Abstract
Background
The role of microRNAs (miRNAs), a key class of regulators of mRNA expression and translation, in patients with eosinophilic esophagitis (EoE) has not been explored.
Objective
We aimed to identify miRNAs dysregulated in patients with EoE and assess the potential of these miRNAs as disease biomarkers.
Methods
Esophageal miRNA expression was profiled in patients with active EoE and those with glucocorticoid-induced disease remission. Expression profiles were compared with those of healthy control subjects and patients with chronic (noneosinophilic) esophagitis. Expression levels of the top differentially expressed miRNAs from the plasma of patients with active EoE and patients with EoE remission were compared with those of healthy control subjects.
Results
EoE was associated with 32 differentially regulated miRNAs and was distinguished from noneosinophilic forms of esophagitis. The expression levels of the most upregulated miRNAs (miR-21 and miR-223) and the most downregulated miRNA (miR-375) strongly correlated with esophageal eosinophil levels. Bioinformatic analysis predicted interplay of miR-21 and miR-223 with key roles in the polarization of adaptive immunity and regulation of eosinophilia, and indeed, these miRNAs correlated with key elements of the EoE transcriptome. The differentially expressed miRNAs were largely reversible in patients who responded to glucocorticoid treatment. EoE remission induced a single miRNA (miR-675) likely to be involved in DNA methylation. Plasma analysis of the most upregulated esophageal miRNAs identified miR-146a, miR-146b, and miR-223 as the most differentially expressed miRNAs in the plasma.
Conclusions
We have identified a marked dysregulated expression of a select group of miRNAs in patients with EoE and defined their reversibility with glucocorticoid treatment and their potential value as invasive and noninvasive biomarkers.
Keywords: Eosinophilic esophagitis, microRNA, glucocorticoid, biomarkers
MicroRNAs (miRNAs) are single-stranded RNA molecules of 19 to 25 nucleotides in length that mediate posttranscriptional gene silencing of target genes.1 MiRNAs have diverse roles in fundamental biological processes, such as cell proliferation, differentiation, apoptosis, stress response, and immune response, among many others.1 To date, few studies have examined the role of miRNAs in human allergic diseases or esophageal diseases, except for the setting of asthma and esophageal cancers.2–8
Eosinophilic esophagitis (EoE) is an emerging worldwide allergic disease characterized by intense eosinophil infiltration of the esophageal mucosal epithelium that is refractory to acid-suppressive therapy and is associated with chronic symptoms from childhood into adulthood.9,10 First described in the late 1970s, the incidence and prevalence of EoE have been on the increase. It is now a global health disease reported in every continent except Africa.9 Multiple studies have demonstrated that EoE is associated with marked changes in gene expression, particularly in the esophagus, where approximately 1% of the human genome has an altered tissue-specific expression pattern collectively referred to as the EoE transcriptome, which is largely but not fully reversible after disease remission with glucocorticoid therapy.11–14 Furthermore, EoE is a unique opportunity to study inflammatory diseases in human subjects, especially in the pediatric population, because obtaining tissue specimens through endoscopy is routine standard of care and the biopsy material is amenable to molecular analysis.9,13 This approach has uncovered the key interplay of the adaptive and innate immune system, including the key role of IL-13–driven epithelial cell gene responses, including eotaxin-3. In addition to acquired gene expression changes in the esophagus, EoE is also an inherited disease that involves a complex interplay of genetic and environmental factors.14 Most studies concerning the regulation of the EoE transcriptome have focused on the induction and regulation of in situ gene expression by cytokines (eg, IL-13), transcription factors, and coactivators (eg, signal transducer and activator of transcription 6 and CREB-binding protein),12,15,16 yet other regulatory processes, such as miRNAs, have not been explored. MiRNAs represent a particularly attractive class of molecules in the regulation of the EoE transcriptome because a single miRNA can target hundreds of genes and can mediate the epigenetic mechanisms underlying gene-environment interactions, which are likely to have a key but currently unexplored role in EoE.17 Lastly, further interest in studying miRNA involvement in patients with EoE is derived from the recent identification of a key role of a specific TH2-associated miRNA (miR-21) in critically regulating TH cell polarization because EoE clearly involves a local polarized TH2 response.7,18,19 Thus, we aimed to elucidate miRNA expression profiles in patients with active EoE, active chronic esophagitis, EoE in remission after glucocorticoid treatment who have a mainly normalized EoE transcriptome,12,20 and healthy control subjects.
METHODS
Human esophageal tissues
Patients were selected without regard to age, race, or sex. Healthy patients had symptoms consistent with gastroesophageal reflux disease or EoE, but endoscopic and histologic appearances were normal, as previously reported.13 The inclusion criteria for patients with active EoE included a clinical diagnosis of EoE and eosinophil counts of 24 or greater per ×400 high-power field in the esophageal biopsy specimens; most patients had lack of response to proton-pump inhibitor therapy (see Table E1 in this article’s Online Repository at www.jacionline.org). The inclusion criteria for patients with active chronic esophagitis include a clinical diagnosis of esophagitis and eosinophil counts of 1 to 15 per ×400 high-power field in esophageal biopsy specimens. Patients with systemic or swallowed topical glucocorticoid use were excluded from the selection of patients with active EoE or patients with active chronic esophagitis. The inclusion criteria for patients with EoE responding to glucocorticoid treatment include a history of EoE, treatment with swallowed topical glucocorticoid, and response as indicated by eosinophil counts of 1 or less per ×400 high-power field and normalization of histologic features of the disease. The patients with EoE not responding to glucocorticoid treatment have a history of EoE, treatment with swallowed topical glucocorticoid, and lack of response, as indicated by esophageal eosinophil counts of 24 or greater per ×400 high-power field in biopsy specimens obtained at follow-up visits after therapy. The patients’ characteristics are listed in Table E1. Eight healthy control subjects, 10 patients with active EoE, 5 patients with chronic esophagitis, and 6 patients with EoE responding to glucocorticoid treatment were included in the microarray analysis. The remaining patients were included in the quantitative PCR studies only. Patients were selected for miRNA microarray analysis and quantitative PCR studies based on systematic enrollment on the basis of their presentation to our clinic between November 2007 and July 2009. Subsequently, RNA samples were selected based on the patients’ inclusion criteria and RNA integrity number of greater than 8, as determined with the Agilent 2100 Bioanalyzer (Agilent Technologies, Santa Clara, Calif). Additional patients enrolled between July 2009 and June 2011 who met the inclusion criteria and have RNA samples that passed the RNA quality analysis were included in the PCR analysis. This study was approved by the Institutional Review Board of the Cincinnati Children’s Hospital Medical Center.
