Abstract
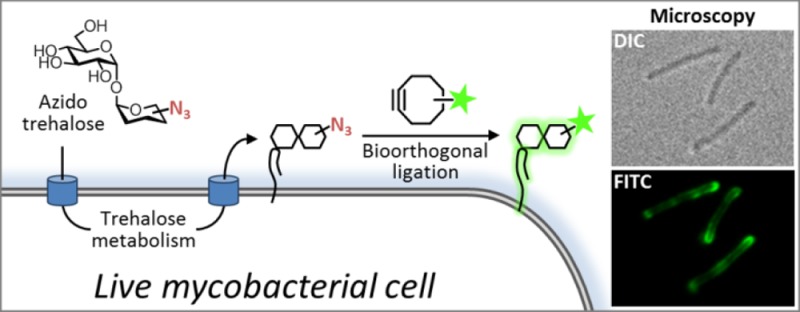
Mycobacteria, including the pathogen Mycobacterium tuberculosis, use the non-mammalian disaccharide trehalose as a precursor for essential cell-wall glycolipids and other metabolites. Here we describe a strategy for exploiting trehalose metabolic pathways to label glycolipids in mycobacteria with azide-modified trehalose (TreAz) analogues. Subsequent bioorthogonal ligation with alkyne-functionalized probes enabled detection and visualization of cell-surface glycolipids. Characterization of the metabolic fates of four TreAz analogues revealed unique labeling routes that can be harnessed for pathway-targeted investigation of the mycobacterial trehalome.
Mycobacterium tuberculosis (Mtb), the causative agent of tuberculosis, currently infects 2 billion people worldwide and causes approximately 2 million deaths annually.1 The success of Mtb as a pathogen is in large part due to its complex cell wall, which is a formidable barrier to antibiotics and residence to many biomolecules that are directly involved in pathogenesis. The mycobacterial cell wall features a unique outer membrane—the “mycomembrane” (MM)—that is composed of long-chain (C60–C90) mycolic acids covalently bound to the underlying peptidoglycan–arabinogalactan polymer, as well as an array of additional free intercalating lipids and glycolipids.2,3
Trehalose-containing glycolipids are abundant, virulence-associated constituents of the MM that have essential roles in cell-wall biosynthesis and disease progression. Trehalose mono- and dimycolate (TMM and TDM), bearing 6-O- and 6,6′-di-O-mycolyl substituents, respectively, are produced by all mycobacterial species (Figure 1A). TMM is an essential mediator of MM biosynthesis4−6 (see Figure 2) and is also the precursor to TDM, an immunomodulatory molecule that promotes Mtb infectivity and survival within host macrophages.7−9 Along with TMM and TDM, numerous other complex metabolites constitute the mycobacterial “trehalome.” Unfortunately, the various roles played by these metabolites in Mtb physiology and pathogenesis are challenging to study using traditional genetic and biochemical methods, which generally require laborious radiolabeling, extraction, and purification procedures that are incompatible with in vivo experimentation.
Figure 1.
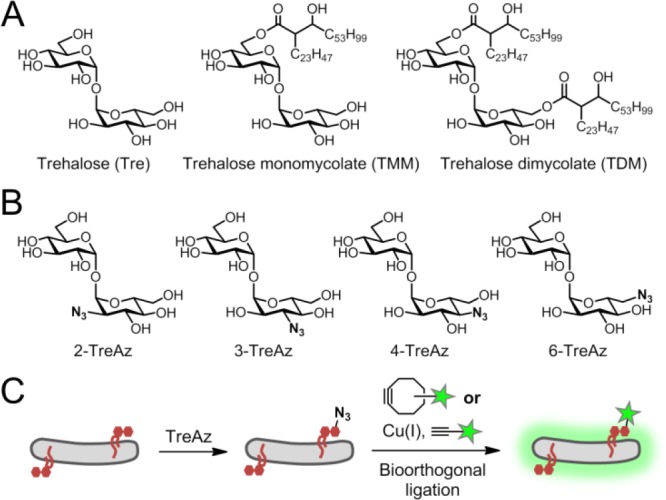
(A) Common trehalose glycolipids in mycobacteria. (B) Synthetic TreAz analogues used in this study. (C) TreAz-based bioorthogonal chemical reporter strategy.
