Abstract
The target of rapamycin (Tor) proteins sense nutrients and control transcription and translation relevant to cell growth. Treating cells with the immunosuppressant rapamycin leads to the intracellular formation of an Fpr1p-rapamycin-Tor ternary complex that in turn leads to translational down-regulation. A more rapid effect is a rich transcriptional response resembling that when cells are shifted from high- to low-quality carbon or nitrogen sources. This transcriptional response is partly mediated by the nutrient-sensitive transcription factors GLN3 and NIL1 (also named GAT1). Here, we show that these GATA-type transcription factors control transcriptional responses that mediate translation by several means. Four observations highlight upstream roles of GATA-type transcription factors in translation. In their absence, processes caused by rapamycin or poor nutrients are diminished: translation repression, eIF4G protein loss, transcriptional down-regulation of proteins involved in translation, and RNA polymerase I/III activity repression. The Tor proteins preferentially use Gln3p or Nil1p to down-regulate translation in response to low-quality nitrogen or carbon, respectively. Functional consideration of the genes regulated by Gln3p or Nil1p reveals the logic of this differential regulation. Besides integrating control of transcription and translation, these transcription factors constitute branches downstream of the multichannel Tor proteins that can be selectively modulated in response to distinct (carbon- and nitrogen-based) nutrient signals from the environment.
The target of rapamycin (Tor) proteins, like other family members of the phosphatidylinositol-3-kinase-related kinases, control numerous aspects of cell function (multifunctional effectors) in what has been termed “horizontal” or “radial” signal transduction (1). The Tor proteins sense nutrients (2), specifically carbon- and nitrogen-quality, and process those signals to generate appropriate outputs (multichannel processors) (3). Defining the molecular bases of multifunctionality and multichannel processing remains a goal of signal transduction research.
Nitrogen regulation in Saccharomyces cerevisiae comprises two distinct systems: one that discriminates the quality of available nitrogen sources, the nitrogen discrimination pathway, and a second that responds to the complete lack of nitrogen (4, 5). Treating cells with rapamycin activates the first but not the second pathway (5). Treatment with rapamycin also causes cells to behave transcriptionally as if they lack a high-quality carbon source such as glucose and instead are growing on a low-quality carbon source such as ethanol (3, 5). The similarity of rapamycin to both low-quality carbon and nitrogen (and not other stress responses such as treatment with DNA-damaging agents) is genome-wide, spanning hundreds of genes in many different functional classes (3).
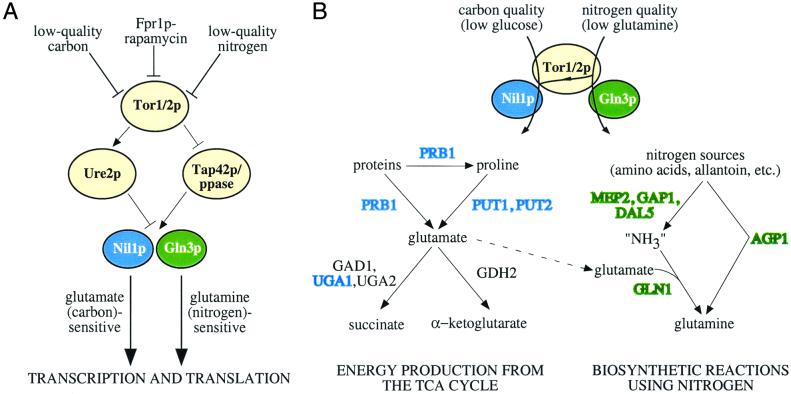
Ure2p and the GATA-type transcription factors Gln3p and Nil1p are critical members of the nitrogen discrimination pathway (6–8). Ure2p anchors Gln3p and Nil1p in the cytoplasm when carbon and nitrogen nutrients are abundant (9). Upon treatment with rapamycin, all three proteins are dephosphorylated, causing Gln3p and Nil1p to enter the nucleus and promote the transcription of genes important in the metabolism of low-quality nitrogen sources (5, 9–11). Nitrogen discrimination pathway target genes can be partitioned into two sets. These sets have: (i) distinct responses to low-quality carbon vs. low-quality nitrogen sources, (ii) differential dependence on GLN3 in a ure2Δ strain, (iii) differential dependence on GLN3 versus NIL1 upon treatment with rapamycin, and (iv) distinct responses to treatment with rapamycin in an mks1Δ strain (3, 12). The expression of one of these sets of genes (PUT1, PUT2, UGA1, NIL1, PRB1) is highly responsive to low-quality carbon sources and has high dependence on Nil1p, whereas the second set (including GAP1, MEP2, DAL5, BAT2, AGP1) is less responsive to low-quality carbon sources and has high dependence on Gln3p (3). The existence of these two sets of genes controlled by Ure2p and Tor1/2p led to the proposal that the Tor proteins are multichannel processors that “intelligently” route information about carbon and nitrogen nutrients and appropriately cause the activation of Gln3p or Nil1p in response to low nitrogen- or carbon-quality, respectively (3).
