Abstract
The bottom-up spatial organization of potential nanoelectronic components is a key intermediate step in the development of molecular electronics. We describe robust 3-space-spanning DNA motifs that are used to organize nanoparticles in 2D. One strand of the motif ends in a gold nanoparticle; only one DNA strand is attached to the particle. By using two of the directions of the motif to produce a two dimensional crystalline array, one direction is free to bind gold nanoparticles. Identical motifs, tailed in different sticky ends enable the 2D periodic ordering of 5 nm and 10 nm diameter gold nanoparticles.
Keywords: DNA self-assembly, 2D DNA arrays, Organizing Matter with DNA, Atomic Force Microscopy, Metallic Nanoparticles, Robust DNA Motifs
Metallic and semiconductor nanoparticles exhibit quantized optical and electronic properties that might be exploited in the design of future nanoelectronic devices.1–3 However, this application requires the deliberate and precise organization of nanoparticles into specific designed structural arrangements. The control of the structure of matter on the finest possible scale entails the successful design of both stiff intramolecular motifs and robust intermolecular interactions. The specificity of DNA base-pairing has provided a 'smart-glue' approach to programming interactions between particles via hybridization of specifically designed linker strands.4,5 Previously, stiff motifs6 based on branched DNA have been used to produce DNA structures with a variety of patterns that are visible in the AFM; these include stripes from double crossover (DX) molecules,7 arrays with tunable cavities from DNA parallelograms,8 and honeycombs from DX triangles.9 DNA-functionalized 1.4 nm gold nanoparticles have been assembled into linear arrays forming parallel stripes on a 2D DNA striped scaffolding by self-assembly during scaffolding formation10 and 6 nm gold nanoparticles with multiple DNA attachments have been fashioned into similar arrays by in situ hybridization to a pre-assembled scaffolding on a striped DX surface.11 Sequence-encoded in situ assembly of 5 nm and 10 nm gold particles in alternating stripes has also been achieved.12 While such linear nanoparticle arrays are of interest for some applications, other periodic arrangements also offer significant potential. Furthermore, control over nanoparticle positions more precise than that afforded by polyvalent functionalization is highly desirable. Yan and his colleagues have used stiff motifs recently to organize individual nanoparticles in 1D and 2D arrays.13,14
Our experience with honeycomb lattices demonstrates that cohesion by two sticky ends on each end of a DX molecule is more robust than a single sticky end; we were unable to obtain the honeycomb arrays if only a single sticky end was used.9 We have built several motifs that span 3-space (e.g., 6-helix bundles15); one of these motifs (termed a 3D-DX triangle) is based on Mao et al.'s tensegrity triangle,16 but contains DX molecules, instead of single helices in each of its three domains (Fig. 1). It is possible to produce 2D lattices with this motif if only two of the linearly independent directions contain cohesive ends. This leaves a third direction not involved in lattice formation, and its blunt end can be used as a site to include a gold nanoparticle. Specificity is increased by using a nanoparticle that contains only a single DNA strand; this strand is one of those that form the motif and its 5' end is on one of the blunt ends. Thus, we have developed a system that combines all of the robustness features of which we are aware, single-stranded attachment of the nanoparticle, DX-cohesion and a 3-space-spanning triangular motif based on stiff DX molecules. To demonstrate the organizational power of this system, we have attached 5 nm nm nanoparticles to one or both of two different 3D-DX triangles. In addition, we have used this system to produce a well-positioned alternating 2D array of 5 nm and 10 nm gold nanoparticles.
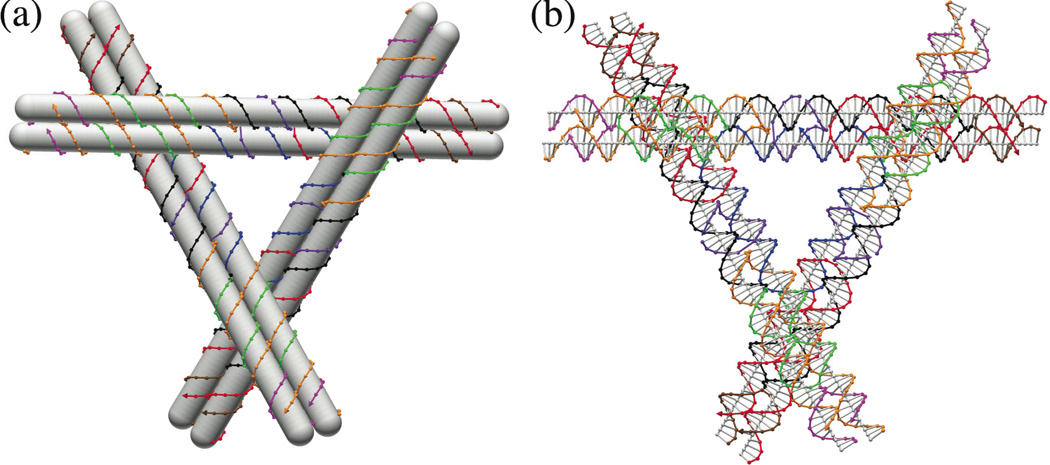
Figure 1. The DNA Motif Used to Build the Arrays.