Figure 2.
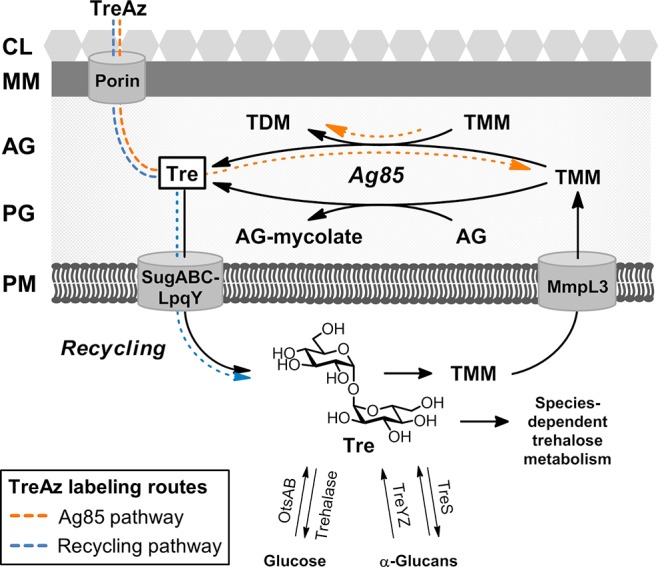
Trehalose metabolism in mycobacteria. Exogenous TreAz can label glycolipids via the Ag85 or recycling pathways. AG, arabinogalactan; CL, capsular layer; MM, mycomembrane; PG, peptidoglycan; PM, plasma membrane. Exact extracellular location of Ag85 is unknown.
Metabolic labeling with unnatural sugar substrates is a powerful alternative for investigating glycoconjugates in living organisms.10 Here we report that trehalose glycolipids can be metabolically labeled with azide-modified trehalose (TreAz) analogues (Figure 1B) in live mycobacteria, enabling bioorthogonal ligation with alkyne-functionalized fluorescent probes (Figure 1C). We capitalized on the conserved pathways for mycobacterial trehalose metabolism shown in Figure 2. While trehalose glycolipids reside in the MM, they originate in the cytoplasm, where free trehalose is synthesized through metabolic cycles involving either glucose (via the OtsAB/trehalase enzymes) or α-glucans (via the TreYZ/TreS enzymes).11−14 Trehalose and mycolic acid combine in the cytoplasm to form TMM, which is then translocated across the plasma membrane by MmpL3.5,6 Subsequently, the antigen 85 (Ag85) complex mediates the transfer of mycolate from TMM to either arabinogalactan, which forms covalently bound mycolates that make up the foundation of the MM, or to another molecule of TMM, which generates TDM.15,16 Both processes release free trehalose, which is recycled by the trehalose-specific transporter SugABC-LpqY.4 Species-dependent metabolic pathways (not shown in detail) can generate a range of additional metabolites in Mtb and other mycobacteria.
In recent work, Backus et al. demonstrated that a fluorescein-conjugated keto-trehalose analogue (FITC-Tre) is incorporated into TDM via the Ag85 complex.17 This observation underscores one possible route by which unnatural substrates might access trehalose glycolipids: after crossing through the MM, likely by a porin-mediated process,18 the unnatural analogue could be processed by Ag85 and incorporated into TMM or TDM outside of the cell (Figure 2, orange dotted arrow). Alternatively, labeling could occur via the trehalose recycling pathway, in which analogues would be internalized by the SugABC-LpqY transporter and incorporated into glycolipids from the inside-out (Figure 2, blue dotted arrow). Access to the recycling pathway has not been reported for any chemical probes to date but is essential for investigating processes that originate in the cytoplasm, such as de novo biosynthesis of glycolipids and MM, as well as species-dependent trehalose metabolism.
