Abstract
In voltage-sensitive phosphatases (VSPs), a transmembrane voltage sensor domain (VSD) controls an intracellular phosphoinositide phosphatase domain, thereby enabling immediate initiation of intracellular signals by membrane depolarization. The existence of such a mechanism in mammals has remained elusive, despite the presence of VSP-homologous proteins in mammalian cells, in particular in sperm precursor cells. Here we demonstrate activation of a human VSP (hVSP1/TPIP) by an intramolecular switch. By engineering a chimeric hVSP1 with enhanced plasma membrane targeting containing the VSD of a prototypic invertebrate VSP, we show that hVSP1 is a phosphoinositide-5-phosphatase whose predominant substrate is PI(4,5)P2. In the chimera, enzymatic activity is controlled by membrane potential via hVSP1’s endogenous phosphoinositide binding motif. These findings suggest that the endogenous VSD of hVSP1 is a control module that initiates signaling through the phosphatase domain and indicate a role for VSP-mediated phosphoinositide signaling in mammals.
Keywords: phosphoinositides, voltage sensor, electrochemical coupling, ion channel, lipid signaling
The recent discovery of voltage-sensitive phosphatases (VSPs) (1) established a novel molecular principle of electrochemical coupling: VSPs directly mediate the degradation of phosphoinositides in response to depolarization of the membrane potential (2, 3). These so-far-unique electro-enzymes consist of a transmembrane voltage sensor domain (VSD) homologous to the voltage sensors of voltage-gated ion channels and an intracellular C-terminal catalytic domain (CD) with high similarity to the tumor suppressor lipid phosphatase, PTEN (Fig. 1A) (1). The enzymatic activity of the CD is controlled by the VSD via an intramolecular conformational switch (4, 5). At typical negative resting voltages, the CD is inactive and is rapidly activated at more-positive (depolarized) voltages. The VSP homologs characterized to date are phosphoinositide 5-phosphatases that degrade the major signaling phospholipids PI(4,5)P2 and PI(3,4,5)P3 (2). Both phosphoinositides have key signaling roles in many cellular processes, including cell proliferation and differentiation, cytoskeletal dynamics, membrane trafficking, and control of ion channels (6, 7). Although little is known to date about the biological functions of VSPs, the ubiquitous roles of phosphoinositides and of electrical signaling suggest a potential impact on a large spectrum of cellular processes.
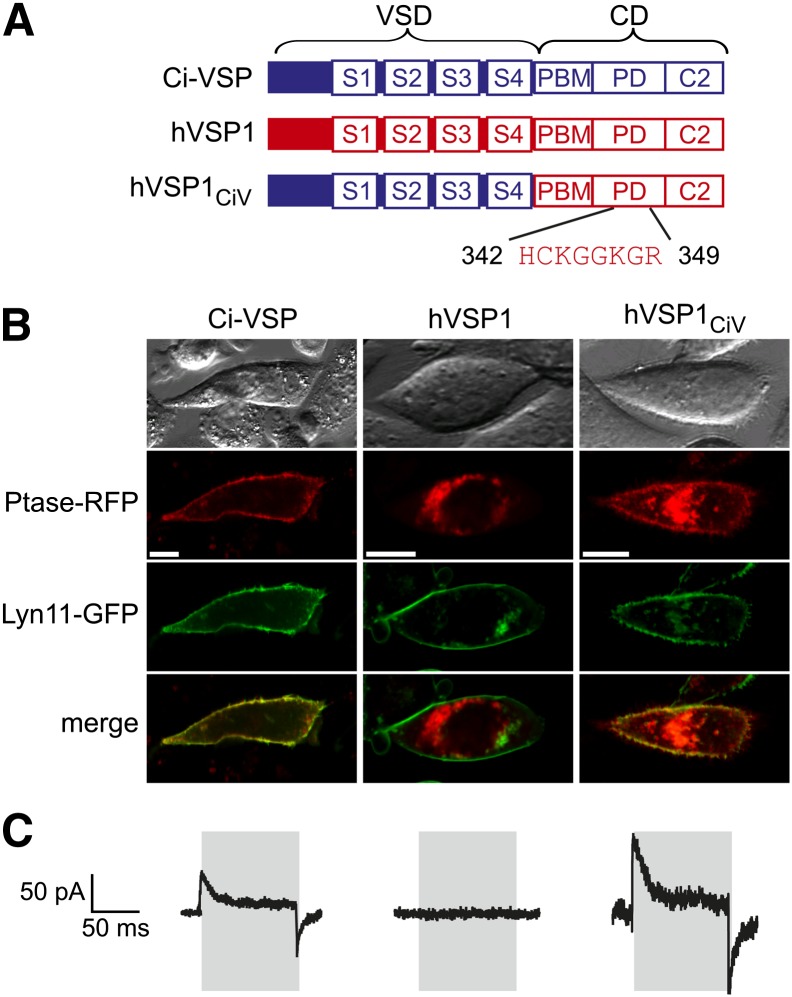
Fig. 1.
Generation of a functional hVSP1 chimera targeted to the plasma membrane. A: Domain structure of Ci-VSP and human VSP1 (full-length variant) and construction of the chimera hVSP1CiV. Inset shows catalytic CX5R motif. S1-S4, transmembrane segments 1–4; PBM, phosphoinositide binding motif; CD, catalytic domain; PD, phosphatase domain; C2, C2 domain. B: Subcellular localization of N-terminally mRFP-fused VSP constructs transfected into CHO together with the plasma membrane marker Lyn11-GFP (37) (scale bar, 10 µm). C: Sensing currents recorded from CHO cells in response to a voltage pulse (timing as indicated in gray) from the holding potential (−60 mV for Ci-VSP and hVSP1 or −100 mV for hVSP1CiV) to +80 mV.
The principle of operation of VSPs was initially demonstrated for Ci-VSP, the prototypic VSP from the invertebrate chordate Ciona intestinalis. Subsequently, functional vertebrate VSPs have been identified in fishes and amphibia (8, 9). The VSP gene is also conserved in mammals (10). In general, there appears to be one VSP homolog in mammalian genomes; in the human genome, however, there are two expressed homologs, TPTE and TPTE2 (also termed TPIP) and additional pseudo-genes (10, 11). TPTE conforms to the architecture of VSPs, but it lacks phosphatase activity due to amino acid exchanges in the catalytic CX5R motif in the P-loop of the phosphatase domain (12). In contrast, the intracellular domain of TPTE2/TPIP has phosphoinositide phosphatase activity in vitro (11). According to its conserved catalytic center, it is apparently the bona-fide ortholog of nonmammalian VSPs and the VSP homologs in other mammals [e.g., mouse PTEN2 (13), also termed mTpte (14)]. For consistent nomenclature, we will thus rename both homologs hVSP1 (for TPIP/TPTE2) and hVSP2 (for human TPTE).