RNA extraction and miRNA microarray analysis
In brief, miRNA expression was profiled with the TaqMan Human MicroRNA Array version 2.0 (Applied Biosystems, Foster City, Calif), and data analyses were carried out with GeneSpring software (Agilent Technologies). Detailed analyses are described in the Methods section in this article’s Online Repository at www.jacionline.org. The microarray data have been deposited into the Array Express database (www.ebi.ac.uk/arrayexpress; accession no. E-MEXP-3298) in compliance with minimum information about microarray experiment (MIAME) standards.
Quantitative RT-PCR for miRNA in the esophagus and correlation with major EoE signature genes
Levels of miRNA expression were measured quantitatively by using TaqMan MicroRNA Assays (Applied Biosystems), according to the manufacturer’s protocol, as described in the Methods section in this article’s Online Repository. The list of EoE signature genes used for correlation analysis is provided in Table E2 in this article’s Online Repository at www.jacionline.org.
Analysis of miR-21 and miR-223 coregulated genes by RNA-Seq and gene expression microarray
Genome-wide expression correlation is described in detail in the Methods section in this article’s Online Repository.
Patient selection for plasma miRNA analysis
For the plasma miRNA level analysis, healthy atopic control subjects (defined as having asthma, allergic rhinitis, and/or eczema) were selected to control for the atopy commonly seen in patients with EoE. The patients with active EoE were selected based on a clinical diagnosis of EoE and eosinophil counts of 24 or greater per ×400 high-power field in the esophageal biopsy specimens. Patients with systemic or swallowed topical glucocorticoid use were excluded from the selection of healthy atopic control subjects or patients with active EoE. The patients with EoE remission were selected based on a history of EoE, treatment with swallowed topical glucocorticoid, and response as indicated by eosinophil counts of 1 or less per ×400 high-power field and normalization of histologic features of the disease. Patients were selected retrospectively from cell-free plasma samples collected between November 2007 and June 2011 based on the selection criteria outlined and the availability of a sufficient quantity of plasma samples for RNA isolation. A total of 13 healthy subjects, 13 patients with EoE, and 14 patients with EoE remission were included in the plasma miRNA analysis. Three healthy subjects, 4 patients with EoE, and 3 patients with EoE remission included in the plasma miRNA analysis overlapped with patients selected for esophageal miRNA microarray analysis. Other patients included in the esophageal miRNA microarray analysis did not have sufficient quantities of plasma samples available for RNA isolation.
RNA isolation from human plasma samples