We hypothesized that TreAz analogues, in which a hydroxyl group is replaced with a relatively small, minimally perturbing azido group, would be well-tolerated by the mycobacterial biosynthetic machinery, allowing unprecedented access to the trehalose recycling pathway. In addition, the modular nature of bioorthogonal ligation affords numerous advantages over other labeling strategies, including choice of bioorthogonal reaction and probe type, temporal control of probe delivery (permitting pulse-chase biological experiments19), and the ability to minimize background signal by use of low probe concentrations.
We synthesized a series of trehalose analogues containing azido groups at all possible positions with native stereochemistry [Figure 1B; refer to the Supporting Information (SI) for schemes and procedures]. The compounds were initially evaluated for metabolic incorporation into glycolipids in the model organism M. smegmatis mc2155 (Msmeg). Briefly, bacteria were cultured with TreAz until logarithmic phase (typically 12–16 h), then reacted with BARAC-Fluor (1 μM, 30 min), a fluorescein-conjugated biarylazacyclooctynone used for rapid Cu-free click chemistry.20 Analysis by flow cytometry revealed that all four TreAz analogues labeled Msmeg, albeit with different efficiencies (Figure 3A). 2- and 6-TreAz labeled cells robustly at low concentrations (5–25 μM), while labeling with 3- and 4-TreAz required higher concentrations (250–500 μM). At these doses, no changes in bacterial growth were observed. The described flow cytometry assay was also used to evaluate the dependence of Msmeg labeling on analogue concentration, culture time, and exogenously added free trehalose (i.e., competition studies; Figures S1–S3).
Figure 3.
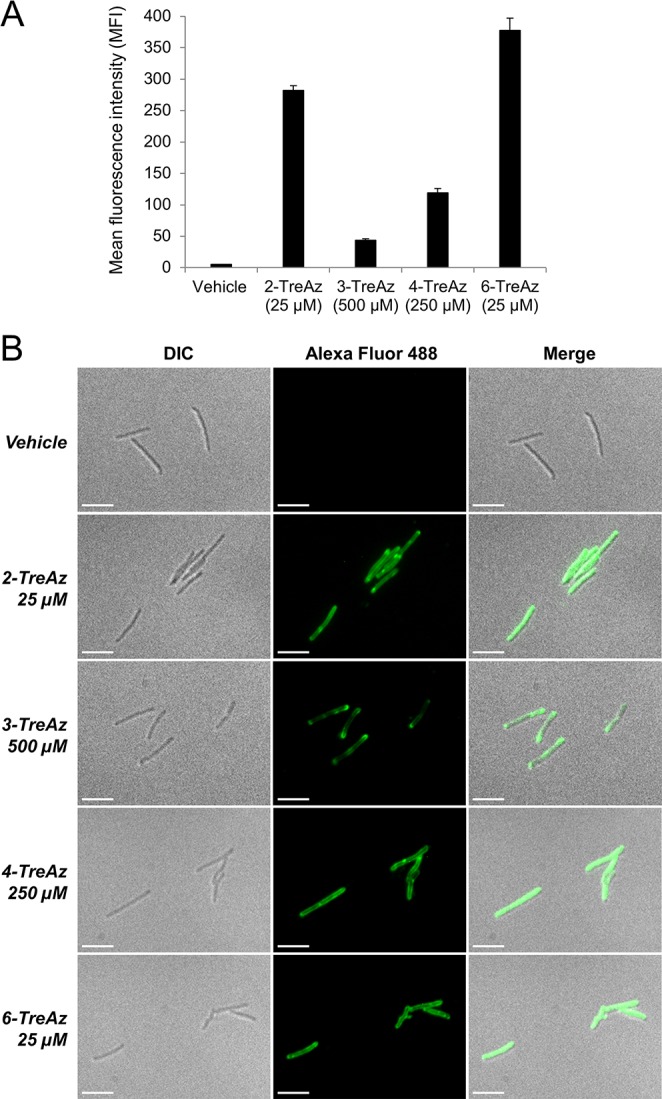
(A) Flow cytometry analysis of TreAz-labeled Msmeg reacted with BARAC-Fluor. Error bars denote the standard deviation of three replicate experiments. (B) Fluorescence microscopy of TreAz-labeled Msmeg reacted with alk-AF488 via CuAAC. Scale bars, 5 μm.