The S. cerevisiae GATA transcription factors (GLN3, NIL1, NIL2, DAL80) bind DNA and regulate the expression of genes that control nitrogen metabolism (13). Although deleting GLN3 or NIL1 leads to rapamycin resistance for growth (9–11), it is not understood how Gln3p or Nil1p has control over this fundamental cell process. Here, we demonstrate that in addition to their roles in transcription of genes relating to nitrogen metabolism, these GATA-type factors are used by Tor1/2p to regulate the transcription of genes that control translation, a critical process for cell growth. Furthermore, carbon and nitrogen quality differentially regulate Gln3p and Nil1p in the control of translation. These data, along with consideration of the functions of genes that these GATA-type factors control, lead to an understanding of how the cell uses Gln3p and Nil1p in the response to low-quality carbon or nitrogen.
Materials and Methods
Strains and Media.
The following S. cerevisiae strains were used in this study. PM38 (MATα leu2-3,112 ura3-52), PM71 (MATα leu2-3,112 ura3-52 gln3Δ5∷LEU2), MS221 (MATα ura3-52 nil1∷hisG), YDR21 (MATα leu2-3,112 ura3-52 gln3∷LEU2 nil1∷hisG), PH2 (MATα leu2-3,112 ura3-52 ure2Δ12∷URA3), and P40-1A strains (MATα leu2-3,112 his4-619 ade2-102 gln3Δ4∷LEU2 ure2Δ11∷LEU2) were kind gifts from Boris Magasanik (Massachusetts Institute of Technology, Cambridge, MA) and Marjorie Brandriss (University of Medicine and Dentistry of New Jersey, Newark, NJ). Strains CY4927 (MATa tap42del∷HIS3 SSD1-v [TAP42 on LEU2/CEN] W303) and CY4928 (MATa tap42Δ∷HIS3 SSD1-v [tap42–11 on LEU2/CEN] W303) were kind gifts from Charles Di Como (Memorial Sloan–Kettering Cancer Center, New York, NY). Yeast extract/peptone/dextrose consists of 20 g of glucose/liter, 20 g of peptone/liter, and 10 g of yeast extract/liter. Synthetic media consist of 1.7 g of yeast nitrogen base without amino acids and without ammonium sulfate, 2% carbon source (wt/vol for glucose and vol/vol for ethanol), 0.1% nitrogen source (wt/vol), and auxotrophic supplements as needed (leucine, 120 mg/liter; uracil, 20 mg/liter).
Electrophoresis and Western Blotting.
For experiments analyzing eIF4G levels, strains were grown to logarithmic phase in yeast extract/peptone/dextrose and incubated for the indicated time in the absence or presence of 50 nM rapamycin. Cell lysates were prepared as described (3). Relative protein contents of the lysates were determined by measuring absorbance at 260 nm. Equal amounts of protein were run on a 7.5% SDS/PAGE gel and transferred to a polyvinylidene fluoride membrane (Millipore). The membrane was blotted with rabbit polyclonal Abs against eIF4G1 generously provided by Alan Sachs (University of California, Berkeley, CA), and the immunocomplex was visualized by an enhanced chemiluminescence detection system (Amersham Pharmacia). A portion of the blot was then Coomassie blue-stained to confirm equal protein loading.
Quantitative Import, Translation, and Transcription Assays.
A culture of a given strain was grown to logarithmic phase and treated as indicated for 30 min. After 30 min, a mixture of 15 tritiated amino acids (Amersham Pharmacia) was added to the cultures and they were grown for an additional 30 min. For import measurements, a portion of the culture was vacuum harvested onto paper filters (Whatman) and washed with the medium in which the culture was grown. Filters were then dried by oven baking. The remainder of the culture was then pelleted, and total protein was trichloroacetic acid precipitated, vacuum harvested onto glass microfiber filters (Whatman), and washed with 10% TCA followed by ethanol. Radioactivity on the two filters containing total protein or whole cells was measured on a scintillation counter (Beckman–Coulter LS 6500). The percentage of remaining translation represents cpm from treated samples divided by cpm from untreated counterparts. Quantitative transcription assays were performed in the same manner except that tritiated adenine (Amersham Pharmacia) was used as a label instead of tritiated amino acids.