Cell-free human plasma samples (750 µL) were mixed with Trizol LS reagent (Invitrogen, Carlsbad, Calif) at a 1:3 ratio with 1 µg of MS2 RNA added as a carrier RNA. The bacteriophage MS2 RNA was selected as a carrier RNA because it does not contain miRNAs. The RNA was then extracted with the miRNEasy Mini Kit (Qiagen, Hilden, Germany), according to the manufacturer’s protocols.
Detection of plasma miRNAs
Plasma (prepared from Na-EDTA tubes) miRNAs were reverse transcribed with MegaPlex RT primers, Human pool set V2.1, with preamplification (Applied Biosystems) according to the manufacturer’s protocols. The preamplified samples were diluted 1:40, and then 1.5 µL of the diluted product was used in 15 µL of PCR reaction with TaqMan miRNA Assays, according to the manufacturer’s protocols.
RESULTS
Expression profiling of miRNA in patients with EoE
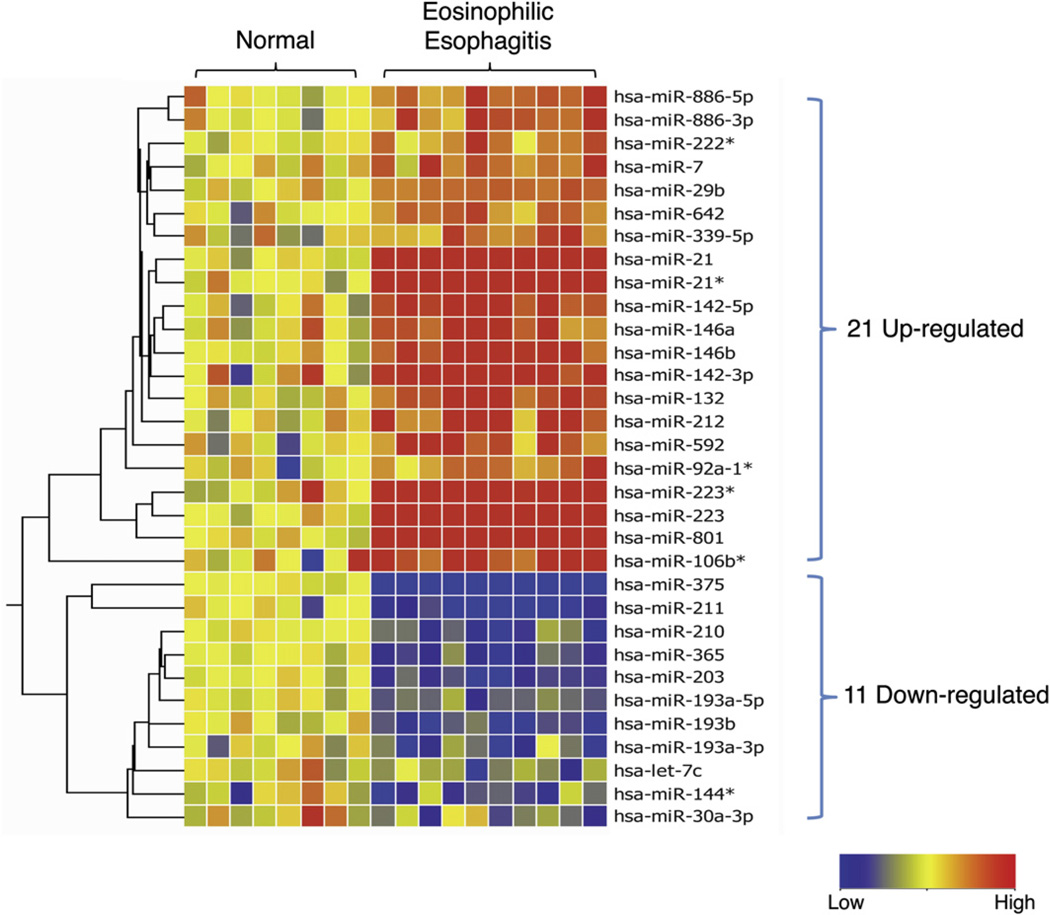
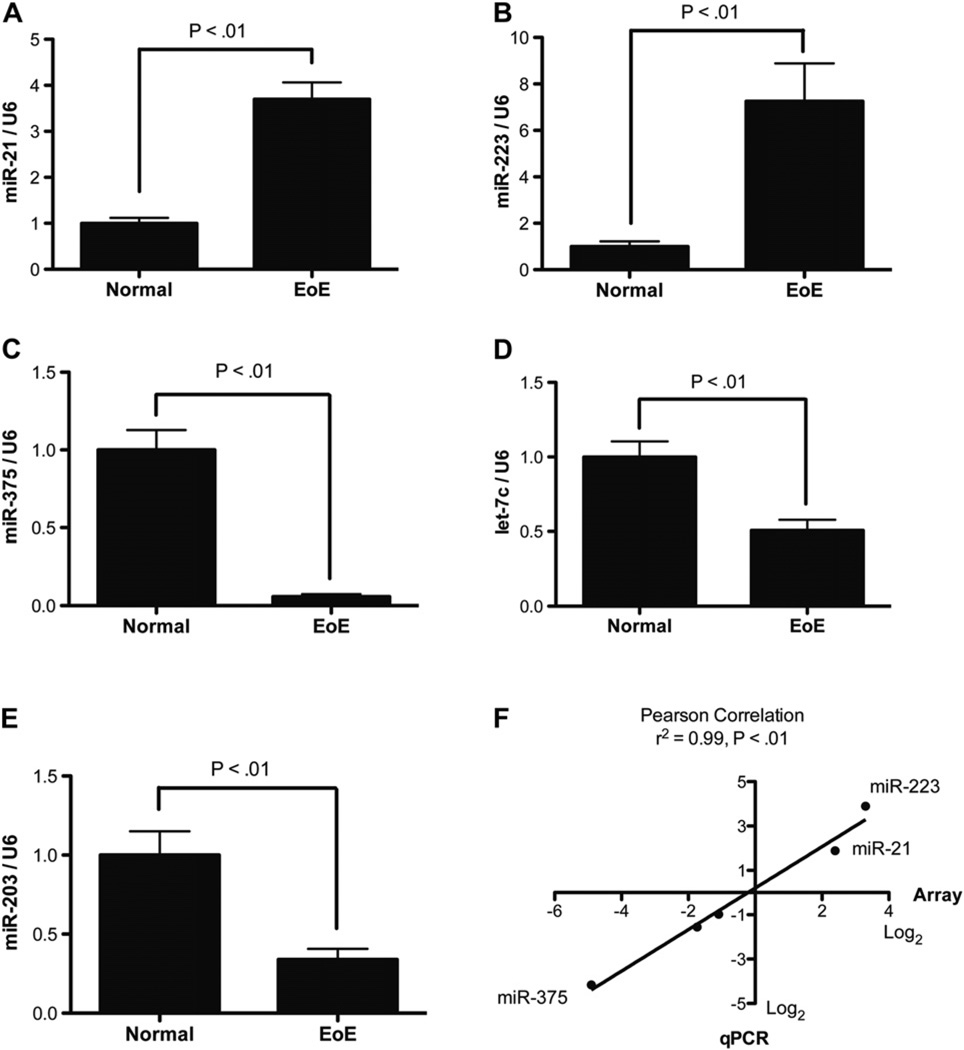
Esophageal biopsy specimens from patients with EoE and healthy control subjects were profiled with the TaqMan Human miRNA Array V2.0, comprising 677 miRNAs, as annotated in version 10 of the miRBase registry, to identify miRNAs differentially expressed in patients with EoE.21 Of the 677 miRNAs assayed, 254 miRNAs were expressed at greater than background levels (see Table E3 in this article’s Online Repository at www.jacionline.org). A comparison between healthy control subjects and patients with EoE identified 21 upregulated and 11 downregulated miRNAs (Fig 1). The most upregulated miRNAs included miR-21 and miR-223, and the most downregulated miRNA was miR-375. To validate the differentially expressed miRNAs, we performed quantitative RT-PCR on a selected set of differentially expressed miRNAs, including miR-21, miR-223, miR-375, let-7c, and miR-203 (Fig 2, A–E). There was a strong correlation between the quantitative RT-PCR and microarray data, with a Pearson correlation coefficient of 0.99 and a P value of less than .01 (Fig 2, F).
FIG 1.
MiRNA expression profile in healthy control subjects and patients with EoE: heat map of 21 upregulated and 11 downregulated miRNAs in patients with EoE compared with healthy control subjects. Red, Upregulated in patients with EoE compared with healthy control subjects; blue, downregulated in patients with EoE compared with healthy control subjects. Asterisks (*) after the name of the miRNA indicate the minor form of the miRNA derived from the passenger strand.
FIG 2.
Quantitative RT-PCR verification of a selected set of differentially expressed miRNAs in patients with EoE. A–E, Expression levels of miR-21 (Fig 2, A), miR-223 (Fig 2, B), miR-375 (Fig 2, C), let-7c (Fig 2, D), and miR-203 (Fig 2, E) were determined in patients with EoE compared with healthy control subjects. The relative expression levels were normalized to U6 small nuclear RNA. F, Correlation of miRNA expression levels between microarray data and quantitative RT-PCR (qPCR) validation (n = 7–17 patients per group). Data are presented as mean ± SEM.