Next, fluorescence microscopy was used to visualize azide-bearing cell-surface glycolipids (Figure 3B). Msmeg was cultured with TreAz, fixed, and then reacted with alkyne-functionalized Alexa Fluor 48821 (alk-AF488) via Cu(I)-catalyzed azide–alkyne cycloaddition (CuAAC).22,23 For all TreAz analogues, azide-specific fluorescence was observed uniformly throughout the bacterial population, and signal was predominantly localized to the cell surface, which is in agreement with glycolipids inhabiting the MM. Interestingly, fluorescence was frequently concentrated at the bacterial poles, consistent with a polar growth model for mycobacteria.24
In addition to fluorescence detection, we directly characterized TreAz-labeled glycolipids by chemical methods. Analysis of partially purified chloroform–methanol lipid extracts by TLC and high-resolution mass spectrometry confirmed that all four TreAz analogues produced azide-labeled glycolipids in Msmeg (Figure S4). To provide further direct evidence that TreAz was incorporated into glycolipids, petroleum ether lipid extracts from Msmeg cultured with 2- or 6-TreAz (or vehicle) were deacylated by treatment with NaOMe at 60 °C. The released free sugars were analyzed by high-pH anion exchange chromatography with pulsed amperometric detection (HPAEC-PAD), which showed peaks for 2- and 6-TreAz in the corresponding samples but not in vehicle-treated or non-deacylated control samples (Figure S5). Comparison of peak integrations indicated that 2- and 6-TreAz replaced approximately 20% and 70% of natural trehalose, respectively, in surface-exposed glycolipids extracted from TreAz-treated cells.
To establish whether TreAz incorporation into glycolipids occurred via the Ag85 or recycling pathway, azide labeling was evaluated in the Msmeg mutants ΔsugC and ΔlpqY, which lack the trehalose transporter,4 by reaction with BARAC-Fluor followed by flow cytometry (Figure 4A). Fluorescence observed for 2-, 4-, and 6-TreAz in the wild-type strain was completely abolished in the mutants, demonstrating that metabolic labeling of glycolipids using these analogues requires uptake into the cytoplasm and proceeds through the recycling pathway. In contrast, labeling with 3-TreAz was not reduced in the transporter mutants, indicating that this compound likely undergoes extracellular incorporation into glycolipids via Ag85, similar to FITC-Tre.17
Figure 4.
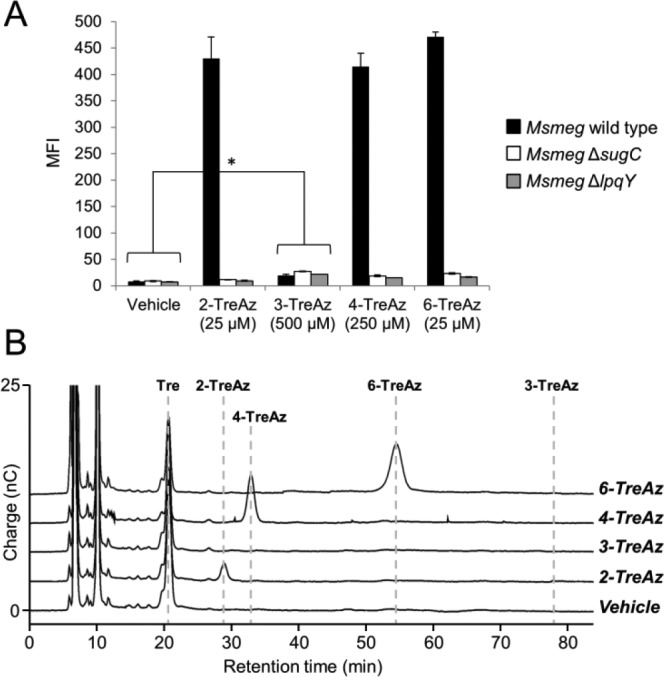
(A) Flow cytometry analysis of TreAz-labeled Msmeg strains. Error bars denote the standard deviation from three replicate experiments. * p < 0.05. (B) HPAEC-PAD analysis of cytosolic extracts from TreAz-treated wild-type Msmeg. Dotted lines represent retention times for authentic standards (see SI for details).