Results
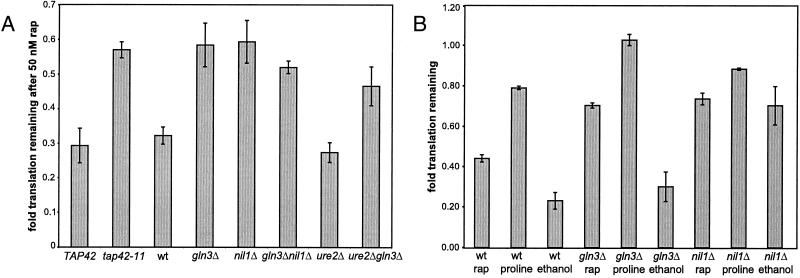
A widely studied effect of rapamycin is its ability to down-regulate translation in organisms ranging from yeast to mammals. We used tritiated amino acids to measure rapamycin-induced translation inhibition in S. cerevisiae strains with mutant or deleted effectors (TAP42, GLN3, NIL1, and URE2) of the Tor proteins. In wild-type cells of two different backgrounds (S288C and W303) growing in a rich medium, rapamycin reduced translation to ≈30% of normal levels, whereas in a tap42-11 strain, a strain resistant to rapamycin-induced growth arrest (14), the effects of rapamycin were significantly diminished (Fig. 1A). Surprisingly, although Gln3p and Nil1p are transcription factors that have not been implicated in the upstream regulation of translation, gln3Δ and nil1Δ cells were as rapamycin resistant as tap42-11 cells (Fig. 1A). GLN3 is epistatic to URE2, a gene encoding a cytoplasmic anchor protein that binds Gln3p, with respect to translation because ure2Δ gln3Δ cells are as rapamycin resistant as gln3Δ cells (Fig. 1A).
Figure 1.
The Tor proteins regulate translation through Gln3p and Nil1p. (A) Strains CY4927 (wild type, wt), CY4928 (tap42-11), PM38 (wild type), PM71 (gln3Δ), MS221 (nil1Δ), PH2 (ure2Δ), and P40-1A (gln3Δ ure2Δ) were treated with 50 nM rapamycin for 30 min and then labeled with tritiated amino acids for another 30 min. After the second 30-min period, total protein was trichloroacetic acid precipitated and counted. Fold translation remaining is defined as counts in the rapamycin-treated samples divided by counts in the untreated samples, normalized for cell density. Error bars span 1 SD about the mean for at least three experiments performed. (B) The indicated strains were grown to logarithmic phase in a synthetic glucose with ammonium sulfate medium (SD + AS), pelleted, and resuspended in water. These suspensions were added to either fresh SD + AS medium, SD + AS medium containing 50 nM rapamycin (rap), synthetic glucose with proline medium (proline), or synthetic ethanol with ammonium sulfate medium (ethanol). The remainder of the assay was performed as in A. Normalized translation was calculated relative to the SD + AS sample and the error bars shown span 1 SD about the mean of three experiments performed.
Shifting from high- to low-quality nitrogen caused a mild decrease in translation while shifting from high- to low-quality carbon caused a strong decrease in translation (Fig. 1B). Notably, gln3Δ cells resist the reduction of translation induced by shifting to low-quality nitrogen (yet have little resistance to the effects of low-quality carbon), whereas nil1Δ cells resist full reduction of translation induced by shifting to low-quality carbon (yet have little resistance to low-quality nitrogen) (Fig. 1B). These data are in agreement with the proposal that low-quality nitrogen and carbon preferentially activate distinct transcriptional branches downstream of the Tor proteins, one involving Gln3p and the other involving Nil1p, respectively (3).
To ensure that varying levels of radiolabeled amino acid import did not confound the translation measurements, we simultaneously measured total cellular uptake of the radioactive label and calculated translation per amino acid imported. These data indicate that rapamycin-induced inhibition of translation is not because of reduction in amino acid import. Of the amino acids entering a cell, >2-fold fewer are incorporated into polypeptides following treatment with rapamycin (Table 1). Although the gln3Δ and nil1Δ strains translated less efficiently per amino acid imported than wild-type cells under normal conditions, this translation was more resistant to treatment with rapamycin or poor nutrients than wild-type cells (Table 1). Also, cells shifted from high- to low-quality carbon experienced a significant reduction in amino acid import, to which nil1Δ cells are resistant (Table 1). The resistance of nil1Δ cells to the reduction in translation caused by shifting from high- to low-quality carbon is due to both elevated import and greater translation per amino acid imported compared with wild type (Table 1).
Table 1.