Specificity of the differentially expressed miRNA in patients with EoE
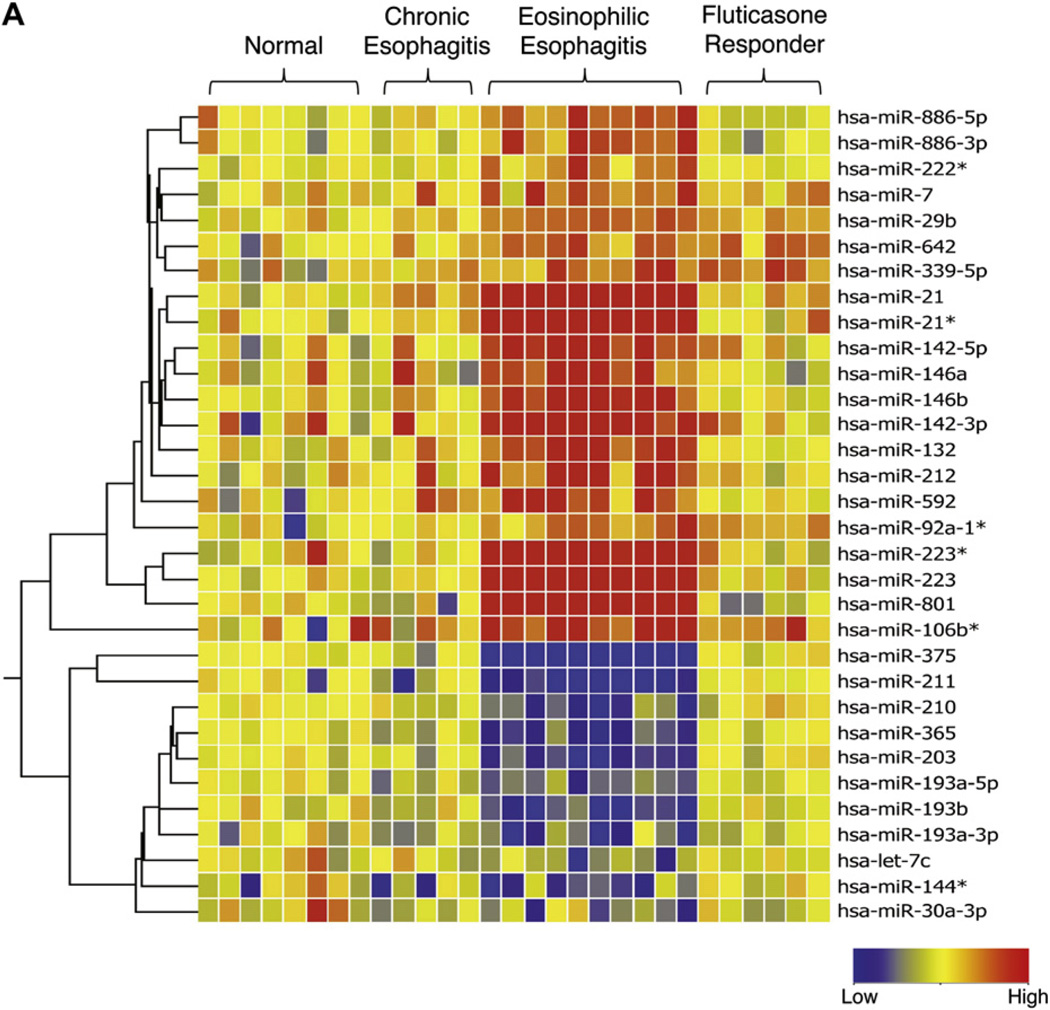
To determine whether the miRNA expression signature is specific to EoE, we compared the miRNA expression profile with that of healthy control subjects, as well as that of patients who presented with symptoms of EoE but were ultimately given a diagnosis of chronic (noneosinophilic) esophagitis. The patients with chronic esophagitis had miRNA expression profiles similar to those of healthy control subjects and distinct from those of patients with EoE (Fig 3, A).We did not identify any differentially regulated miRNAs between healthy control subjects and patients with chronic esophagitis.
FIG 3.
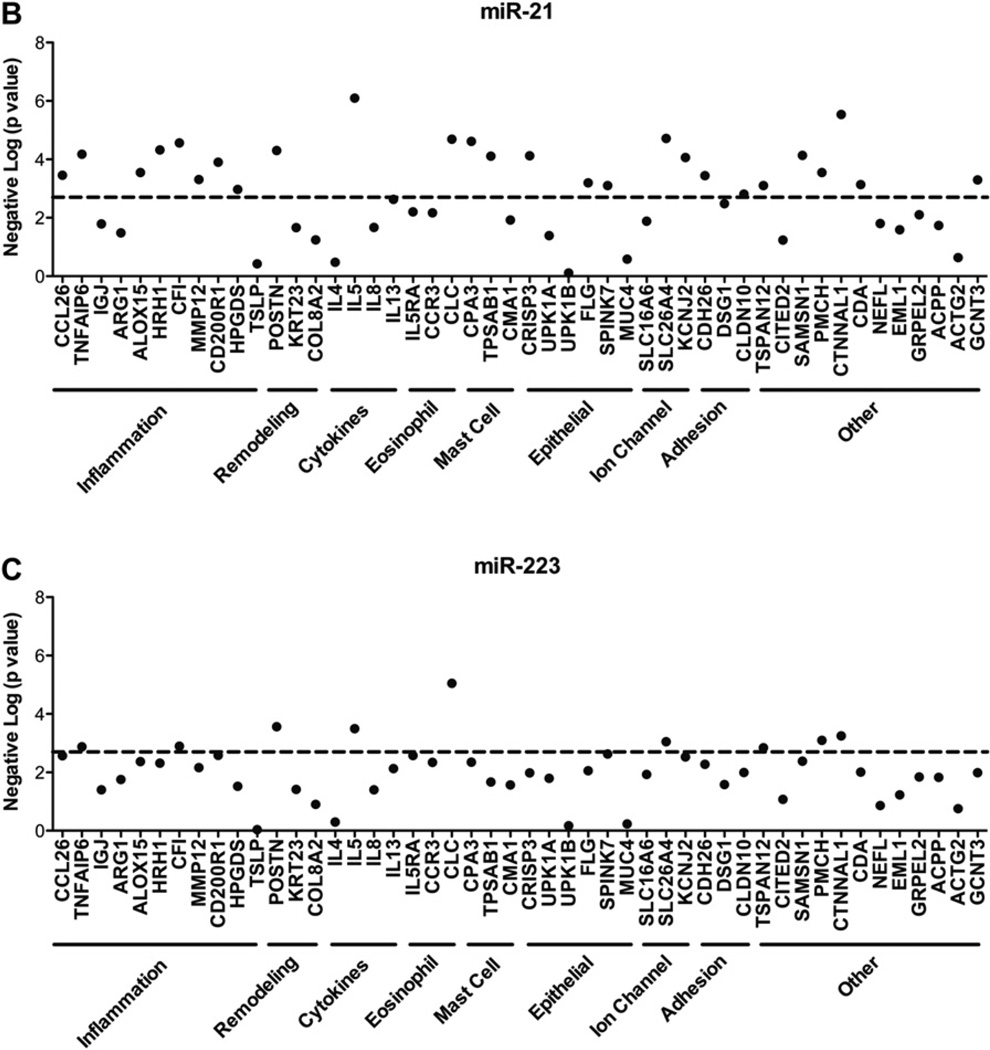
MiRNA expression profile in healthy control subjects and patients with EoE compared with patients with chronic esophagitis and patients with EoE responding to glucocorticoid therapy and correlation of miR-21 and miR-223 with EoE signature genes. A, Heat map showing the expression level of 32 differentially expressed miRNAs in patients with EoE compared with patients with chronic esophagitis and patients with EoE in remission after glucocorticoid therapy. Red, Upregulated in comparison with healthy control subjects; blue, downregulated in comparison with healthy control subjects. Asterisks (*) after the name of the miRNA indicate the minor form of the miRNA derived from the passenger strand. B and C, Correlation of EoE signature genes with miR-21 (Fig 3, B) and miR-223 (Fig 3, C). The significance of the correlation was plotted as the negative log of P values for each gene. The dashed line represents the significance level after false discovery rate correction.