To confirm that 2-, 4-, and 6-TreAz enter the cytoplasm, HPAEC-PAD was used to assess the presence of these analogues in cytosolic extracts. As expected, 2-, 4-, and 6-TreAz were observed in extracts from wild-type Msmeg, while 3-TreAz was not (Figure 4B); identically prepared samples from the ΔsugC mutant showed no cytosolic TreAz (Figure S6). Taken together, these data reveal that our TreAz analogues have the capacity to selectively label trehalose-containing metabolites via the Ag85 or recycling pathways, and thus provide a platform for pathway-targeted probing of the trehalome.
Our finding that 2-, 4-, and 6-TreAz enter the cell through the recycling pathway in Msmeg raises the possibility that these compounds might also be processed by trehalase or TreS (Figure 2). We sought address whether these enzymes affect cell-surface labeling, either by diverting TreAz into cell-surface α-glucan (via TreS) or by degrading TreAz (via trehalase). Msmeg ΔtreS14 and ΔMSMEG_4535 (trehalase) mutants were generated (see SI) and evaluated for TreAz labeling compared to wild-type Msmeg (Figures S7–S11). For the most part, no significant differences were observed in the mutants, ruling out any effects on cell-surface labeling by these enzymes. However, we did observe a TreS-dependency for 4-TreAz labeling in Msmeg (Figure S8). We are currently investigating the intriguing possibility that TreAz analogues are incorporated into cell-surface α-glucan, an experimentally elusive25 cell-wall structure that has been implicated in immune evasion.14,26
Finally, we tested the TreAz analogues in pathogenic mycobacteria. Mtb H37Rv and the closely related avirulent species M. bovis BCG-Pasteur (BCG) were assessed by flow cytometry as described above for Msmeg, and all four compounds led to significant TreAz-dependent fluorescence in both species (Figure 5A). We also generated a BCG mutant ΔlpqY-sugC missing the trehalose transporter and its corresponding complemented strain (see SI) to test the route of TreAz labeling (Figure 5B). Consistent with the results from Msmeg, 2- and 6-TreAz labeling was substantially reduced in the mutant, suggesting that a recycling pathway mechanism for these analogues may be conserved across mycobacterial species. Also in agreement with data from Msmeg, 3-TreAz labeling was unaffected in the mutant, indicating that selective incorporation of this analogue by Ag85 is also conserved. 4-TreAz labeling, which proceeded via the recycling pathway in Msmeg, instead appeared to primarily use Ag85 in BCG. These results confirmed that pathway-targeted TreAz labeling can be extended to other mycobacteria, including pathogenic Mtb.
Figure 5.