Relative amino acid import and transcription per amino acid imported in wild-type, gln3, and nil1 strains
| Strain and condition | Import remaining, % | t/i, % |
|---|---|---|
| wt nt | — | 91 ± 5 |
| wt rap | 100 ± 6 | 40 ± 1 |
| wt proline | 108 ± 7 | 68 ± 7 |
| wt ethanol | 49 ± 9 | 42 ± 5 |
| gln3Δ nt | — | 66 ± 11 |
| gln3Δ rap | 87 ± 11 | 53 ± 5 |
| gln3Δ proline | 97 ± 11 | 72 ± 14 |
| gln3Δ ethanol | 54 ± 13 | 39 ± 10 |
| nil1Δ nt | — | 74 ± 8 |
| nil1Δ rap | 106 ± 8 | 52 ± 5 |
| nil1Δ proline | 114 ± 5 | 58 ± 8 |
| nil1Δ ethanol | 104 ± 19 | 51 ± 3 |
Relative amino acid import and the percentage of amino acids translated per amino acid imported (t/i) were calculated by measuring cellular import with the same cells used in Fig. 1B. Ranges given span 1 SD about the mean of three experiments performed. wt, wild type; rap, rapamycin.
To ensure that different mRNA levels in the strains did not confound the translation measurements, either in the steady state or after treatment with rapamycin, mRNA levels were measured in control and treated cells (30 and 60 min after addition of rapamycin) in wild-type, gln3Δ, and nil1Δ cells. This serves to discriminate changes in translation being due to changes in initiation or elongation vs. changes in total cellular mRNA content. To quantitate mRNA levels, we purified mRNA using beads linked to poly(dT) and measured the amount of RNA that bound to the beads after washing. Because total mRNA levels were comparable in the steady state across strains and were essentially unchanged after treatment with rapamycin (data not shown), we conclude that the resistance of gln3Δ and nil1Δ with respect to the translational repression (Fig. 1A) is not due to elevated levels of mRNA, but due to differences in translation itself. These data are also consistent with the fact that treatment with rapamycin leads to the transcriptional repression of components involving RNA polymerase (Pol) I and Pol III, but not Pol II (3).
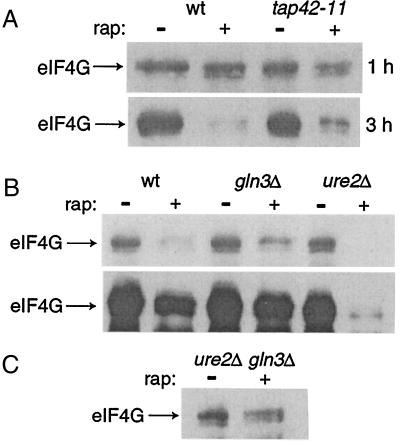
Although other translation factors maintain constant protein levels, eIF4G (encoded by the TIF4631 and TIF4632 genes) is degraded after treatment with rapamycin (15). The synthesis of eIF4G is also repressed after treatment with rapamycin (3, 16). This dual regulation serves as a means to repress translation by decreasing the abundance of the translation initiation complex. Because Tap42p and GATA-type transcription factors control translation (Fig. 1A), we reasoned that these proteins might also control eIF4G levels. As hypothesized, the loss of eIF4G upon treatment with rapamycin is impaired in tap42-11 cells (Fig. 2A). We examined eIF4G down-regulation in gln3Δ and ure2Δ strains and observed that a gln3Δ strain was also partially resistant to rapamycin-induced eIF4G loss (Fig. 2B Upper). This pattern therefore mirrors the sensitivity of translation to rapamycin. Overexposure of the same blot showed that a ure2Δ strain was hypersensitive to rapamycin-induced eIF4G loss (Fig. 2B Lower). Cells that lack URE2 are hypersensitive to rapamycin with respect to growth inhibition (10, 11). To test whether the same epistatic relationship seen in translation holds, we examined eIF4G loss in the strain harboring the double deletion ure2Δ gln3Δ. As before, GLN3 is epistatic to URE2 because the ure2Δ gln3Δ strain also showed resistance to rapamycin-induced eIF4G loss (Fig. 2C). Tap42p and Gln3p regulate eIF4G (TIF4631) transcription (ref. 3 and data not shown) and these data likely reflect diminished transcriptional repression of TIF4631 in tap42-11 or gln3Δ strains. Tap42p and Gln3p may also regulate eIF4G degradation since eIF4G is thought be degraded in the vacuole (15) and the induction of several vacuolar proteases after treatment with rapamycin requires TAP42 and GLN3 (3, 17).
Figure 2.
The Tor proteins regulate levels of eIF4G through Gln3p and Nil1p. (A) Strains CY4927 (wild type, wt) and CY4928 (tap42-11) were grown to logarithmic phase and treated with 50 nM rapamycin (rap) for either 1 or 3 h. Cells were lysed and total protein isolated was quantitated. Equal amounts of protein were separated by SDS/PAGE and visualized by immunoblotting with anti-eIF4G Abs. (B) Strains PM38 (wild type), PM71 (gln3Δ), and PH2 (ure2Δ) were grown to logarithmic phase and treated with 50 nM rapamycin for 3 h and processed as in A. (Upper) The resistance of a gln3Δ strain to rapamycin-induced eIF4G loss. (Lower) An overexposure of the identical blot to illustrate the hypersensitivity of a ure2Δ strain to rapamycin-induced eIF4G loss. (C) The double deletion strain gln3Δ ure2Δ (P40-1A) was treated and processed as in B.