Various splice variants of hVSP1 have been reported that mainly differ in the VSD-homologous transmembrane domain. Because all splice variants examined so far in expression systems lacked localization to the plasma membrane, it is unknown whether enzymatic activity is controlled by the VSD-homologous domain and, specifically, whether it is sensitive to membrane potential. Consequently the prevalence of VSP-mediated signaling in mammals has remained unknown. In human and mouse, VSPs are expressed mainly in the testis (10, 13) but also in brain and stomach (11). Cellular expression has been detected in secondary spermatocytes and early spermatids in human and mouse (10, 13), whereas in the tunicate, Ciona intestinalis, Ci-VSP, is also found in mature sperm (1). Thus, a role of VSPs in phosphoinositide signaling in mammalian spermatogenesis or sperm physiology is an exciting possibility.
Here, we address the enzymatic properties and the molecular mechanism of activation of hVSP1 using fluorescent sensors for phosphoinositides in living cells. Because hVSP1 localized predominantly to intracellular compartments when transfected into culture cells, we generated a chimeric hVSP1 containing the N-terminus from Ci-VSP that robustly targets to the plasma membrane. Functional assays showed that hVSP1 is a phosphoinositide 5-phosphatase that can be activated via the N-terminal VSD. These findings support a role for VSP-mediated phosphoinositide signaling in mammalian spermatocyte differentiation or sperm function.
MATERIALS AND METHODS
Cloning of the hVSP1CiV chimera and full-length hVSP1
mRFP-tagged hVSP1CiV was constructed by amplifying the coding sequence for mRFP and amino acids 1 to 239 of Ci-VSP from a pRFP-C1-Ci-VSP plasmid (2). A HindIII restriction site was added using reverse primer 5′-ATA TAA GCT TGT TGA TGG GAA TAA AAT ATT C-3′. The resulting amplicon was digested for 30 min at 37°C with fast-digest HindIII, BamHI, and DpnI (Fermentas, St. Leon-Rot, Germany) to avoid carryover of template DNA. Amino acids 120 to STOP of hVSP1-3 [TPIPα (11)] were amplified using forward primer 5′-GAC AAG CTT GAA AAG CTG ATG AGA AGG-3′ to insert a guanine to form a HindIII restriction site. The resulting fragment was digested with HindIII and BamHI and ligated to the corresponding mRFP-Ci-VSP fragment. The additional guanine was deleted from the fusion construct by mutagenesis PCR using PfuUltra DNA Polymerase AD (Stratagene, Waldbronn, Germany). The complete coding sequence of the resulting construct was verified by sequencing.
Full-length hVSP1 (previously termed TPTE2-1 or TPIPγ, UniProt accession # Q6XPS3) was cloned from a commercial RNA preparation from human testis (Invitrogen; Darmstadt, Germany). Briefly, hVSP1 cDNA was obtained by RT-PCR using semi-nested PCR as described elsewhere (10). PCR products were subcloned in pJET2.1/blunt vector (Fermentas) and sequenced. Two novel hVSP1 splice variants were identified, lacking either aa 40 to 59 or aa 434 to 464 (numbering according to the full-length sequence). The latter variant was subcloned into pRFP-C, yielding an mRFP-hVSP1 fusion construct. Finally, the C terminus was replaced with the full-length C terminus of hVSP1 isoform 3, yielding the full-length hVSP1 clone. The hVSP1-mRFP fusion construct was obtained by subcloning full-length hVSP1 into pRFP-N vector. The hVSP1hV2-3N chimera was constructed from amino acids 1–50 from hVSP2-3 (TPTEγ, acc. # P56180-3) in pRFP-C vector and amino acids 71–522 from hVSP1. A KpnI restriction site was added at the site of fusion of both constructs using forward mutagenesis primer 5′-GCA TTC AAT GGT ACC ATC CTT TGC ATT TGG-3′. The KpnI restriction site was removed from the fusion construct by mutagenesis PCR.
Plasmids and transfection of mammalian cells
The following plasmids were used: Ci-VSP (UniProt acc. # Q4W8A1) or hVSP variants in pRFP-C or pRFP-N vector; PLCδ1-PH (P51178), Btk-PH (Q06187), OSBP-PH (P22059) in pEGFP-N1 vector; TAPP1-PH (Q9HB21) in FUGW vector (contains eGFP); Bovine phosphatidylinositol 3-kinase p110α (constitutively active mutant K227E; P32871); Lyn11-GFP (P07948) in pcDNA3; and KCNQ4 (Kv7.4, P56696-1) in pBK-CMV.
CHO cells plated on glass coverslips or glass-bottom dishes were transfected with these plasmids using JetPEI transfection reagent as described (2, 15). VSPs were transfected individually or cotransfected with either Lyn11-GFP, phosphoinositide sensors, or KCNQ4 potassium channel. For experiments on 3-phosphorylated phosphoinositides, a constitutively active PI-3-kinase [p110α(K227E)] was additionally cotransfected (2). Experiments were done 24–48 h posttransfection, with cells selected for robust expression of the VSP constructs on the basis of mRFP fluorescence.
Whole-cell patch clamp
CHO cells were whole-cell voltage clamped with an EPC-10 (HEKA; Lambrecht, Germany) or Axopatch 200B amplifier (Molecular Devices; Sunnyvale, CA). Sensing currents were isolated using a P/-10 protocol. Intracellular solution contained (in mM): KCl, 135; MgCl2, 2.5; CaCl2, 2.41; EGTA, 5; HEPES, 5; and Na2ATP, 3, pH 7.3 (with KOH). Extracellular solution, also used for imaging experiments (mM): NaCl, 144; KCl, 5.8; NaH2PO4, 0.7; glucose, 5.6; CaCl2, 1.3; MgCl2, 0.9; and HEPES, 10; pH 7.4 (with NaOH). Patch pipettes were pulled from borosilicate glass (Science Products; Hofheim, Germany). KCNQ4 (Kv7.4) current recordings were sampled at 5 kHz and low-pass-filtered at 2 kHz. Series resistance in these experiments typically was below 5 MΩ and was compensated by 80–90%.
Fluorescence microscopy
Subcellular localization was imaged in living cells with a Zeiss LSM710 confocal microscope (Carl Zeiss AG; Jena, Germany) equipped with a W-Plan Apochromat 20×/1.0 DIC M27 objective. Laser lines used were 561 nm for mRFP and 488 nm for GFP, and detection wavelength ranges were 582–754 nm and 493–582 nm, respectively.
TIRF imaging was done as described previously (2). Briefly, a BX51WI upright microscope equipped with a TIRF-condenser (Olympus) and a 488 nm laser was used. Images were acquired with a CCD camera using TILLvision software (TILL-photonics; Gräfelfing, Germany). Where applicable, the electrophysiology setup was synchronized to the imaging setup. The frame interval was 3 s. TIRF imaging data were analyzed using TILLvision and IgorPro (Wavemetrics; Lake Oswego, OR). Regions of interest (ROIs) encompassed the footprint of a single cell, excluding cell margins to avoid movement artifacts. Normalized fluorescence intensity (F/F0) was calculated pixelwise from the TIRF signal intensity F and the initial fluorescence intensity F0, which was calculated as the average over the baseline interval. Background correction was applied in this process. The F/F0 trace was then calculated by framewise averaging over the ROI. F/F0 traces were corrected for bleaching according to monoexponential fits to the baseline interval as described before (15).