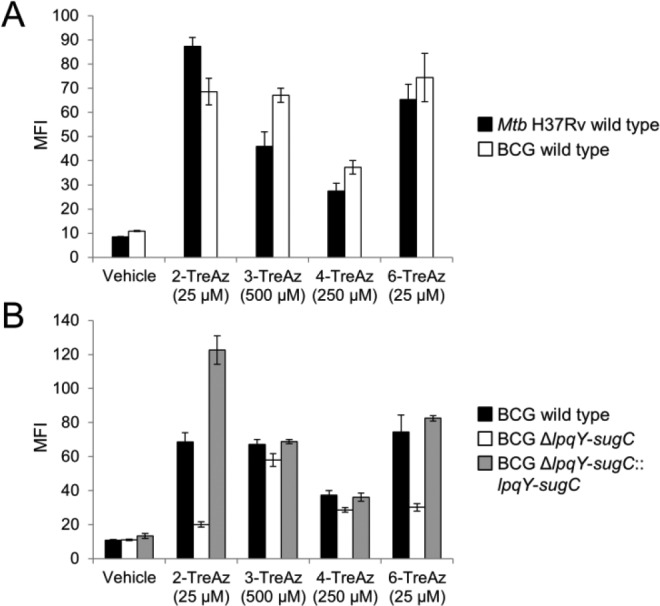
Flow cytometry analysis of TreAz-labeled Mtb and BCG strains. (A) Wild-type Mtb H37Rv and BCG; (B) wild-type BCG, BCG ΔlpqY-sugC, and the corresponding complemented strain. Error bars denote the standard deviation from three replicate experiments.
In summary, metabolic labeling with TreAz enables interrogation of the trehalome in live mycobacteria. We showed that all four TreAz analogues were effective in Msmeg, BCG, and Mtb, and distinct routes of TreAz metabolism—mostly conserved across species—were elucidated through genetic and chemical techniques. This strategy can be used for imaging glycolipid distribution, trafficking, and dynamics as well as metabolite profiling and discovery. As well, the compounds may be employed to assess the effects of various perturbations (e.g., environmental stress, antibiotic treatment, genetic manipulation) on trehalose glycolipids and their associated biosynthetic pathways. We also expect that TreAz analogues will be metabolized in other mycobacterial species given the highly conserved nature of the involved biosynthetic machinery. Importantly, the absence of trehalose metabolism in mammals invites the application of this chemical tool to investigate the trehalome during mycobacterial infection in host cells and model organisms.
Acknowledgments
We thank M. Boyce, M. Breidenbach, and S. Canham for helpful discussions and for critical reading of the manuscript. S. Bauer is acknowledged for providing assistance with HPAEC-PAD instrumentation. This work was supported by a grant to C.R.B. from the NIH (AI51622). B.M.S., J.C.J., and M.S.S. were supported by postdoctoral fellowships from the American Cancer Society, and B.M.S. was also supported by a fellowship from the Center for Emerging and Neglected Diseases. C.M.H. was supported by the UC Davis Office of the Vice Chancellor for Research. R.K. acknowledges support from the Juergen Manchot Foundation.
Supporting Information Available
Experimental procedures, characterization data, supporting figures, schemes, and tables. This material is available free of charge via the Internet at http://pubs.acs.org.
Author Present Address
# Department of Chemistry and Biochemistry, University of Arizona, Tucson, AZ 85721
The authors declare no competing financial interest.
Funding Statement
National Institutes of Health, United States
Supplementary Material
References
- Dye C. Lancet 2006, 367, 938. [DOI] [PubMed] [Google Scholar]
- Brennan P. J.; Crick D. C. Curr. Top. Med. Chem. 2007, 7, 475. [DOI] [PubMed] [Google Scholar]
- Hoffmann C.; Leis A.; Niederweis M.; Plitzko J. M.; Engelhardt H. Proc. Nat. Acad. Sci. U.S.A. 2008, 105, 3963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalscheuer R.; Weinrick B.; Veeraraghavan U.; Besra G. S.; Jacobs W. R. Proc. Nat. Acad. Sci. U.S.A. 2010, 107, 21761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grzegorzewicz A. E.; Pham H.; Gundi V. A.; Scherman M. S.; North E. J.; Hess T.; Jones V.; Gruppo V.; Born S. E.; Kordulakova J.; Chavadi S. S.; Morisseau C.; Lenaerts A. J.; Lee R. E.; McNeil M. R.; Jackson M. Nat. Chem. Biol. 2012, 8, 334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tahlan K.; Wilson R.; Kastrinsky D. B.; Arora K.; Nair V.; Fischer E.; Barnes S. W.; Walker J. R.; Alland D.; Barry C. E. III; Boshoff H. I. Antimicrob. Agents Chemother. 2012, 56, 1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryll R.; Kumazawa Y.; Yano I. Microbiol. Immunol. 2001, 45, 801. [DOI] [PubMed] [Google Scholar]