When cells lack a high-quality nitrogen source or are treated with rapamycin, Gln3p and Nil1p promote the transcription of nitrogen discrimination pathway target genes. When cells lack a high-quality carbon/nitrogen source or are treated with rapamycin, they also transcriptionally down-regulate many genes involved in translation (3). Using data from whole-genome transcription profiling (3), we examined the down-regulation of these genes in gln3Δ or nil1Δ strains. To increase the statistical power of our comparisons, we computed the geometric means of gene repressions within a functional class (ranging from 7 to 33 genes), as defined by the Munich Information Center for Protein Sequences classification scheme (18). Interestingly, the normal repressions of genes after treatment with rapamycin, such as those involved in translation initiation, ribosomal biogenesis, rRNA processing, RNA Pol I, and RNA Pol III, were diminished in a gln3Δ or nil1Δ strain (Table 2). As a control, steady-state levels of these transcripts were examined in wild-type, gln3Δ, or nil1Δ strains and no significant differences were found (Table 2). These data are consistent with Gln3p and Nil1p regulating translation through transcription.
Table 2.
Transcriptional repression of genes involved in translation in wild-type, gln3, and nil1 strains
| Functional class | wt vs. wt + rap | gln3Δ vs. gln3Δ + rap | nil1Δ vs. nil1Δ + rap |
|---|---|---|---|
| Translation initiation (27 genes) | −2.2 | −2.0 (−1.0) | −1.5 (+1.2) |
| Ribosome biogenesis (12 genes) | −5.8 | −3.5 (−1.1) | −3.2 (+1.1) |
| rRNA processing (33 genes) | −4.8 | −3.5 (−1.1) | −3.1 (+1.1) |
| RNA Pol I (7 genes) | −8.2 | −5.0 (−1.1) | −6.1 (+1.1) |
| RNA Pol III (10 genes) | −3.2 | −2.5 (+1.0) | −2.1 (+1.3) |
Using publicly available, downloadable data (3), the geometric means of the sets listed were computed for PM38 (wild-type), PM71 (gln3), and MS221 (nil1) strains treated with 50 nM rapamycin for 30 min. The number of genes in each set is indicated beside the functional class in parentheses and the gene names within each set were previously described (3). To control for differences between the strains, the steady-state differences in transcript levels (the same geometric mean calculation as above) of the gln3 and nil1 strains were computed relative to wild-type and listed in parentheses besides the fold change caused by treatment with rapamycin. wt, wild type; rap, rapamycin.
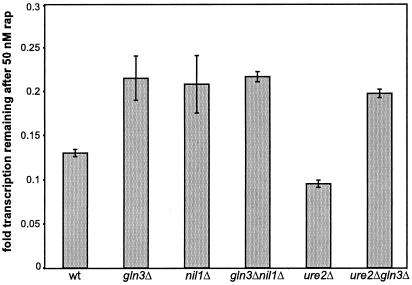
If Gln3p and Nil1p play a role in processes like Pol I or Pol III gene transcription, as the transcription profiling data suggest, then biochemical measurements of Pol I and Pol III activity should be affected in a gln3Δ or nil1Δ strain treated with rapamycin compared with wild type treated with rapamycin. It has previously been shown that the Tor proteins control Pol I- and Pol III-dependent transcription (16, 19), which accounts for the majority of transcription in a logarithmic-phase cell. Using tritiated adenine as a label for transcription, we determined that in rich media, rapamycin decreases bulk transcription to ≈15% of normal levels (Fig. 3). This is therefore an even more pronounced effect than seen with translation, which decreases to ≈30% of normal levels upon treatment with rapamycin. We examined this effect in gln3Δ, nil1Δ, and ure2Δ strains and found a pattern similar to that seen with translation and eIF4G degradation. Cells that are gln3Δ or nil1Δ are resistant to down-regulation of Pol I/III activity (Fig. 4A). These effects were also confirmed in a synthetic medium (data not shown). The resistance is only partial, because treatment of gln3Δ or ure2Δ gln3Δ cells with rapamycin reduced transcription to ≈25% of normal levels, above the 13% seen with wild-type cells (Fig. 3). Although complexing the Tor proteins with Fpr1p-rapamycin leads to Gln3p/Nil1p- and Pol II-dependent activation of some genes over 20-fold (5, 10, 11), the same ternary complexes result in Gln3p/Nil1p-dependent repression of Pol I/III transcription.
Figure 3.