Reversibility of the differentially expressed miRNA in patients with EoE
We next asked whether the EoE miRNA expression signature was fixed or reversible in patients who responded to glucocorticoid treatment and had normalization of esophageal histology, including eosinophil counts. Comparing patients with active EoE with patients with EoE who responded to fluticasone propionate therapy, 27 of the 32 differentially expressed miRNAs were normalized. The reversible miRNAs include all of the top upregulated and downregulated miRNAs (Fig 3, A). Interestingly, 5 upregulated miRNAs were still dysregulated in glucocorticoid-responsive patients. These include miR-7, miR-29b, miR-642, miR-339-5p, and miR92a-1.
Correlation of miRNA expression levels and esophageal eosinophil counts
We aimed to determine whether the magnitude of miRNA expression changes correlated to the eosinophil counts in the biopsy specimens of patients with EoE. Of the 32 differentially regulated miRNAs in patients with EoE, levels of 24 significantly correlated with esophageal eosinophil counts (Table I). Interestingly, the most upregulated miRNAs, miR-21 and miR-223, also had the strongest correlation of their expression level to esophageal eosinophil counts. We further correlated the expression of miR-21 and miR-223 to previously identified EoE signature genes. MiR-21 significantly correlated with the esophageal expression of genes involved in inflammation, including CCL26 (eotaxin-3); remodeling, including POSTN (periostin); eosinophilia, including IL5; and cell-specific markers for eosinophils (CLC [Charcot-Leyden crystal protein]) and mast cells (CPA3 [carboxypeptidase A3] and TPSAB1 [tryptase alpha/beta 1]; Fig 3, B). In addition, miR-21 significantly correlated with the gene CTNNAL1 (alpha-catulin), which has been implicated in cell growth/proliferation and wound repair (Fig 3, B).22 MiR-223 had the highest correlation with POSTN, IL5, and CLC expression (Fig 3, C).
TABLE I.
Correlation between miRNA expression levels and peak eosinophil counts in patients’ biopsy specimens
| P value | R2 | |
|---|---|---|
| hsa-miR-21 | .0002 | 0.663 |
| hsa-miR-223 | .0002 | 0.657 |
| hsa-miR-132 | .0003 | 0.642 |
| hsa-miR-223* | .0005 | 0.617 |
| hsa-miR-146b | .0005 | 0.615 |
| hsa-miR-801 | .0005 | 0.617 |
| hsa-miR-29b | .0005 | 0.614 |
| hsa-miR-21* | .0006 | 0.606 |
| hsa-miR-375 | .0015 | 0.552 |
| hsa-miR-212 | .0015 | 0.551 |
| hsa-miR-203 | .0019 | 0.538 |
| hsa-miR-142-5p | .0052 | 0.463 |
| hsa-miR-211 | .0060 | 0.452 |
| hsa-let-7c | .0065 | 0.447 |
| hsa-miR-642 | .0085 | 0.425 |
| hsa-miR-210 | .012 | 0.396 |
| hsa-miR-222* | .012 | 0.393 |
| hsa-miR-365 | .013 | 0.390 |
| hsa-miR-142-3p | .021 | 0.348 |
| hsa-miR-886-3p | .021 | 0.346 |
| hsa-miR-193b | .022 | 0.343 |
| hsa-miR-92a-1* | .023 | 0.339 |
| hsa-miR-339-5p | .034 | 0.302 |
| hsa-miR-886-5p | .035 | 0.298 |
| hsa-miR-146a | .051 | 0.261 |
| hsa-miR-592 | .060 | 0.246 |
| hsa-miR-106b* | .066 | 0.236 |
| hsa-miR-193a-5p | .068 | 0.234 |
| hsa-miR-193a-3p | .150 | 0.152 |
| hsa-miR-7 | .240 | 0.104 |
| hsa-miR-30a-3p | .810 | 0.005 |
| hsa-miR-144* | .999 | 0.000 |
Asterisks (*) after the name of the miRNA indicate the minor form of the miRNA derived from the passenger strand.
