, Hu H, Jordan R, George A, Katenhusen R, Buchowiecka A, Arciero C, Brzeski H, Hooke J, Shriver C. Proteomics. 2003;3:1863–1873. doi: 10.1002/pmic.200300560. [DOI] [PubMed] [Google Scholar]
- 13.Heijne WHM, Stierum RH, Slijper M, van Bladeren PJ, van Ommen B. Biochem Pharmacol. 2003;65:857–875. doi: 10.1016/s0006-2952(02)01613-1. [DOI] [PubMed] [Google Scholar]
- 14.Somiari RI, Somiari S, Russell S, Shriver CD. J Chromatogr B Analyt Technol Biomed Life Sci. 2005;815:215–225. doi: 10.1016/j.jchromb.2004.11.012. [DOI] [PubMed] [Google Scholar]
- 15.Zhuang Z, Lee YS, Zeng W, Furuta M, Valyi-Nagy T, Johnson MD, Vnencak-Jones CL, Woltjer RL, Weil RJ. Neurology. 2004;62:2316–2319. doi: 10.1212/wnl.62.12.2316. [DOI] [PubMed] [Google Scholar]
- 16.Furuta M, Weil RJ, Vortmeyer AO, Huang S, Lei J, Huang T-N, Lee Y-S, Bhowmick DA, Lubensky IA, Oldfield EH, Zhuang Z. Oncogene. 2004;23:6806–6814. doi: 10.1038/sj.onc.1207770. [DOI] [PubMed] [Google Scholar]
- 17.Vogel TW, Zhuang Z, Li J, Okamoto H, Furuta M, Lee Y-S, Zeng W, Oldfield EH, Vortmeyer AO, Weil RJ. Clin Cancer Res. 2005;11:3624–3632. doi: 10.1158/1078-0432.CCR-04-2115. [DOI] [PubMed] [Google Scholar]
- 18.Li J, Zhuang Z, Okamoto H, Vortmeyer AO, Park DM, Furuta M, Lee YS, Oldfield EH, Zeng W, Weil RJ. Neurology. 2006;66:733–736. doi: 10.1212/01.wnl.0000201270.90502.d0. [DOI] [PubMed] [Google Scholar]
- 19.Zhuang Z, Huang S, Kowalak JA, Shi Y, Lei J, Furuta M, Lee Y-S, Lubensky IA, Rodgers GP, Cornelius AS, Weil RJ, Teh BT, Vortmeyer AO. Int J Oncol. 2006;28:103–110. [PubMed] [Google Scholar]
- 20.Li C, Hong Y, Tan Y-X, Zhou H, Ai J-H, Li S-J, Zhang L, Xia Q-C, Wu J-R, Wang H-Y, Zeng R. Mol Cell Proteomics. 2004;3:399–409. doi: 10.1074/mcp.M300133-MCP200. [DOI] [PubMed] [Google Scholar]
- 21.Zhang J, Hu H, Gao M, Yang P, Zhang X. Electrophoresis. 2004;25:2374–2383. doi: 10.1002/elps.200405956. [DOI] [PubMed] [Google Scholar]
- 22.DeSouza L, Diehl G, Rodrigues MJ, Guo J, Romaschin AD, Colgan TJ, Siu KWM. J Proteome Res. 2005;4:377–386. doi: 10.1021/pr049821j. [DOI] [PubMed] [Google Scholar]
- 23.Cagney G, Park S, Chung C, Tong B, O'Dushlaine C, Shields DC, Emili A. J Proteome Res. 2005;4:1757–1767. doi: 10.1021/pr0500354. [DOI] [PubMed] [Google Scholar]
- 24.Wu S-L, Hancock WS, Goodrich GG, Kunitake ST. Proteomics. 2003;3:1037–1046. doi: 10.1002/pmic.200300382. [DOI] [PubMed] [Google Scholar]
- 25.Zang L, Palmer Toy D, Hancock WS, Sgroi DC, Karger BL. J Proteome Res. 2004;3:604–612. doi: 10.1021/pr034131l. [DOI] [PubMed] [Google Scholar]
- 26.Baker H, Patel V, Molinolo AA, Shillitoe EJ, Ensley JF, Yoo GH, Meneses-Garcia A, Myers JN, El-Naggar AK, Gutkind JS, Hancock WS. Oral Oncol. 2005;41:183–199. doi: 10.1016/j.oraloncology.2004.08.009. [DOI] [PubMed] [Google Scholar]