The Tor proteins regulate Pol I/III transcription through Gln3p and Nil1p. Strains PM38 (wild type, wt), PM71 (gln3Δ), MS221 (nil1Δ), PH2 (ure2Δ), and P40-1A (gln3Δ ure2Δ) were treated with 50 nM rapamycin for 30 min and then labeled with tritiated adenine for another 30 min. After the second 30-min period, total RNA was trichloroacetic acid precipitated and counted. Fold translation remaining is defined as counts in the rapamycin-treated samples divided by counts in the untreated samples, normalized for cell density. Error bars span 1 SD about the mean for at least three experiments performed.
Figure 4.
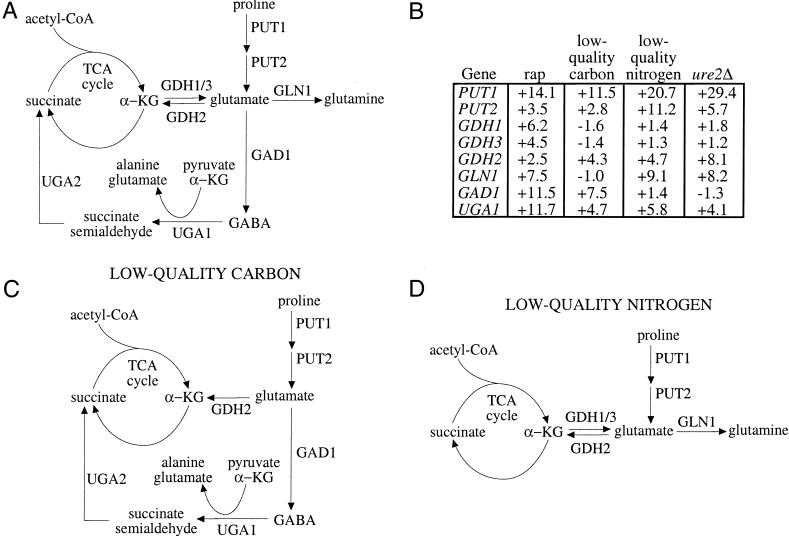
A diagram for the flux of metabolites around the TCA cycle under different nutrient conditions. (A) Shown is a partial map of the TCA cycle, with intermediates succinate and α-ketoglutarate (α-KG). Reactions that produce metabolites that can flow into the TCA cycle or glutamine synthesis are shown. (B) Some of the genes depicted in A are listed in a table along with their induction values after treatment with 50 nM rapamycin for 30 min, low-quality carbon (ethanol) for 30 min, low-quality nitrogen (proline) for 30 min, or in a ure2Δ strain compared with wild type in steady state. Data are taken from publicly available, downloadable whole-genome transcription profiling experiments (3). (C) Based on the gene inductions from B, a flow of metabolites is depicted where positive gene inductions are interpreted as increased flow through the reaction catalyzed by that gene. Shown is the proposed flow of metabolites when cells are shifted from glucose to ethanol, a high- to low-quality carbon source shift, using data from B. (D) Shown is the proposed flow of metabolites when cells are shifted from glutamine to proline, a high- to low-quality nitrogen source shift, using data from B. GABA, γ-aminobutyric acid.
Discussion
This study identifies the GATA-type transcription factors Gln3p and Nil1p as effectors of the Tor proteins in controlling translation (Fig. 1). It is evident that these transcription factors, which have been studied chiefly in their regulation of nitrogen metabolism, have broader functions than previously understood. Transcriptional changes mediated by activated forms of these proteins result in the repression of translation, consistent with the fact that these transcription factors are activated when environmental conditions are harsh for the organism. This translational repression probably occurs secondary to a wide variety of transcriptional effects, such as repression of Pol I/III and translation initiation factor genes. (It is formally possible that the GATA-type factors also directly interact with the translation apparatus, but this seems unlikely.) Our data offer a hypothesis addressing why deleting GLN3 or NIL1 can confer rapamycin resistance for growth. Because these proteins are endowed with control over translation and because a reduction in translation has been shown to be the mechanism of the inhibitory effects of rapamycin on cell proliferation (2), Gln3p and Nil1p can control cell growth. These data further suggest that the many gene deletions which confer rapamycin resistance (20) should be examined with regard to rapamycin-induced translation repression. It may be that translation is the process uniting many seemingly disparate genes to Tor protein function.
Our data further suggest that Gln3p and Nil1p are preferentially activated by distinct nutrient signals, supporting the idea that the Tor proteins are multichannel processors, selectively activating different effectors depending on the precise nutrient input signal. This is consistent with the proposal that GATA-type transcription factors control distinct (although overlapping) sets of genes and that low-quality carbon or nitrogen activates Nil1p or Gln3p differentially (3). This therefore represents a biochemical demonstration of the existence of two branches downstream of Ure2p that were originally identified transcriptionally. It is also consistent with the proposal that Gln3p activity is repressed by intracellular glutamine and that Nil1p activity is repressed by intracellular glutamate (21). Combining these observations, a model emerges that offers a rationale for why the Tor proteins inhibit different GATA-type transcription factors with distinct functions.