(a) A Schematic of the Motif. In this schematic diagram, double helices are shown as opaque rods around which the individual strands are wrapped. Note the 3-space-spanning character of the three DX domains. (b) A Detailed Molecular Structure. Each nucleotide is shown in a representation that illustrates a color-coded backbone virtual atom connected to its neighbors along the helix. A gray line representing the base is drawn from this atom to the helix axis, which is also drawn. Both diagrams drawn by the program Gideon.18.
Two 3D-DX triangles were designed to produce a rhombic lattice arrangement when combined (Figure 2a). The sequences of the triangles are shown in the Supporting Information (Figure S1). The edges of triangles contain 84 nucleotide pairs (8 turns of 10.5-fold DNA, ~27.2 nm) in each of their helices, and two of the directions terminate in a pair of 5’ sticky ends, four nucleotides in length. The particles are attached to the triangles following two-step electrophoretic isolation processes. In the first step, thiolated single-stranded DNA (ssDNA) is reacted directly with 5 nm or 10 nm Au nanoparticles. Discrete bands of low mobility that appear in the same lane (Figure S2) on an agarose gel correspond to a defined number of strands per particle.4 The band corresponding to nanoparticles bearing one ssDNA was isolated from the other bands, and recovered as described elsewhere.4 The highly purified DNA/Au conjugates were subsequently added to the solution containing all other component strands, to form the 3D-DX triangle. The gel (Figure S3) shows that the 3D-DX triangle/Au complex appears as a band with lower mobility. Following a similar isolation procedure, the collected 3D-DX triangle-Au conjugates were mixed with the complementary 3D-DX triangles, to form a 2-triangle array.
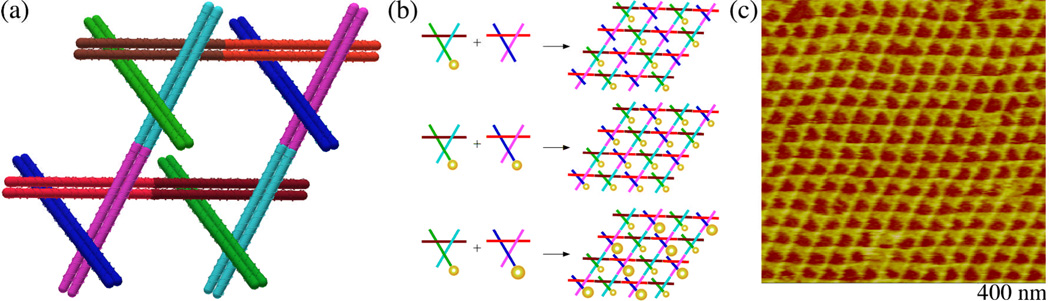
Figure 2. The 2D Arrays Assembled Here.
(a) A Schematic Showing the Formation of a Two-Component Array. Four triangles of two species in which each domain has been color coded are shown connected. The view parallel to the three-fold axes shows how only two domains (cyan bonding to magenta and brown bonding to red) are involved in array formation, while the end of the third domain (blue or green) is free to be involved in scaffolding operations. (b) Diagrams Showing the Attachment of Nanoparticles. The color-coding is the same as in (a). This view is perpendicular to the rhombic surfaces of the 2D array. Its three panels show, top to bottom, 5 nm particles attached to only one of the two triangular tiles, 5 nm particles attached to both of the tiles, and 5 nm particles attached to one of the tiles and 10 nm particles attached to the other tile. (c) A Tapping-Mode Atomic Force Micrograph of an Underivatized Array. Note that despite a pseudo-trigonal appearance, the features in the lighter portion of the array are all parallel to each other, and there is no threefold axis at the centers of the triangles, nor at their vertices. There is a prominent high feature in the lower-left-to-upper-right direction that we interpret as the domain not involved in lattice formation.
Figure 2b contains schematics perpendicular to the 2D array, with the derivatized DX domain indicated and shown foreshortened. Figure 2c illustrates an AFM image of an array that has not been derivatized. Although pseudo-trigonal in aspect, the parallelogram-like structure of the designed arrangements is evident from close inspection of the image. The triangle motifs can be seen to associate with each another to form the 2D lattice, with a periodic repeat of 27.4 nm (estimated from the autocorrelation function).