- 27.Hood BL, Darfler MM, Guiel TG, Furusato B, Lucas DA, Ringeisen BR, Sesterhenn IA, Conrads TP, Veenstra TD, Krizman DB. Mol Cell Proteomics. 2005;4:1741–1753. doi: 10.1074/mcp.M500102-MCP200. [DOI] [PubMed] [Google Scholar]
- 28.Crockett DK, Lin Z, Vaughn CP, Lim MS, Elenitoba-Johnson KSJ. Lab Invest. 2005;85:1405–1415. doi: 10.1038/labinvest.3700343. [DOI] [PubMed] [Google Scholar]
- 29.Palmer-Toy DE, Krastins B, Sarracino DA, Nadol JB, Merchant SN. J Proteome Res. 2005;4:2404–2411. doi: 10.1021/pr050208p. [DOI] [PubMed] [Google Scholar]
- 30.Rahimi F, Shepherd CE, Halliday GM, Geczy CL, Raftery MJ. Anal Chem. 2006;78:7216–7221. doi: 10.1021/ac060294s. [DOI] [PubMed] [Google Scholar]
- 31.Hwang SI, Thumar J, Lundgren DH, Rezaul K, Mayya V, Wu L, Eng J, Wright ME, Han DK. Oncogene. 2007;26:65–76. doi: 10.1038/sj.onc.1209755. [DOI] [PubMed] [Google Scholar]
- 32.Bagnato C, Thumar J, Mayya V, Hwang S-I, Zebroski H, Claffey KP, Haudenschild C, Eng JK, Lundgren DH, Han DK. Mol Cell Proteomics. 2007;6:1088–1102. doi: 10.1074/mcp.M600259-MCP200. [DOI] [PubMed] [Google Scholar]
- 33.Wang Y, Rudnick PA, Evans EL, Li J, Zhuang Z, Devoe DL, Lee CS, Balgley BM. Anal Chem. 2005;77:6549–6556. doi: 10.1021/ac050491b. [DOI] [PubMed] [Google Scholar]
- 34.Wang W, Guo T, Rudnick PA, Song T, Li J, Zhuang Z, Zheng W, Devoe DL, Lee CS, Balgley BM. Anal Chem. 2007;79:1002–1009. doi: 10.1021/ac061613i. [DOI] [PubMed] [Google Scholar]
- 35.Balgley BM, Wang W, Devoe DL, Lee CS. Personalized Medicine. 2007;4:45–58. doi: 10.2217/17410541.4.1.45. [DOI] [PubMed] [Google Scholar]
- 36.Shi S-R, Liu C, Balgley BM, Lee C, Taylor CR. J Histochem Cytochem. 2006;54:739–743. doi: 10.1369/jhc.5B6851.2006. [DOI] [PubMed] [Google Scholar]
- 37.Guo T, Wang W, Rudnick PA, Song T, Li J, Zhuang Z, Weil RJ, DeVoe DL, Lee CS, Balgley BM. J Histochem Cytochem. 2007;55:763–772. doi: 10.1369/jhc.7A7177.2007. [DOI] [PubMed] [Google Scholar]
- 38.Xu H, Yang L, Wang W, Shi S-R, Liu C, Liu Y, Fang X, Taylor CR, Lee CS, Balgley BM. J Proteome Res. 2008;7:1098–1108. doi: 10.1021/pr7006768. [DOI] [PubMed] [Google Scholar]
- 39.An Y, Cooper JW, Balgley BM, Lee CS. Electrophoresis. 2006;27:3599–3608. doi: 10.1002/elps.200600093. [DOI] [PubMed] [Google Scholar]
- 40.Fang X, Yang L, Wang W, Song T, Lee C, Devoe D, Balgley B. Anal Chem. 2007;79:5785–5792. doi: 10.1021/ac070611a. [DOI] [PubMed] [Google Scholar]
- 41.Fang X, Wang W, Yang L, Chandrasekaran K, Kristian T, Balgley BM, Lee CS. Electrophoresis. 2008;29:2215–2223. doi: 10.1002/elps.200700609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wolters DA, Washburn MP, Yates JR. Anal Chem. 2001;73:5683–5690. doi: 10.1021/ac010617e. [DOI] [PubMed] [Google Scholar]
- 43.Washburn MP, Wolters D, Yates JR. Nat Biotechnol. 2001;19:242–247. doi: 10.1038/85686. [DOI] [PubMed] [Google Scholar]