Steady-state fluorescence voltage relations were derived as reported previously (2). Data obtained from individual cells were fitted with a Boltzmann function F = Fmin + (Fmax-Fmin)/[1 + exp((V-V1/2)/s))], where F is the recorded TIRF intensity, V is the holding potential, V1/2 is the voltage at the half-maximal response, and s describes the steepness of the curve. Data were normalized to Fmax-Fmin as derived from the fit for each individual cell.
Oocyte electrophysiology
For expression in Xenopus oocytes, hVSPCiV was cloned into the pBSTA vector. Point mutations were made by PCR using mismatched primers containing the mutation. For RNA synthesis, DNA was linearized with NotI and transcribed using T7 RNA polymerase. KCNQ2 (Kv7.2) and KCNQ3 (Kv7.3) constructs for expression in Xenopus oocytes (vector pTLN; kindly provided by T. Jentsch) were linearized with MluI and HpaI, respectively, and RNA was transcribed using SP6 RNA polymerase (Ambion, Frankfurt, Germany). For coexpression of KCNQ channels with potassium currents, Xenopus oocytes were injected with 10 ng of RNA of each construct and incubated for 2–3 days at 12–18°C in a solution containing 100 mM NaCl, 2 mM KCl, 1 mM MgCl2, 2 mM CaCl2, and 10 mM HEPES, pH 7.5. For sensing currents, Xenopus oocytes were injected with 20–50 ng of RNA, and electrophysiological measurements were performed after 3 days of incubation at 15°C.
Potassium currents mediated by KCNQ channels were recorded by two-electrode voltage clamp. The extracellular recording solution contained 4–8 mM KOH, 112–116 mM NMG, 120 mM MeSO3 (methanesulfonate), 10 mM HEPES, and 2 mM CaCl2, pH 7.4. Sensing currents were measured 3–4 days after injection with the cut-open oocyte voltage clamp technique as described previously (16). The external recording solutions contained 120 mM NMG-MeSO3 (methanesulfonate), 10 mM HEPES, and 2 mM CaCl2, pH 7.4, whereas internal solutions contained 120 mM NMG-MeSO3, 10 mM HEPES, and 2 mM EGTA, pH 7.4. Currents were measured in response to voltage steps (10 s interval) from a holding potential of −60 mV, without leak subtraction during acquisition. Capacitance transient currents were compensated using the amplifier's compensation circuit. Recordings were performed using a homemade acquisition software based on the graphical programming environment LabVIEW (National Instruments; Austin, TX). The software controlled an NI USB-6251 BNC interface (National Instruments).
All experiments were performed at room temperature. Data are given as mean ± standard error of the mean (± SE).
RESULTS
hVSP1 may be subject to extensive splicing yielding differentially truncated VSD domains (10). The described N-terminal truncations appeared incompatible with the function of the domain as a voltage sensor. Moreover, truncated splice variants were not targeted to the plasma membrane (see supplementary Fig. I) (11). We therefore cloned the full-length variant (hVSP1-1) that corresponds to the canonical isoform according to UniProt (accession # Q6XPS3) (17). When transfected into CHO cells (Fig. 1B) or HEK293 cells (see supplementary Fig. IB), localization of even this full-length protein was restricted to intracellular compartments, possibly the Golgi apparatus, as suggested previously for mouse VSP1 (mPTEN2) (13). This subcellular localization was not the result of N-terminal mRFP fusion, because hVSP1-1 with a C-terminal mRFP showed the same localization (see supplementary Fig. IB). Accordingly, electrophysiological examination did not reveal sensing currents, the electrical signature of a functional VSD as found in Ci-VSP (Fig. 1C) (1). Moreover, the intracellular localization of hVSP1 in the expression system prohibited a direct assessment of voltage-dependent activity of this putative lipid phosphatase. Interestingly, a short splice variant of the paralog, hVSP2-3 (TPTEγ), targets robustly to the plasma membrane (11), although functional examination showed absence of sensing currents, indicating a nonfunctional VSD in hVSP2 (see supplementary Fig. IC). Given the distinct subcellular localization of both hVSPs, we considered a possible role of their distinct cytoplasmic N-termini in membrane targeting. However, replacement of the hVSP1 N-terminus with the corresponding terminus of hVSP2-3 also failed to localize the protein to the plasma membrane (see supplementary Fig. IA, B).
We therefore adopted an approach that we developed previously to examine the function of phosphoinositide phosphatases (15). Thus, we generated a chimeric hVSP1 variant by replacing its entire VSD with the highly homologous domain of the prototypic Ci-VSP (Fig. 1A, lower panel). We will refer to this chimera as hVSP1CiV. hVSP1CiV was robustly targeted to the plasma membrane (Fig. 1B). Depolarizing voltage steps revealed sensing currents similar to Ci-VSP, indicating intact functionality of the VSD in the chimeric hVSP1 (Fig. 1C). Expression of hVSP1CiV in Xenopus oocytes confirmed functionality and allowed for a detailed examination of sensing currents, revealing the sigmoidal dependence on membrane voltage that characterizes voltage sensor domains (see supplementary Fig. II).
Enzymatic activity and specificity of hVSP1
We tested for lipid phosphatase activity of hVSP1CiV in vivo, using GFP-fused phosphoinositide binding protein domains as fluorescent phosphoinositide sensors (18). Association of these probes to the membrane reports on the concentration of specific phosphoinositide species, and was measured quantitatively by TIRF microscopy (2, 15).
In cells coexpressing hVSP1CiV and the PI(4,5)P2 sensor PHPLCδ1-GFP, depolarization of the membrane potential via whole-cell patch clamping resulted in the reversible dissociation of the probe from the membrane (Fig. 2A, B) indicating depletion of PI(4,5)P2. This depletion resulted from phosphatase activity of hVSP1, because effects on PHPLCδ1 localization and PI(4,5)P2 concentration were abolished when an inactivating mutation (C363S) was introduced into the catalytic CX5R motif (Fig. 2B; cf. Refs. 15, 19). Thus, hVSP1 has phosphoinositide phosphatase activity in vivo, and, specifically, dephosphorylates PI(4,5)P2. Moreover, the depletion of PI(4,5)P2 in response to depolarization shows that in hVSP1CiV, enzymatic activity is regulated by membrane voltage via the attached VSD.