- Indrigo J.; Hunter R. L.; Actor J. K. Microbiology 2003, 149, 2049. [DOI] [PubMed] [Google Scholar]
- Ishikawa E.; Ishikawa T.; Morita Y. S.; Toyonaga K.; Yamada H.; Takeuchi O.; Kinoshita T.; Akira S.; Yoshikai Y.; Yamasaki S. J. Exp. Med. 2009, 206, 2879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sletten E. M.; Bertozzi C. R. Angew. Chem., Int. Ed. 2009, 48, 6974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Smet K. A. L.; Weston A.; Brown I. N.; Young D. B.; Robertson B. D. Microbiology 2000, 146, 199. [DOI] [PubMed] [Google Scholar]
- Woodruff P. J.; Carlson B. L.; Siridechadilok B.; Pratt M. R.; Senaratne R. H.; Mougous J. D.; Riley L. W.; Williams S. J.; Bertozzi C. R. J. Biol. Chem. 2004, 279, 28835. [DOI] [PubMed] [Google Scholar]
- Murphy H. N.; Stewart G. R.; Mischenko V. V.; Apt A. S.; Harris R.; McAlister M. S. B.; Driscoll P. C.; Young D. B.; Robertson B. D. J. Biol. Chem. 2005, 280, 14524. [DOI] [PubMed] [Google Scholar]
- Kalscheuer R.; Syson K.; Veeraraghavan U.; Weinrick B.; Biermann K. E.; Liu Z.; Sacchettini J. C.; Besra G.; Bornemann S.; Jacobs W. R. Nat. Chem. Biol. 2010, 6, 376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sathyamoorthy N.; Takayama K. J. Biol. Chem. 1987, 262, 13417. [PubMed] [Google Scholar]
- Belisle J. T.; Vissa V. D.; Sievert T.; Takayama K.; Brennan P. J.; Besra G. S. Science 1997, 276, 1420. [DOI] [PubMed] [Google Scholar]
- Backus K. M.; Boshoff H. I.; Barry C. S.; Boutureira O.; Patel M. K.; D’Hooge F.; Lee S. S.; Via L. E.; Tahlan K.; Barry C. E. III; Davis B. G. Nat. Chem. Biol. 2011, 7, 228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niederweis M.; Danilchanka O.; Huff J.; Hoffmann C.; Engelhardt H. Trends Microbiol. 2010, 18, 109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laughlin S. T.; Baskin J. M.; Amacher S. L.; Bertozzi C. R. Science 2008, 320, 664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jewett J. C.; Sletten E. M.; Bertozzi C. R. J. Am. Chem. Soc. 2010, 132, 3688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- AF488 was used instead of fluorescein for imaging experiments due to its improved photostability.
- Rostovtsev V. V.; Green L. G.; Fokin V. V.; Sharpless K. B. Angew. Chem., Int. Ed. 2002, 41, 2596. [DOI] [PubMed] [Google Scholar]
- Tornøe C. W.; Christensen C.; Meldal M. J. Org. Chem. 2002, 67, 3057. [DOI] [PubMed] [Google Scholar]
- Hett E. C.; Rubin E. J. Microbiol. Mol. Biol. Rev. 2008, 72, 126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sani M.; Houben E. N. G.; Geurtsen J.; Pierson J.; de Punder K.; van Zon M.; Wever B.; Piersma S. R.; Jiménez C. R.; Daffé M.; Appelmelk B. J.; Bitter W.; van der Wel N.; Peters P. J. PloS Pathog. 2010, 6, e1000794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambou T.; Dinadayala P.; Stadthagen G.; Barilone N.; Bordat Y.; Constant P.; Levillain F.; Neyrolles O.; Gicquel B.; Lemassu A.; Daffé M.; Jackson M. Mol. Microbiol. 2008, 70, 762. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