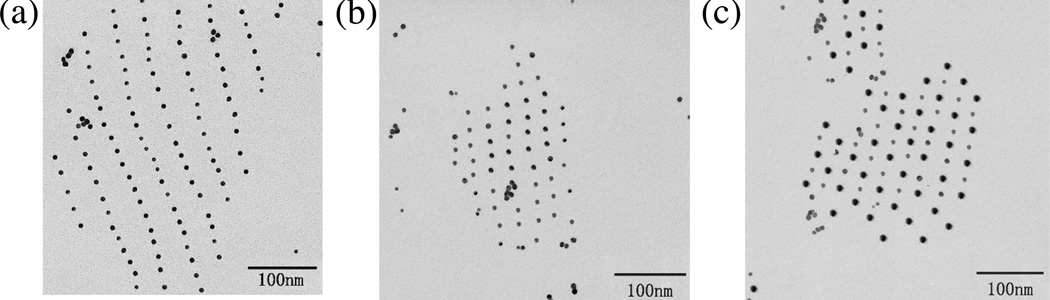
Figure 3 shows TEM images of the designs in Figure 2b. Panel (a) illustrates an A-B array in which the A-triangles contain 5 nm gold particles and the B-triangles do not. The spacing is twice as large in one direction as in the other. In panel (b), both triangles contain 5 nm gold particles, and the spacings are the same (the short direction of (a) in both cases. In panel (c), the B-triangles contain 10 nm gold particles, and the A-triangles contain 5-nm gold particles. The alternation in size with two-dimensional regularity is evident in this image. The Au nanoparticles are evenly spaced, separated on average by 27.6±0.5 nm in (b) and (c), and by 25.4±0.7 in the short direction while 54.9±0.9 nm in the long direction of (a); these values are in good agreement with the expected values of 27.2 and 54.4 nm, respectively.
Figure 3. Transmission Electron Micrographs of 2D Arrays of Organized Gold Nanoparticles.
(a) An array where one tile contains 5 nm particles. It is clear that this arrangement results in one short distance and one long distance. Sometimes a particle is missing. (b) An array where both tiles contain 5 nm particles. The distances between particles are seen to be equal here. (c) An array where one tile contains a 5 nm particle and the other tile contains a 10 nm particle. The alternation of 5 nm particles and 10 nm particles is evident from this image. Note that the spacings are precise in both directions, and that the pattern mimics the rhombic pattern of the tile array.
It is evident from these results that the combination of robust motifs, robust cohesion and specific attachment of particles makes it possible to incorporate different metallic nanoparticles into a highly precise 2D periodic pattern. These arrays are somewhat smaller than those normally obtained with this motif, which we attribute to the presence of the nanoparticles. It is clear that more complex periodic patterns could also be generated in 2D by using more tile species. In a similar fashion, 2D algorithmic assembly17 might be used to produce aperiodic patterns of nanoparticles. It is likely that the methods described here could be used to produce ordered 2D arrays of species smaller than those used here, such as biological macromolecules or organic molecules.
Supplementary Material
ACKNOWLEDGMENTS
This research has been supported by grants GM-29554 from NIGMS, grants, EIA-0086015, CCF-0432009, CCF-0523290 and CTS-0103002 from the NSF, 48681-EL from ARO and NTI-001 from Nanoscience Technologies, Inc. to N.C.S., DMI-0210844 from the NSF to RAK and a DARPA contract to APA.
Footnotes
Supporting Information Available: The sequences of the molecules used and experimental methods. This material is available free of charge via the Internet at http://pubs.acs.org/.
Contributor Information
Jiwen Zheng, Email: jz24@nyu.edu.
Pamela E. Constantinou, Email: pec214@nyu.edu.
Christine Micheel, Email: cmicheel@uclink4.berkeley.edu.
A. Paul Alivisatos, Email: alivis@uclink4.berkeley.edu.
Richard A. Kiehl, Email: kiehl@ece.umn.edu.
Nadrian C. Seeman, Email: ned.seeman@nyu.edu.