- 44.Peng J, Elias JE, Thoreen CC, Licklider LJ, Gygi SP. J Proteome Res. 2003;2:43–50. doi: 10.1021/pr025556v. [DOI] [PubMed] [Google Scholar]
- 45.Park DM, Li J, Okamoto H, Akeju O, Kim SH, Lubensky I, Vortmeyer A, Dambrosia J, Weil RJ, Oldfield EH, Park JK, Zhuang Z. Cell Cycle. 2007;6:467–470. doi: 10.4161/cc.6.4.3856. [DOI] [PubMed] [Google Scholar]
- 46.Tang W, Harrata AK, Lee CS. Anal Chem. 1997;69:3177–3182. doi: 10.1021/ac970015o. [DOI] [PubMed] [Google Scholar]
- 47.Yang L, Lee CS, Hofstadler SA, Smith RD. Anal Chem. 1998;70:4945–4950. doi: 10.1021/ac980223w. [DOI] [PubMed] [Google Scholar]
- 48.Liu T, Qian W-J, Gritsenko MA, Xiao W, Moldawer LL, Kaushal A, Monroe ME, Varnum SM, Moore RJ, Purvine SO, Maier RV, Davis RW, Tompkins RG, Camp DG, Smith RD. Mol Cell Proteomics. 2006;5:1899–1913. doi: 10.1074/mcp.M600068-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Geer LY, Markey SP, Kowalak JA, Wagner L, Xu M, Maynard DM, Yang X, Shi W, Bryant SH. J Proteome Res. 2004;3:958–964. doi: 10.1021/pr0499491. [DOI] [PubMed] [Google Scholar]
- 50.Elias JE, Haas W, Faherty BK, Gygi SP. Nat Methods. 2005;2:667–675. doi: 10.1038/nmeth785. [DOI] [PubMed] [Google Scholar]
- 51.Balgley BM, Laudeman T, Yang L, Song T, Lee CS. Mol Cell Proteomics. 2007;6:1599–1608. doi: 10.1074/mcp.M600469-MCP200. [DOI] [PubMed] [Google Scholar]
- 52.Elias JE, Gygi SP. Nat Methods. 2007;4:207–214. doi: 10.1038/nmeth1019. [DOI] [PubMed] [Google Scholar]
- 53.Bao S, Wu Q, McLendon RE, Hao Y, Shi Q, Hjelmeland AB, Dewhirst MW, Bigner DD, Rich JN. Nature. 2006;444:756–760. doi: 10.1038/nature05236. [DOI] [PubMed] [Google Scholar]
- 54.Piccirillo SGM, Reynolds BA, Zanetti N, Lamorte G, Binda E, Broggi G, Brem H, Olivi A, Dimeco F, Vescovi AL. Nature. 2006;444:761–765. doi: 10.1038/nature05349. [DOI] [PubMed] [Google Scholar]
- 55.MacCoss MJ, Wu CC, Yates JR. Anal Chem. 2002;74:5593–5599. doi: 10.1021/ac025826t. [DOI] [PubMed] [Google Scholar]
- 56.Liu H, Sadygov RG, Yates JR. Anal Chem. 2004;76:4193–4201. doi: 10.1021/ac0498563. [DOI] [PubMed] [Google Scholar]
- 57.Rappsilber J, Ryder U, Lamond AI, Mann M. Genome Res. 2002;12:1231–1245. doi: 10.1101/gr.473902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ishihama Y, Oda Y, Tabata T, Sato T, Nagasu T, Rappsilber J, Mann M. Mol Cell Proteomics. 2005;4:1265–1272. doi: 10.1074/mcp.M500061-MCP200. [DOI] [PubMed] [Google Scholar]
- 59.Lu P, Vogel C, Wang R, Yao X, Marcotte EM. Nat Biotechnol. 2007;25:117–124. doi: 10.1038/nbt1270. [DOI] [PubMed] [Google Scholar]
- 60.Lin B, White JT, Lu W, Xie T, Utleg AG, Yan X, Yi EC, Shannon P, Khrebtukova I, Lange PH, Goodlett DR, Zhou D, Vasicek TJ, Hood L. Cancer Res. 2005;65:3081–3091. doi: 10.1158/0008-5472.CAN-04-3218. [DOI] [PubMed] [Google Scholar]