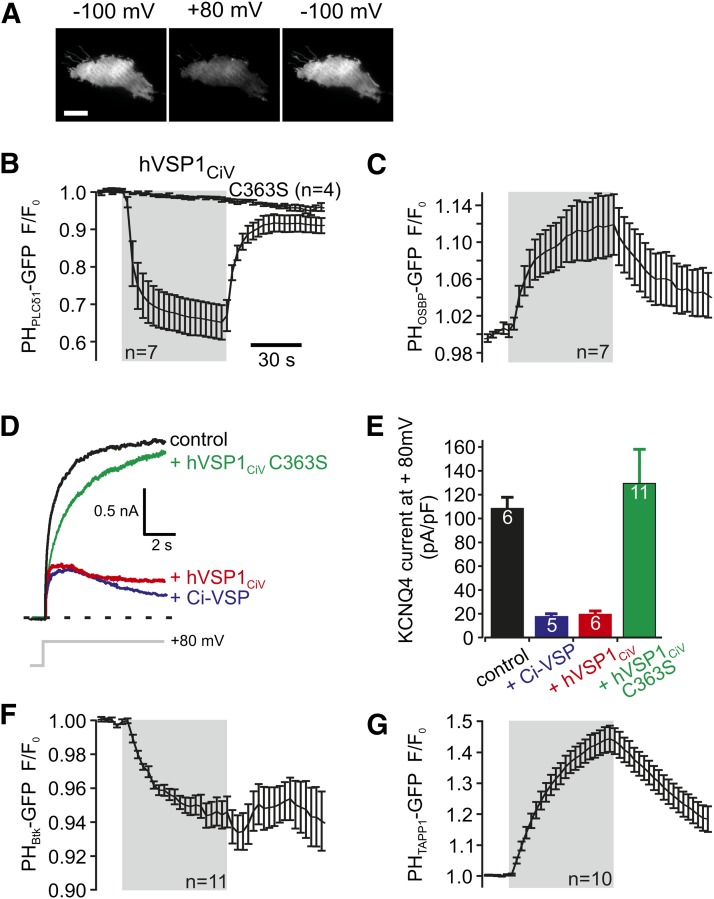
Fig. 2.
Phosphatase activity and substrate specificity of hVSPCiV. A: TIRF images of a CHO cell coexpressing mRFP-hVSP1CiV and PHPLCδ1-GFP acquired before (left), during (middle) and after (right) depolarization by whole-cell voltage clamp (scale bar, 10 µm). B, C: TIRF signals in response to step depolarization (−100 to +80 mV, gray shading) from cells expressing RFP-hVSP1CiV with the PI(4,5)P2 sensor PHPLCδ1-GFP (B) or the PI(4)P sensor PHOSBP-GFP (C). Decrease and increase of fluorescence signals indicate dissociation and association of the probes from and to the membrane, respectively. Voltage-dependent sensor translocation was not observed with the hVSP1CiV(C363S) mutant. D: Representative currents mediated by KCNQ4 channels with or without coexpression of the VSPs indicated. Holding potential was −60 mV (controls and coexpression of Ci-VSP) or −100 mV (hVSP1CiV). E: Mean KCNQ4 current amplitudes from experiments as in D. F, G: Depolarization-induced changes of TIRF intensities in cells cotransfected with hVSP1CiV(D136N) and the PI(3,4,5)P3 probe PHBtk (F) or the PI(3,4)P2 sensor PHTAPP1 (G) (−60 to +80 mV depolarization, gray shading). D136N mutant that activates at more-positive voltages (see Fig. 3) was used to avoid constitutive depletion of slowly synthesized 3-phosphorylated substrates prior to the experiments. Similar results were obtained with the wild-type VSD (see supplementary Fig. IV). Error bars indicate standard error of the mean.
Initial in vitro work on the isolated CD suggested that hVSP1 has 3-phosphatase activity (11), which would be incompatible with the observed consumption of cellular PI(4,5)P2 but should instead produce PI(4,5)P2 from PI(3,4,5)P3 (15). We therefore performed a detailed analysis of the enzymatic specificity of hVSP1, using fluorescent probes with distinct phosphoinositide specificities.
Activation of hVSP1CiV by depolarization resulted in an increase of the PI(4)P concentration, as shown by translocation of PHOSBP-GFP from the cytosol to the plasma membrane (Fig. 2C). Quantitative considerations indicate that the additional PI(4)P results from dephosphorylation of PI(4,5)P2. Thus, PI(4,5)P2 and PI(4)P together make up the bulk of phosphoinositides in the plasma membrane, whereas all other phosphoinositides have much lower steady-state concentrations in this compartment (6, 20, 21). Therefore, PI(4,5)P2 is the only available source for a substantial increase of PI(4)P by phosphatase activity. In conjunction with the observed depletion of PI(4,5)P2, this finding directly indicates that hVSP1 is a 5-phosphatase that converts PI(4,5)P2 to PI(4)P. To independently confirm the results obtained with fluorescent sensors, we tested the effect of hVSP1CiV on another type of PI(4,5)P2 biosensor, the KCNQ (Kv7) potassium channels. These channels specifically require PI(4,5)P2, but not PI(4)P, for their activity (22). In cells coexpressing KCNQ4 with hVSP1CiV, activation of the channels upon depolarization was strongly reduced and the currents showed time-dependent deactivation (Fig. 2D, E). Such channel deactivation was not observed in cells expressing KCNQ4 only or the catalytically inactivated mutant hVSP1CiV(C363S). Similar results were obtained with Xenopus oocytes expressing heteromeric KCNQ2/3 channels together with hVSP1CiV (see supplementary Fig. III). These findings are consistent with depletion of PI(4,5)P2 by hVSP1.
Measurements using fluorescent probes for 3-phosphorylated phosphoinositides further revealed that hVSP1 can also use PI(3,4,5)P3 as an alternative substrate, again cleaving the phosphate at position 5. Thus, activation of hVSP1CiV reduced the concentration of PI(3,4,5)P3, as measured with the PI(3,4,5)P3 probe, PHBtk-GFP (Fig. 2F), and at the same time increased the membrane's PI(3,4)P2 concentration as reported by increased membrane binding of the specific PI(3,4)P2 probe PHTAPP1-GFP (Fig. 2G).
Together, the changes in the various PI isoforms and the inhibition of PI(4,5)P2-sensitive KCNQ channels demonstrate that in vivo hVSP1 is a 5-phosphatase that can dephosphorylate both PI(4,5)P2 and PI(3,4,5)P3. In other words, hVSP1 has the same enzymatic properties as previously determined for the prototypic nonmammalian VSP, Ci-VSP.
Control of activity by the VSD
Next, we addressed the regulation of enzymatic activity in hVSP1. Is hVSP1 a functional mammalian VSP? Because its subcellular localization precluded the direct analysis of the native protein in the expression system, we analyzed hVSP1CiV to derive information on the regulation of hVSP1.
First, we note that the enzymatic activity is switched on rapidly (i.e., in less than a second) upon depolarization of the membrane potential. Similarly, hVSP1CiV rapidly returns to an inactive state upon repolarization, as reported by recovery of the PI(4,5)P2 level (Fig. 2B). This recovery results from cellular resynthesis of PI(4,5)P2 by endogenous PI-5-kinase activity (2, 21). Second, a detailed analysis using PHPLCδ1-GFP to monitor PI(4,5)P2 concentrations showed that phosphatase activity increased with the degree of depolarization in a sigmoidal, saturating manner (Fig. 3). This tight dependence of activity on voltage indicates close coupling of enzymatic activity to the dynamics of the VSD characterized by the sensing currents (see supplementary Fig. II). Direct functional coupling between both domains also predicts that altering the voltage dependence of the VSD affects the activation of the CD by membrane potential (3). In the prototypic Ci-VSP, the mutation D136N in the S2 helix of the VSD shifted its voltage dependence to more-depolarized voltages by about +50 mV. Accordingly, activation of the CD required stronger depolarization (Fig. 3B, C). Activation of hVSP1CiV occurred at substantially more hyperpolarized potentials than activation of Ci-VSP (cf Ref. 2). However, in hVSP1CiV, the D136N mutation also shifted voltage dependence of phosphatase activity by +50 mV (Fig. 3A, C), supporting the same close coupling between VSD and CD in hVSP1CiV as found in Ci-VSP.