In this model, the cell responds to low-quality carbon (which implies a future energy-poor state) by making intermediates that can serve as substrates for the tricarboxylic acid (TCA) cycle. This occurs because when cells lack a high-quality carbon source, intracellular glutamate concentration is thought to be reduced. Although this remains to be measured, this reduction can be anticipated as the cell oxidizes alternative fuels such as α-ketoglutarate (derived from glutamate) and succinate to meet energy needs. According to an earlier proposal, a low intracellular glutamate concentration would cause Nil1p activation (21). The set of genes that are controlled by Nil1p includes PUT1, PUT2, UGA1, NIL1, and PRB1 (3). Put1p (proline oxidase) and Put2p (P5C dehydrogenase) convert proline to glutamate (Fig. 4A). Glutamate can be deaminated by Gdh2p to produce α-ketoglutarate or can be decarboxylated by Gad1p to produce α-aminobutyric acid (Fig. 4A). (GAD1 transcription is up-regulated by treatment with rapamycin but not by deleting URE2 (Fig. 4B), indicating that the Tor proteins likely regulate this gene via a distinct pathway.) Uga1p (4-aminobutyrate aminotransferase), whose expression is Nil1p dependent, deaminates γ-aminobutyric acid to succinate semialdehyde. Succinate semialdehyde is further metabolized by Uga2p (also called Uga5p) to produce succinate (Fig. 4A) (22). When cells are deprived of glucose, these genes are induced according to a pattern strikingly consistent with production of the TCA cycle intermediates, αketoglutarate and succinate (Fig. 4 B and C).
When cells lack a high-quality nitrogen source, intracellular glutamine concentration is low, which causes Gln3p activation (21). The genes with high Gln3p dependence are aimed at the production of glutamine and not at providing TCA cycle intermediates (Fig. 4D). Gln3p-dependent genes like GLN1 are induced by low-quality nitrogen and not by low-quality carbon (Fig. 4B). This is in contrast with GAD1, which is induced by low-quality carbon and not by low-quality nitrogen (Fig. 4B). Such fulcrum points could serve to change the flux of metabolites to generate either glutamine or TCA cycle intermediates. Low-quality nitrogen induces genes that are also up-regulated by low-quality carbon (3), perhaps to synthesize glutamate that Gln1p will convert to glutamine (23). Notably, treatment with rapamycin induces genes up-regulated by low-quality nitrogen but not by low-quality carbon (such as GLN1) as well as genes up-regulated by low-quality carbon but not by low-quality nitrogen (such as GAD1), consistent with previous findings that rapamycin resembles a superposition of the low-quality carbon and nitrogen states (3).
In summary, it appears as if carbon- and nitrogen-quality signal preferentially through the multiprocessing Tor proteins to Nil1p and Gln3p, respectively. This has two main effects—the first being translational control, likely secondary to transcriptional effects on components of the translational apparatus (Table 2 and Fig. 5A). Second, low-quality carbon and nitrogen result in distinct patterns of gene inductions that resemble feedback loops to the cellular states that regulate Nil1p and Gln3p activity—energy (glutamate) and nitrogen (glutamine) status, respectively (Fig. 5B). Studies of the molecular basis of the signal processing performed by the Tor proteins will be essential to elucidating this nutrient-sensing network.
Figure 5.
(A) A model for how the Tor proteins regulate Nil1p and Gln3p using the cytoplasmic anchor protein Ure2p and Tap42p/phosphatase. This regulation is differential, depending on the quality of available carbon and nitrogen sources. (B) When Nil1p is activated, the genes that are up-regulated suggest that the cell is trying to generate energy via production of TCA cycle intermediates. (PRB1 is a vacuolar, broad-specificity protease that is envisioned to supply amino acids like proline or glutamate.) When Gln3p is activated, the genes that are up-regulated suggest that the cell is trying to collect alternative nitrogen sources to synthesize glutamine. Low-quality nitrogen up-regulates genes regulated by Nil1p, perhaps as a source of glutamate from which to make glutamine. Thus, the sets of genes controlled by Nil1p and Gln3p are overlapping. Shown in blue or green are genes primarily dependent on Nil1lp or Gln3p, respectively.
Acknowledgments
We are grateful to Nilesh Kumar and Rebecca Butcher for helpful comments. We thank Charles Di Como, Alan Sachs, and Hans Trachsel for yeast strains and antibodies. F.G.K. was supported by the National Institutes of Health Medical Scientist Training Program. A.F.S. was supported by the Harvard College Research Program. S.L.S. is an Investigator at the Howard Hughes Medical Institute. This research was funded by National Institute of General Medical Sciences Grant GM-38627.