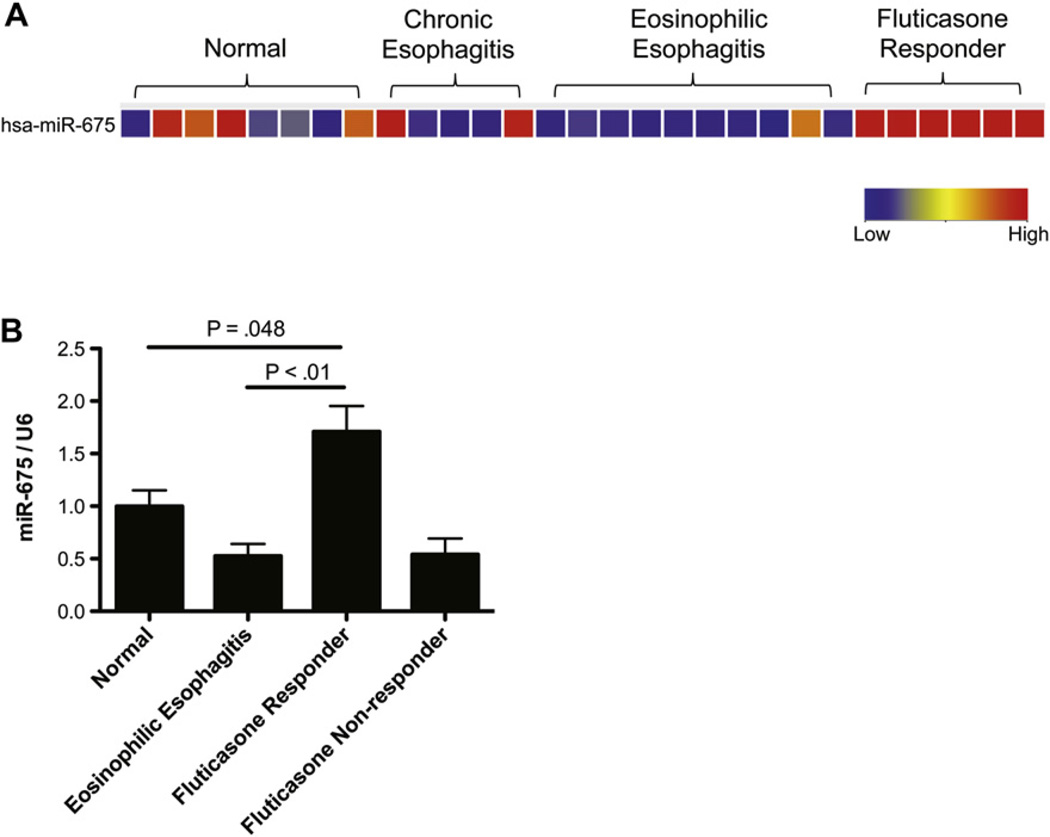
MiR-675 is a disease remission–induced miRNA in patients with EoE
We aimed to determine whether any miRNA was differentially regulated in response to treatment. Interestingly, we identified 1 miRNA, miR-675, that was upregulated in glucocorticoid-responsive patients compared with healthy control subjects, patients with EoE, or patients with chronic esophagitis (Fig 4, A). To determine whether miR-675 is a glucocorticoid-induced or EoE remission–induced miRNA, we measured miR-675 expression levels in healthy control subjects, patients with EoE, patients with EoE who responded to glucocorticoid treatment, and patients with EoE who did not respond to glucocorticoid treatment. We found that miR-675 was induced in patients who responded to glucocorticoid treatment and not induced in patients who did not respond to glucocorticoid treatment (Fig 4, B).
FIG 4.
MiRNAs differentially regulated in patients with EoE in remission. A, Heat map showing the expression level of miR-675, which is the only miRNA differentially regulated in patients with EoE who responded to glucocorticoid therapy, compared with that seen in healthy control subjects, patients with chronic esophagitis, and patients with EoE. Red, Upregulated in comparison with healthy control subjects; blue, downregulated in comparison with healthy control subjects. B, Relative expression level of miR-675 in healthy control subjects, patients with EoE, fluticasone propionate–responsive patients, and fluticasone propionate– nonresponsive patients. Relative expression levels determined by means of quantitative RT-PCR was normalized to U6 (n = 7–11 patients per group). Data are presented as mean ± SEM.
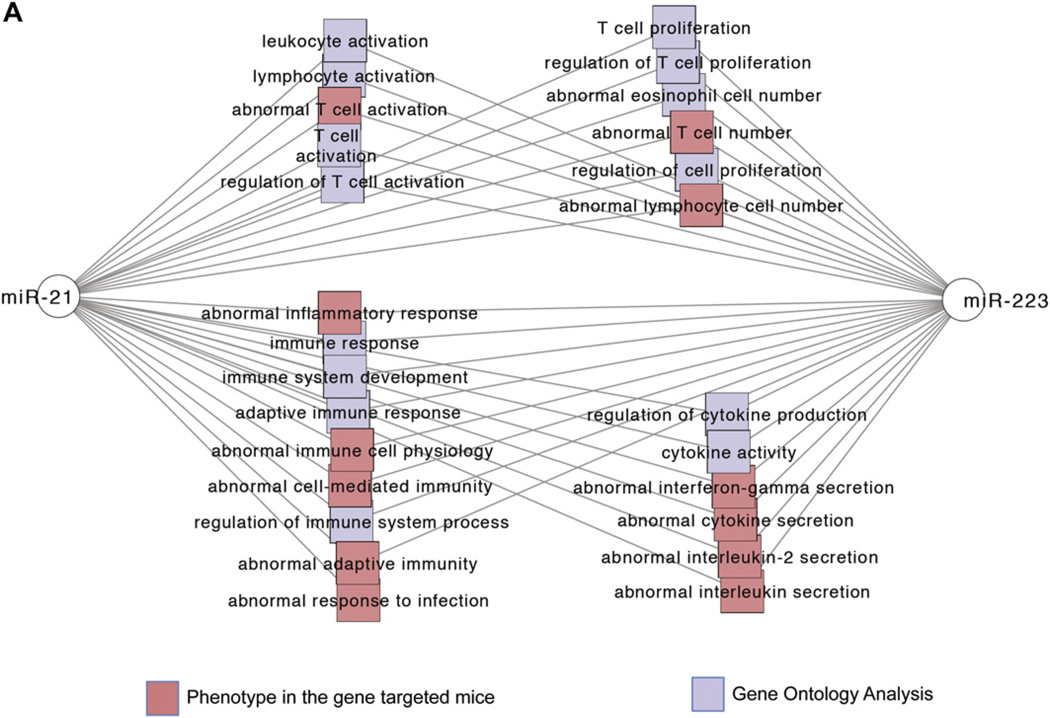
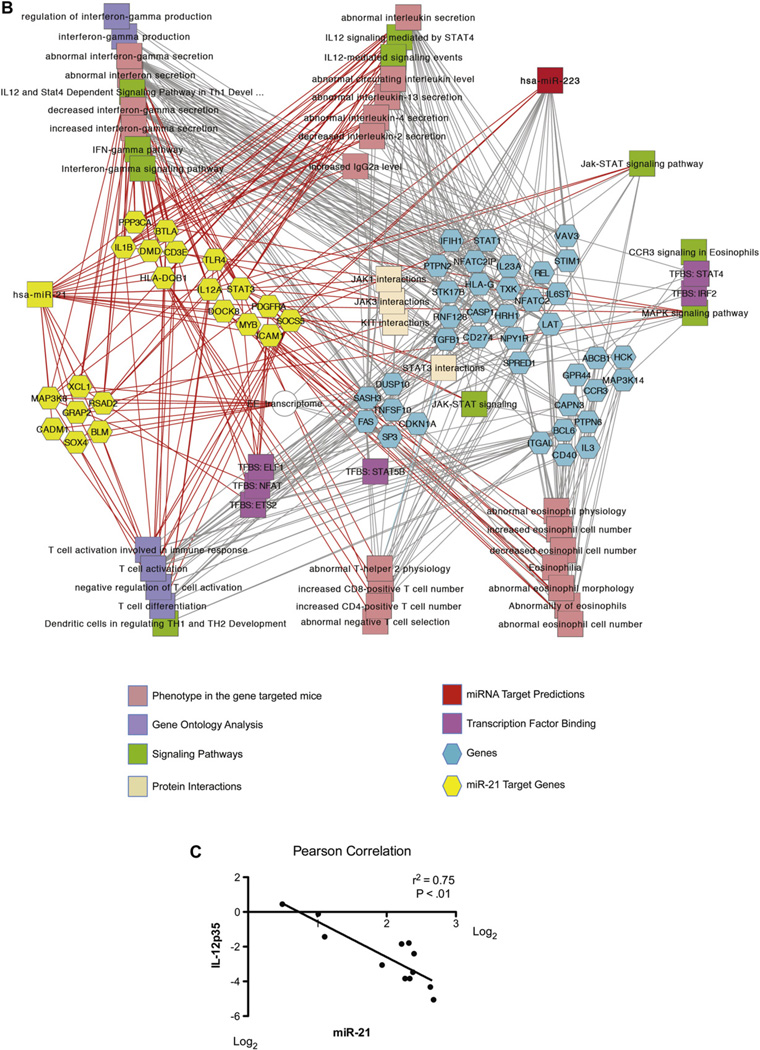
MiR-21 and miR-223 coregulated genes are significantly enriched in pathways involved in adaptive immunity and regulation of eosinophils