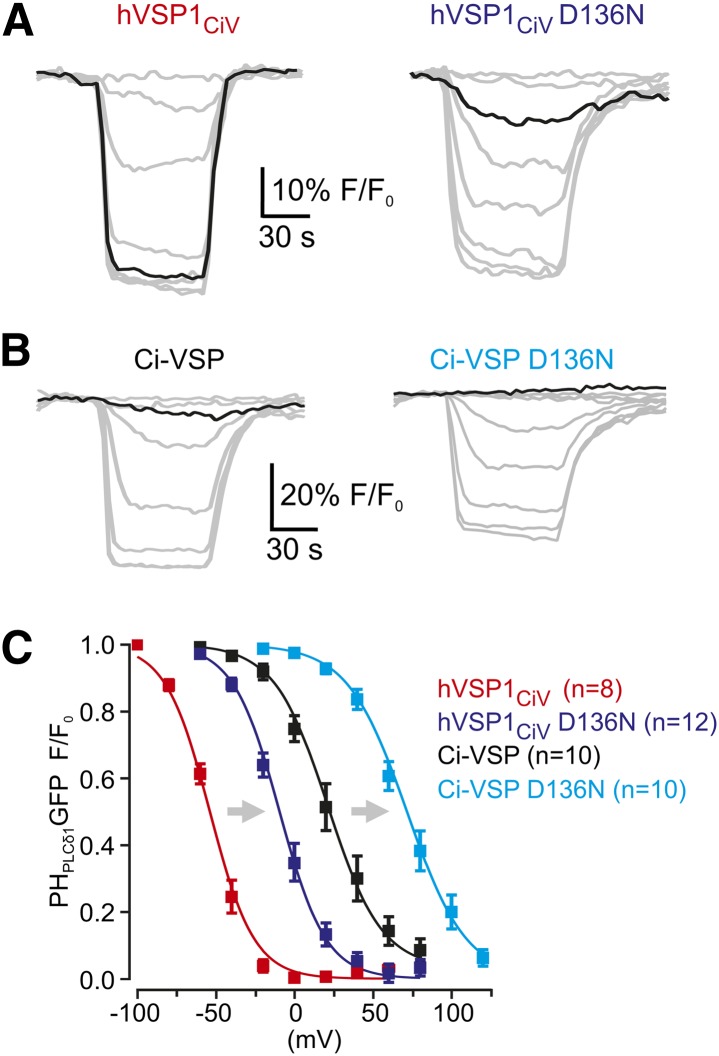
Fig. 3.
Activity of the intracellular CD is controlled by the VSD. A: Representative depolarization-induced TIRF fluorescence responses from cells expressing PHPLCδ1-GFP together with mRFP-hVSPCiV or its D136N mutant. Membrane potential was subjected to a family of depolarizing voltage pulses of increasing amplitude (20 mV increment). Black traces were obtained at depolarization to −20 mV. B: Depolarization-induced TIRF fluorescence changes obtained as in A from cells expressing Ci-VSP or Ci-VSP(D136N). Black traces were obtained at depolarization to −20 mV. C: Normalized steady-state fluorescence-voltage relationship from recordings as in A and B. Voltage at half-maximal fluorescence decrease (V1/2) was calculated by fitting a Boltzmann function (continuous line) to the mean data. V1/2 (obtained from fits to data of individual experiments) was −60.2 ± 5.3 mV for hVSP1CiV, −9.4 ± 3.0 for hVSP1CiV(D136N), 21.3 ± 5.3 mV for Ci-VSP, and 71.6 ± 5.1 mV for Ci-VSP(D136N).
Electrochemical coupling in Ci-VSP is mediated by a phosphoinositide binding motif (PBM), also referred to as the linker (5, 23, 24), connecting the VSD and the CD (Fig. 4A). It has been suggested that the VSD controls the activity of the CD by modulating the binding of the PBM to the membrane (5, 15) or its interaction with a “gating loop” that controls substrate access to the active site of the CD (23, 24). Thus, mutation of conserved cationic residues in the PBM uncouples enzymatic activity from membrane potential in Ci-VSP (4, 5). The basic PBM residues previously identified as critical for electrochemical coupling in Ci-VSP are fully conserved in hVSP1 (Fig. 4A), indicating that the same mechanism may mediate activation of the CD hVSP1.
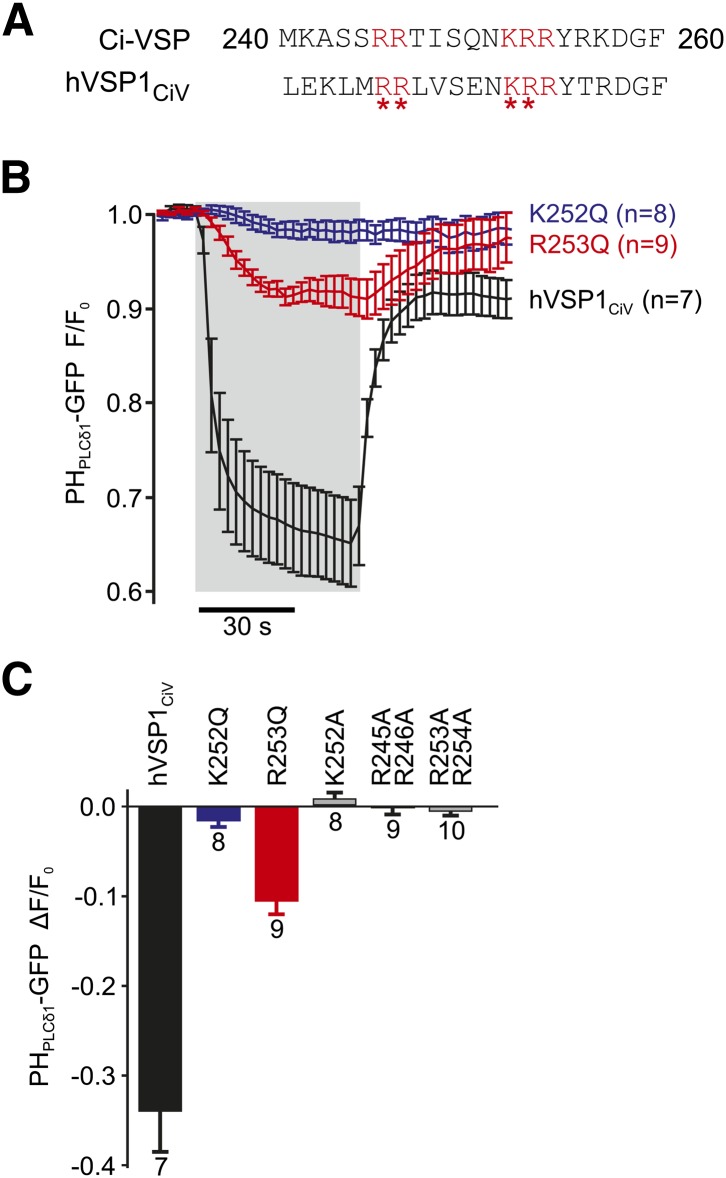
Fig. 4.