Abbreviations
- Tor
target of rapamycin
- Pol
RNA polymerase
- TCA
tricarboxylic acid
References
- 1.Kuruvilla F G, Schreiber S L. Chem Biol. 1999;6:R129–R36. doi: 10.1016/S1074-5521(99)80070-2. [DOI] [PubMed] [Google Scholar]
- 2.Barbet N C, Schneider U, Helliwell S B, Stansfield I, Tuite M F, Hall M N. Mol Biol Cell. 1996;7:25–42. doi: 10.1091/mbc.7.1.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shamji A F, Kuruvilla F G, Schreiber S L. Curr Biol. 2000;10:1574–1581. doi: 10.1016/s0960-9822(00)00866-6. [DOI] [PubMed] [Google Scholar]
- 4.Park H D, Beeser A E, Clancy M J, Cooper T G. Yeast. 1996;12:1135–1151. doi: 10.1002/(sici)1097-0061(19960915)12:11<1135::aid-yea11>3.0.co;2-l. [DOI] [PubMed] [Google Scholar]
- 5.Hardwick J S, Kuruvilla F G, Tong J K, Shamji A F, Schreiber S L. Proc Natl Acad Sci USA. 1999;96:14866–14870. doi: 10.1073/pnas.96.26.14866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Coschigano P W, Magasanik B. Mol Cell Biol. 1991;11:822–832. doi: 10.1128/mcb.11.2.822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cooper T G, Ferguson D, Rai R, Bysani N. J Bacteriol. 1990;172:1014–1018. doi: 10.1128/jb.172.2.1014-1018.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stanbrough M, Magasanik B. J Bacteriol. 1995;177:94–102. doi: 10.1128/jb.177.1.94-102.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Beck T, Hall M N. Nature (London) 1999;402:689–692. doi: 10.1038/45287. [DOI] [PubMed] [Google Scholar]
- 10.Cardenas M E, Cutler N S, Lorenz M C, Di Como C J, Heitman J. Genes Dev. 1999;13:3271–3279. doi: 10.1101/gad.13.24.3271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bertram P G, Choi J H, Carvalho J, Ai W, Zeng C, Chan T F, Zheng X F. J Biol Chem. 2000;275:35727–35733. doi: 10.1074/jbc.M004235200. [DOI] [PubMed] [Google Scholar]
- 12.Xu S, Falvey D A, Brandriss M C. Mol Cell Biol. 1995;15:2321–2330. doi: 10.1128/mcb.15.4.2321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hofman-Bang J. Mol Biotechnol. 1999;12:35–73. doi: 10.1385/MB:12:1:35. [DOI] [PubMed] [Google Scholar]
- 14.Di Como C J, Arndt K T. Genes Dev. 1996;10:1904–1916. doi: 10.1101/gad.10.15.1904. [DOI] [PubMed] [Google Scholar]
- 15.Berset C, Trachsel H, Altmann M. Proc Natl Acad Sci USA. 1998;95:4264–4269. doi: 10.1073/pnas.95.8.4264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Powers T, Walter P. Mol Biol Cell. 1999;10:987–1000. doi: 10.1091/mbc.10.4.987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Coffman J A, Cooper T G. J Bacteriol. 1997;179:5609–5613. doi: 10.1128/jb.179.17.5609-5613.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mewes H W, Heumann K, Kaps A, Mayer K, Pfeiffer F, Stocker S, Frishman D. Nucleic Acids Res. 1999;27:44–48. doi: 10.1093/nar/27.1.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zaragoza D, Ghavidel A, Heitman J, Schultz M C. Mol Cell Biol. 1998;18:4463–4470. doi: 10.1128/mcb.18.8.4463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chan T F, Carvalho J, Riles L, Zheng X F. Proc Natl Acad Sci USA. 2000;97:13227–13232. doi: 10.1073/pnas.240444197. . (First Published November 14, 2000, 10.1073/pnas.240444197) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stanbrough M, Rowen D W, Magasanik B. Proc Natl Acad Sci USA. 1995;92:9450–9454. doi: 10.1073/pnas.92.21.9450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Coleman S T, Fang T K, Rovinsky S A, Turano F J, Moye-Rowley W S. J Biol Chem. 2001;276:244–250. doi: 10.1074/jbc.M007103200. [DOI] [PubMed] [Google Scholar]
- 23.Komeili A, Wedaman K P, O'Shea E K, Powers T. J Cell Biol. 2000;151:863–878. doi: 10.1083/jcb.151.4.863. [DOI] [PMC free article] [PubMed] [Google Scholar]