Because miR-21 and miR-223 are the top 2 upregulated miRNAs in patients with EoE, we further identified mRNA expression patterns that significantly correlated with miR-21 and miR-223 expression from esophageal RNA samples subjected to RNA-Seq analysis, as well as previously published mRNA microarray experiments.11,13 MiR-21 and miR-223 interactively regulated many similar pathways, including leukocyte proliferation, leukocyte activation, cytokine production, and immune response (Fig 5, A). The coregulated target genes of miR-21 and miR-223 were involved in adaptive immune system polarization, IFN-γ signaling, and regulation of eosinophilia (Fig 5, B, and see Fig E1 in this article’s Online Repository at www.jacionline.org). The significantly enriched pathways regulated by miR-21 include regulation of interleukin secretion, interferon production and signaling, and T-cell differentiation and activation (Fig 5, B). Esophageal IL-12p35 levels showed a strong inverse correlation with esophageal miR-21 levels (Fig 5, C), supporting our previous finding that miR-21 regulates TH1 versus TH2 balance by targeting IL-12p35 expression.7,8
FIG 5.
Gene enrichment analysis of miR-21 and miR-223 coregulated genes in patients with EoE. Extensive enrichment of genes with functional features associated with T-cell polarization, IFN-γ signaling, and regulation of eosinophilia among genes coregulated by miR-21 and miR-223 in esophageal biopsy specimens. Networks are shown as Cytoscape graph networks generated from ToppCluster network analysis. A, Abstracted interactions between miR-21 and miR-223 coregulated target genes. B, Expanded network of miR-21 and miR-223 coregulated genes. MiR-21 targets that are significantly correlated with miR-21 expression or in the EoE transcriptome are shown as yellow hexagons, and their respective feature and function relationships are shown connected through red edges. The features that are highly significant for other genes in the cluster are connected by gray edges. For example, miR-21 coregulated targets are significantly enriched in genes that regulate interleukin production. C, Pearson correlation of miR-21 and IL-12p35 expression levels in esophageal biopsy specimens from patients with EoE.