Functional coupling of the enzymatic domain to the VSD is mediated by the PBM. A: Critical basic residues (red) are conserved between the PBMs of Ci-VSP and hVSP1. Asterisks indicate positions mutated to glutamine or alanine. B: Charge-neutralizing mutations K252Q and R253Q in the PBM of hVSP1 greatly reduce voltage-dependent activation of hVSP1CiV. Experiments were performed as in (Fig 1A, B), and data from cells expressing wt hVSP1CiV are replotted from Fig. 1B for comparison. Depolarization to 80 mV is indicated by shaded area [N = 8 (K252Q) or 9 (R253Q) cells]. C: Average depolarization-activated enzymatic activity for the PBM mutants indicated. Activity is quantified as the degree of dissociation of PHPLCδ1-GFP from the membrane, measured as in B.
To test this hypothesis, we mutated K252 or R253 to glutamine. These mutations have previously been shown to drastically reduce voltage-dependent activity of Ci-VSP by uncoupling the VSD and the PD, while leaving the enzymatic activity of the isolated CD largely intact (4). In hVSP1CiV, these mutants showed strongly reduced depolarization-dependent activity (Fig. 4B, C). Additionally, further combinatorial alanine replacement of cationic residues within the PBM that are essential for activity of Ci-VSP (5) fully abrogated phosphatase activity of hVSP1CiV upon depolarization, as shown in Fig. 4C.
These findings indicate that the activation of the enzymatic domain of hVSP1 involves the endogenous PBM, and proceeds via a mechanism similar to electrochemical coupling in the prototypic Ci-VSP.
DISCUSSION
Enzymatic properties of mammalian VSPs
Here, we show and characterize for the first time activity of a mammalian VSP in living cells and demonstrate that this activity is controlled by a VSD-homologous domain. Specifically, we demonstrate that hVSP1 is a phosphoinositide 5-phosphatase that degrades the major signaling phosphoinositides PI(4,5)P2 and PI(3,4,5)P3.
This finding may be unexpected, given that previous in vitro experiments with the isolated intracellular CD of human or mouse VSP1 suggested 3-phosphatase properties (11, 13). However, our conclusion is based on several lines of evidence obtained in living cells. Notably, the same responses of phosphoinositide biosensors are found with the phosphoinositide 5-phosphatase Ci-VSP (2, 3). For comparison, rapid activation of the bona-fide 3-phosphatase, PTEN, results in a completely different pattern of phosphoinositide concentration changes when examined using the same approach. Thus, biosensor responses to PTEN activation showed synchronous decrease of PI(3,4,5)P3 and PI(3,4)P2, increase of PI(4,5)P2, but no change in PI(4)P (15).
Moreover, in line with our conclusion, previous functional assays in living cells did not support a role in 3-phosphoinositide signaling (11), and recent in vitro data are fully consistent with the 5-phosphatase activity (25). The apparently different enzymatic specificity previously observed in vitro might therefore point to the importance of the native membrane environment in determining the stereochemistry of interaction of the phosphatase with its phospholipid substrate.
Unexpectedly, a recent report suggested an additional 3-phosphatase activity of Ci-VSP, because in Xenopus oocytes, overexpressed Ci-VSP induced a dissociation of the PI(3,4)P2 probe TAPP1-PH from the membrane at strongly depolarized potentials (25). We have not observed such apparent depletion of PI(3,4)P2 by Ci-VSP in mammalian cells previously (2). Additional experiments with strong (+80 mV) and prolonged (>5 min) depolarization also failed to indicate any PI(3,4)P2 hydrolysis either by Ci-VSP or by hVSPCiV (data not shown). Therefore, it remains to be seen whether the apparent 3-phosphatase activity is specific for the oocyte expression system.
Alternatively, it might be argued that the exogenous VSD in hVSPCiV caused changes in the active site, resulting in altered catalytic selectivity. However, for Ci-VSP it has been shown that deletion of the VSD does not change the 5-phosphatase nature of its CD (26). Likewise, we have shown previously that attaching the VSD of Ci-VSP to PTEN does not affect the catalytic properties of this phosphatase (15). We thus conclude that it is unlikely that attaching the VSD of Ci-VSP to the CD of hVSP1 causes a switch from a 3- to a 5-phosphatase.
Given the striking structural similarity of the intracellular CD of VSPs to PTEN (15, 27), the distinct enzymatic activities may appear surprising. However, the catalytic CX5R motif of hVSP (CKGGKGR) differs in a single position from PTEN (CKAGKGR). In Ci-VSP, this amino acid contributes strongly to the 5-phosphatase specificity (26, 27). Additional sites, including glutamate 411 in the catalytic gating loop of Ci-VSP, are also important for determining substrate selectivity (23, 25). Further differences in the catalytic center may also contribute to the distinct enzymatic specificities; this remains to be addressed. Despite this complexity, all CX5R-type 5-phosphatases examined to date display the critical G variant motif (2, 8, 9, 26), suggesting that not only hVSP1, but also yet-uncharacterized VSP homologs from other mammalian species, are 5-phosphatases, because all share the G variant.
Functional role of hVSP1
Our results on the enzymatic properties of hVSP1 indicate that mammalian VSPs are involved in PI(4,5)P2 or PI-3 signaling pathways or contribute to the homeostasis of these phosphoinositides. The range of possible biological functions of hVSP1 is constrained by its cellular expression pattern and subcellular localization. Little is known about the expression pattern of mammalian VSPs, but available data on mRNA abundance suggest that it is confined to specific organs (testis, stomach, brain) (11, 13, 14). Consequently, VSPs may serve cell type-specific functions rather than mediating general cellular mechanisms. However, expression may be more widespread during development (28), as observed for the nonvertebrate Ci-VSP (29) and the homolog in birds (30).
Notably, the intracellular Golgi-like localization of hVSPs is in contrast to the plasma membrane localization of all previously examined nonmammalian VSPs (1, 9, 26). However, the localization in native cells has not been determined. Meanwhile, it remains possible that the localization observed is an artifact of overexpression in culture cells. VSPs localized to the plasma membranes can impact on the many signaling roles of PI(4,5)P2 and PI(3,4,5)P3/PI(3,4)P2. In contrast, a role of this 5-phosphatase at the Golgi membranes is less obvious, inasmuch as the main phosphoinositide in this compartment is PI(4)P, whereas there is little PI(4,5)P2 or PI(3,4,5)P3 (31, 32). Accordingly, PI(4)P is a key player, essential for Golgi function, whereas PI(4,5)P2 seems less important (31). This consideration may also argue in favor of plasma membrane localization of endogenously expressed hVSP1 in native cells. On the other hand, Golgi-localized VSP may function to ensure low PI(4,5)P2 levels, as has been suggested for the 5-phosphatase OCRL that is also associated with the Golgi (33).