REFERENCES
- 1.(a) Alivisatos AP. J. Phys. Chem. 1996;100:13226–13239. [Google Scholar]; (b) Redl FX, Cho KS, Murray CB, O'Brien S. Nature. 2003;423:978–971. doi: 10.1038/nature01702. [DOI] [PubMed] [Google Scholar]
- 2.Kiehl RA. J. Nanopart. Res. 2000;2:331–332. [Google Scholar]
- 3.(a) Likharev KK. Electronics Below 10 nm. In: Greer J, Korkin A, Labanowski J, editors. Nano and Giga Challenges in Microelectronics. Amsterdam: Elsevier; 2003. pp. 27–68. [Google Scholar]; (b) Maier SA, Brongersma ML, Kik PG, Meltzer S, Requicha AAG, Atwater HA. Adv. Mater. 2001;13:1501–1505. [Google Scholar]; (c) Shipway AN, Katz E, Willner I. ChemPhysChem. 2000;1:18–52. doi: 10.1002/1439-7641(20000804)1:1<18::AID-CPHC18>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- 4.(a) Alivisatos A, Johnsson K, Peng X, Wilson T, Lowth C, Bruchez M, Schultz P. Nature. 1996;382:609–611. doi: 10.1038/382609a0. [DOI] [PubMed] [Google Scholar]; (b) Loweth C, Caldwell W, Peng X, Alivisatos P, Schultz P. Angew. Chem. Int. Ed. 1999;38:1808–1812. doi: 10.1002/(SICI)1521-3773(19990614)38:12<1808::AID-ANIE1808>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]; (c) Zanchet D, Micheel C, Parak W, Alivisatos P. Nano Lett. 2001;1:32–35. [Google Scholar]; (d) Zanchet D, Micheel C, Parak W, Alivisatos P. J. Phys. Chem. B. 2002;106:11758–11763. [Google Scholar]
- 5.(a) Mucic R, Storhoff J, Letsinger R, Mirkin C. Nature. 1996;382:607–609. doi: 10.1038/382607a0. [DOI] [PubMed] [Google Scholar]; (b) Storhoff J, Elghenian R, Mucic R, Mirkin C. J. Am. Chem. Soc. 1998;120:1959–1964. [Google Scholar]; (c) Jin R, Wu G, Li Z, Mirkin C, Schatz G. J. Am. Chem. Soc. 2003;125:1643–1654. doi: 10.1021/ja021096v. [DOI] [PubMed] [Google Scholar]; (d) Anderson CJ, Sykes TJ, Kornberg RD. Proc. Nat. Acad. Sci. (USA) 2005;102:13383–13385. doi: 10.1073/pnas.0506290102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.(a) Li X, Yang X, Qi J, Seeman NC. J. Am. Chem. Soc. 1996;118:6131–6140. [Google Scholar]; (b) Sa-Ardyen P, Vologodskii AV, Seeman NC. Biophys. J. 2003;84:3829–3837. doi: 10.1016/S0006-3495(03)75110-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Winfree E, Liu F, Wenzler LA, Seeman NC. Nature. 1998;394:539–544. doi: 10.1038/28998. [DOI] [PubMed] [Google Scholar]
- 8.Mao C, Sun W, Seeman NC. J. Am. Chem. Soc. 1999;121:5437–5443. [Google Scholar]
- 9.Ding B, Sha R, Seeman NC. J. Am. Chem. Soc. 2004;126:10230–10231. doi: 10.1021/ja047486u. [DOI] [PubMed] [Google Scholar]
- 10.Xiao S, Liu F, Rosen A, Hainfeld J, Seeman N, Kiehl R. J. Nanoparticle Resear. 2002;4:313–317. [Google Scholar]
- 11.Le JD, Pinto Y, Seeman NC, Musier-Forsyth K, Taton TA, Kiehl RA. Nano Lett. 2004;4:2343–2347. doi: 10.1021/nl0515495. [DOI] [PubMed] [Google Scholar]
- 12.Pinto YY, Le JD, Seeman NC, Musier-Forsyth K, Taton TA, Kiehl RA. Nano Lett. 2005;5:2399–2402. doi: 10.1021/nl0515495. [DOI] [PubMed] [Google Scholar]
- 13.Zhang J, Liu Y, Ke Y, Yan H. NanoLett. 2006;6:248–251. doi: 10.1021/nl052210l. [DOI] [PubMed] [Google Scholar]
- 14.Sharma J, Chhabra R, Liu Y, Ke Y, Yan H. Angew. Chem. Int. Ed. 2006;45:730–735. doi: 10.1002/anie.200503208. [DOI] [PubMed] [Google Scholar]
- 15.Mathieu F, Liao S, Mao C, Kopatsch J, Wang T, Seeman NC. NanoLett. 2005;5:661–665. doi: 10.1021/nl050084f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu D, Wang M, Deng Z, Walulu R, Mao C. J. Am Chem. Soc. 2004;126:2324–2325. doi: 10.1021/ja031754r. [DOI] [PubMed] [Google Scholar]
- 17.Winfree E. On the computational power of DNA annealing and ligation. In DNA Based Computing ed. EJ Lipton, EB Baum. 199–219. Providence: Am. Math. Soc. 1996:219. [Google Scholar]
- 18.Birac JJ, Sherman WB, Kopatsch J, Constantinou PE, Seeman NC. J. Mol. Graphics & Modeling. 2006 doi: 10.1016/j.jmgm.2006.03.005. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.