The issue of subcellular distribution of hVSP1 also raises an important question as to the stimulus that activates native hVSP1. Although our current observations were obtained using the voltage sensor chimera hVSP1CiV, the findings strongly suggest that in native hVSP1, the enzymatic activity is regulated by an intramolecular interaction of the CD with the endogenous VSD-homologous transmembrane module. Thus, activity was readily controlled by the exogenous VSD, and the coupling is mediated through the endogenous PBM of hVSP1, indicating that all molecular requirements for intramolecular switching by the VSD are present in mammalian VSPs. Thus, we conclude that the native VSD domain of hVSP1 is a sensor domain that controls the signaling activity of the VSP. Its homology to VSDs from bona fide voltage-sensitive VSPs suggests that it may act as a voltage sensor, which, however, remains to be examined experimentally.
Recently, Sutton et al. (34) addressed the function of the VSDs of hVSP1 and hVSP2 using chimeric constructs in which the S3-S4 segments of the VSDs of both hVSPs were transplanted into the VSD of Dr-VSP. In these constructs, sensing currents suggested functionality of the chimeric VSD. In contrast, we found that the endogenous VSD of hVSP2-3 [human TPTEγ (10)] did not produce sensing currents, despite robust targeting of this isoform to the plasma membrane (see supplementary Fig. IC). This finding may indicate that in hVSP2, the full-length VSD does not act as a voltage sensor in the physiological range of membrane potentials. The fact that the VSD is largely conserved between the hVSP1 and hVSP2 full-length splice forms (93% sequence identity) might also point toward a lack of voltage sensitivity in hVSP1. In support of this idea, the alignment of VSP primary sequences reveals two relevant differences in the VSDs of Ci-VSP versus hVSP. First, the homolog residue to R232 within the putative S4 helix of Ci-VSP is a histidine in the hVSPs. This residue has been proposed as critical for electrochemical coupling (1). Second, the hVSPs feature additional positively charged residues (R171 and R177) close to the extracellular end of the S4 segment. It was previously observed that in Ci-VSP, neutralization of a positive charge (R217) located in the homologous sequence stretch causes a strong shift in voltage dependence toward negative potentials (35). Vice versa, the additional positive charges present in the hVSPs may position the voltage dependence at very positive potentials beyond the range that we can assay.
Therefore, it may be considered that alternative cellular signals such as pH or lipid composition could operate the mammalian VSPs. In fact, such hypothetical activation stimuli may be consistent with Golgi localization observed in transfected cells, because changes in membrane potential may not be prevalent in this organelle.
Beyond their as yet enigmatic physiological role, nonmammalian VSPs have received attention as powerful experimental tools for the analysis of PI(4,5)P2-dependent cellular processes (e.g., 20, 36). So far, patch-clamp or similar electrophysiological techniques have been used to rapidly switch VSPs and thereby deplete and replenish PI(4,5)P2 levels. However, to access a broader range of cell biological processes, it may be advantageous to operate VSPs without the need for specialized equipment, e.g., by depolarization via elevation of extracellular K+ (15). The left-shifted activation range of hVSPCiV in comparison to Ci-VSP may render this chimera particularly suitable for this approach, inasmuch as its activation range fits well with the range of membrane potentials that can easily be achieved by altering the extracellular K+ concentration.
In summary, our current results suggest that mammalian VSPs are functional phosphoinositide phosphatases that mediate membrane-delimited signaling by 5-phosphoinositides at intracellular or plasma membranes and that may be involved in controlling the phosphoinositide composition of intracellular compartments. To date, expression of hVSP1 has been observed in testis, brain, and stomach (11). In particular, the striking finding of hVSP1 expression in secondary spermatocytes and in spermatids (10, 13) suggests specific roles for VSP-mediated 5-phosphoinositide signaling in mammalian reproductive physiology.
Supplementary Material
Footnotes
Abbreviations:
- CD
- catalytic domain
- PBM
- phosphoinositide binding motif
- PD
- phosphatase domain
- ROI
- region of interest
- VSD
- voltage sensor domain
- VSP
- voltage-sensitive phosphatase
The authors thank O. Ebers and G. Fischer for expert technical assistance. cDNAs encoding hVSP1-3 and Ci-VSP were kindly provided by Drs. Nick Leslie (Dundee, UK) and Yasushi Okamura (Osaka, Japan), respectively. This work was supported by a grant of the University Medical Center Giessen and Marburg (UKGM 32/2011 MR) to C.R.H. and by Grant SFB 593 TPA12 from the Deutsche Forschungsgemeinschaft to D.O.
The online version of this article (available at http://www.jlr.org) contains supplementary data in the form of four figures.
REFERENCES
- 1.Murata Y., Iwasaki H., Sasaki M., Inaba K., Okamura Y. 2005. Phosphoinositide phosphatase activity coupled to an intrinsic voltage sensor. Nature. 435: 1239–1243 [DOI] [PubMed] [Google Scholar]
- 2.Halaszovich C. R., Schreiber D. N., Oliver D. 2009. Ci-VSP is a depolarization-activated phosphatidylinositol-4,5-bisphosphate and phosphatidylinositol-3,4,5-trisphosphate 5′-phosphatase. J. Biol. Chem. 284: 2106–2113 [DOI] [PubMed] [Google Scholar]
- 3.Murata Y., Okamura Y. 2007. Depolarization activates the phosphoinositide phosphatase Ci-VSP, as detected in Xenopus oocytes coexpressing sensors of PIP2. J. Physiol. 583: 875–889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kohout S. C., Bell S. C., Liu L., Xu Q., Minor D. L., Jr, Isacoff E. Y. 2010. Electrochemical coupling in the voltage-dependent phosphatase Ci-VSP. Nat. Chem. Biol. 6: 369–375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Villalba-Galea C. A., Miceli F., Taglialatela M., Bezanilla F. 2009. Coupling between the voltage-sensing and phosphatase domains of Ci-VSP. J. Gen. Physiol. 134: 5–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Di Paolo G., De Camilli P. 2006. Phosphoinositides in cell regulation and membrane dynamics. Nature. 443: 651–657 [DOI] [PubMed] [Google Scholar]
- 7.Suh B. C., Hille B. 2008. PIP2 is a necessary cofactor for ion channel function: how and why? Annu. Rev. Biophys. 37: 175–195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hossain M. I., Iwasaki H., Okochi Y., Chahine M., Higashijima S., Nagayama K., Okamura Y. 2008. Enzyme domain affects the movement of the voltage sensor in ascidian and Zebrafish voltage-sensing phosphatases. J. Biol. Chem. 283: 18248–18259 [DOI] [PubMed] [Google Scholar]
- 9.Ratzan W. J., Evsikov A. V., Okamura Y., Jaffe L. A. 2011. Voltage sensitive phosphoinositide phosphatases of Xenopus: their tissue distribution and voltage dependence. J. Cell. Physiol. 226: 2740–2746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tapparel C., Reymond A., Girardet C., Guillou L., Lyle R., Lamon C., Hutter P., Antonarakis S. E. 2003. The TPTE gene family: cellular expression, subcellular localization and alternative splicing. Gene. 323: 189–199 [DOI] [PubMed] [Google Scholar]
- 11.Walker S. M., Downes C. P., Leslie N. R. 2001. TPIP: a novel phosphoinositide 3-phosphatase. Biochem. J. 360: 277–283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Leslie N. R., Yang X., Downes C. P., Weijer C. J. 2007. PtdIns(3,4,5)P(3)-dependent and -independent roles for PTEN in the control of cell migration. Curr. Biol. 17: 115–125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wu Y., Dowbenko D., Pisabarro M. T., Dillard-Telm L., Koeppen H., Lasky L. A. 2001. PTEN 2, a Golgi-associated testis-specific homologue of the PTEN tumor suppressor lipid phosphatase. J. Biol. Chem. 276: 21745–21753 [DOI] [PubMed] [Google Scholar]
- 14.Guipponi M., Tapparel C., Jousson O., Scamuffa N., Mas C., Rossier C., Hutter P., Meda P., Lyle R., Reymond A., et al. 2001. The murine orthologue of the Golgi-localized TPTE protein provides clues to the evolutionary history of the human TPTE gene family. Hum. Genet. 109: 569–575 [DOI] [PubMed] [Google Scholar]
- 15.Lacroix J., Halaszovich C. R., Schreiber D. N., Leitner M. G., Bezanilla F., Oliver D., Villalba-Galea C. A. 2011. Controlling the activity of a phosphatase and tensin homolog (PTEN) by membrane potential. J. Biol. Chem. 286: 17945–17953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Villalba-Galea C. A., Sandtner W., Starace D. M., Bezanilla F. 2008. S4-based voltage sensors have three major conformations. Proc. Natl. Acad. Sci. USA. 105: 17600–17607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jain E., Bairoch A., Duvaud S., Phan I., Redaschi N., Suzek B. E., Martin M. J., McGarvey P., Gasteiger E. 2009. Infrastructure for the life sciences: design and implementation of the UniProt website. BMC Bioinformatics. 10: 136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Varnai P., Balla T. 2006. Live cell imaging of phosphoinositide dynamics with fluorescent protein domains. Biochim. Biophys. Acta. 1761: 957–967 [DOI] [PubMed] [Google Scholar]
- 19.Maehama T., Taylor G. S., Dixon J. E. 2001. PTEN and myotubularin: novel phosphoinositide phosphatases. Annu. Rev. Biochem. 70: 247–279 [DOI] [PubMed] [Google Scholar]
- 20.Falkenburger B. H., Jensen J. B., Hille B. 2010. Kinetics of PIP2 metabolism and KCNQ2/3 channel regulation studied with a voltage-sensitive phosphatase in living cells. J. Gen. Physiol. 135: 99–114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Leslie N. R., Downes C. P. 2002. PTEN: the down side of PI 3-kinase signalling. Cell. Signal. 14: 285–295 [DOI] [PubMed] [Google Scholar]
- 22.Suh B-C., Inoue T., Meyer T., Hille B. 2006. Rapid chemically induced changes of PtdIns(4,5)P2 gate KCNQ ion channels. Science. 314: 1454–1457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu L., Kohout S. C., Xu Q., Muller S., Kimberlin C. R., Isacoff E. Y., Minor D. L., Jr 2012. A glutamate switch controls voltage-sensitive phosphatase function. Nat. Struct. Mol. Biol. 19: 633–641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hobiger K., Utesch T., Mroginski M. A., Friedrich T. 2012. Coupling of Ci-VSP modules requires a combination of structure and electrostatics within the linker. Biophys. J. 102: 1313–1322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kurokawa T., Takasuga S., Sakata S., Yamaguchi S., Horie S., Homma K. J., Sasaki T., Okamura Y. 2012. 3′ Phosphatase activity toward phosphatidylinositol 3,4-bisphosphate [PI(3,4)P2] by voltage-sensing phosphatase (VSP). Proc. Natl. Acad. Sci. USA. 109: 10089–10094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Iwasaki H., Murata Y., Kim Y., Hossain M. I., Worby C. A., Dixon J. E., McCormack T., Sasaki T., Okamura Y. 2008. A voltage-sensing phosphatase, Ci-VSP, which shares sequence identity with PTEN, dephosphorylates phosphatidylinositol 4,5-bisphosphate. Proc. Natl. Acad. Sci. USA. 105: 7970–7975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Matsuda M., Takeshita K., Kurokawa T., Sakata S., Suzuki M., Yamashita E., Okamura Y., Nakagawa A. 2011. Crystal structure of the cytoplasmic phosphatase and tensin homolog (PTEN)-like region of Ciona intestinalis voltage-sensing phosphatase provides insight into substrate specificity and redox regulation of the phosphoinositide phosphatase activity. J. Biol. Chem. 286: 23368–23377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Reymond A., Marigo V., Yaylaoglu M. B., Leoni A., Ucla C., Scamuffa N., Caccioppoli C., Dermitzakis E. T., Lyle R., Banfi S., et al. 2002. Human chromosome 21 gene expression atlas in the mouse. Nature. 420: 582–586 [DOI] [PubMed] [Google Scholar]
- 29.Ogasawara M., Sasaki M., Nakazawa N., Nishino A., Okamura Y. 2011. Gene expression profile of Ci-VSP in juveniles and adult blood cells of ascidian. Gene Expr. Patterns. 11: 233–238 [DOI] [PubMed] [Google Scholar]
- 30.Neuhaus H., Hollemann T. 2009. Kidney specific expression of cTPTE during development of the chick embryo. Gene Expr. Patterns. 9: 568–571 [DOI] [PubMed] [Google Scholar]
- 31.Mayinger P. 2009. Regulation of Golgi function via phosphoinositide lipids. Semin. Cell Dev. Biol. 20: 793–800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Watt S. A., Kular G., Fleming I. N., Downes C. P., Lucocq J. M. 2002. Subcellular localization of phosphatidylinositol 4,5-bisphosphate using the pleckstrin homology domain of phospholipase C delta1. Biochem. J. 363: 657–666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pirruccello M., De Camilli P. 2012. Inositol 5-phosphatases: insights from the Lowe syndrome protein OCRL. Trends Biochem. Sci. 37: 134–143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sutton K. A., Jungnickel M. K., Jovine L., Florman H. M. Evolution of the voltage sensor domain of the voltage-sensitive phosphoinositide phosphatase, VSP/TPTE, suggests a role as a proton channel in eutherian mammals. Mol. Biol. Evol. 29: 2147–2155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kohout S. C., Ulbrich M. H., Bell S. C., Isacoff E. Y. 2008. Subunit organization and functional transitions in Ci-VSP. Nat. Struct. Mol. Biol. 15: 106–108 [DOI] [PubMed] [Google Scholar]
- 36.Lindner M., Leitner M. G., Halaszovich C. R., Hammond G. R., Oliver D. 2011. Probing the regulation of TASK potassium channels by PI(4,5)P2 with switchable phosphoinositide phosphatases. J. Physiol. 589: 3149–3162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Inoue T., Heo W. D., Grimley J. S., Wandless T. J., Meyer T. 2005. An inducible translocation strategy to rapidly activate and inhibit small GTPase signaling pathways. Nat. Methods. 2: 415–418 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.