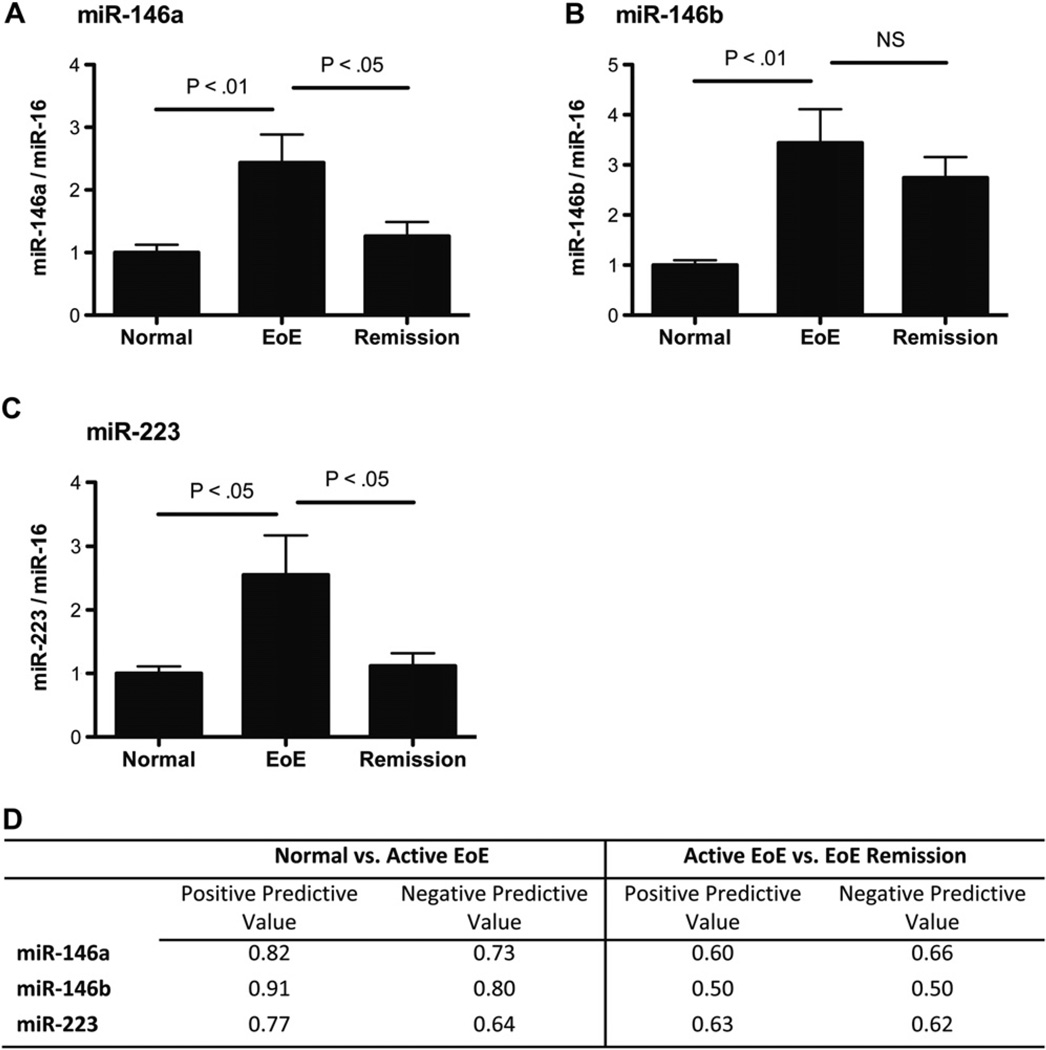
Identification of differentially expressed miRNAs from plasma samples of patients with EoE
MiRNAs have recently been reported to be present in plasma samples in a stable form protected from endogenous RNAse activities.23 This suggests the potential for plasma miRNAs to be used as noninvasive biomarkers.23–25We hypothesized that a subset of differentially regulated miRNA in the esophageal biopsy specimens of patients with EoE is also differentially regulated in the patients’ plasma samples. Using plasma samples from patients with EoE and healthy control subjects, we determined the expression levels of the 10 most differentially regulated miR-NAs associated with EoE. This analysis included 6 upregulated miRNAs (miR-21, miR-132, miR-142-3p, miR-146a, miR-146b, and miR-223) and 4 downregulated miRNAs (miR-203, miR-210, miR-365 and miR-375). We did not detect any of the downregulated miRNAs in the plasma samples from patients with EoE or healthy control subjects after 40 cycles of PCR. For the upregulated miRNAs, we detected expression of miR-142-3p, miR-146a, miR-146b, and miR-223 in all the plasma samples. Expression of miR-21 and miR-132 was only detected in some of the samples at a low level and was not analyzed further. To determine differential expression of the miRNAs between plasma samples from patients with EoE and healthy control subjects, we used miR-16 as an endogenous control because miR-16 has been reported to have a constant expression level in plasma samples.25 There were no significant differences in the average cycle threshold value of miR-16 between healthy control subjects and patients with EoE (25.5 ± 0.4 vs 26.0 ± 1.1, respectively). Comparing patients with EoE with healthy control subjects, we found upregulation of miR-146a, miR-146b, and miR-223 in plasma samples from patients with EoE (Fig 6, A–C). Although miR-146a and miR-223 levels returned to baseline levels in patients with EoE remission, miR-146b levels remained increased (Fig 6, A–C). Using a 1.5-fold change cutoff, we calculated the positive and negative predicative values of healthy control subjects versus patients with active EoE and patients with active EoE versus patients with EoE in remission (Fig 6, D). The levels of miR-142-3p were not changed between any of the groups (see Fig E2 in this article’s Online Repository at www.jacionline.org).
FIG 6.
MiRNAs differentially expressed in the plasma of patients with active EoE and patients with EoE remission compared with healthy control subjects. A–C, Expression levels of miR-146a (Fig 6, A), miR-146b (Fig 6, B), and miR-223 (Fig 6, C) were determined in plasma samples from patients with active EoE and patients with EoE remission compared with those seen in healthy control subjects. The relative expression levels were normalized to miR-16 (n = 13–14 plasma samples per group). Data are presented as mean ± SEM. NS, Not significant (P > .05). D, Positive and negative predictive values of differentially regulated plasma miRNAs.
DISCUSSION
Herein we provide a comprehensive analysis of global miRNA expression in the esophageal tissues of patients with EoE. Specifically, we identified 21 upregulated and 11 downregulated miRNAs in patients with active EoE, including miR-21 and miR-223 as the most upregulated miRNAs and miR-375 as the most downregulated miRNA in patients with EoE. This EoE-associated miRNA signature correlated with the degree of tissue eosinophilia and was distinct from patients with chronic (noneosinophilic) esophagitis. Furthermore, the differentially expressed miRNAs were largely reversible in patients who responded to glucocorticoid therapy. To the best of our knowledge, this is the first study to demonstrate dynamic expression of miRNAs in a human allergic disease and the potential role for tissue and blood miRNAs as biomarkers that provide insight into disease diagnosis, response to therapy, and the degree of allergic inflammation.
EoE is currently considered a TH2-associated disease.12,18,19 We found miR-21 to be one of the most upregulated miRNAs in patients with EoE. MiR-21 has been shown to regulate IL-12 expression and the balance of TH1 versus TH2 responses in mice.7,8 The high level of species conservation of the miR-21 binding site in the 3' untranslated region of IL12p35 suggests that miR-21 might have a similar role in human allergic inflammation.7 Herein, we have provided the first set of human data that substantiate that miR-21 likely has a similar role in human allergic inflammation. Upregulation of miR-21 in patients with EoE could partially explain the increased TH2 cytokine levels and TH2 responses seen in patients with EoE. Indeed, we found that esophageal miR-21 levels inversely correlated with esophageal IL-12p35 levels. Coregulated miR-21 target genes in the patients with EoE were significantly enriched in the regulation of T-cell polarization and IFN-γ production. Direct analysis of a myriad of esophageal transcripts for their correlation with miR-21 demonstrated impressive correlations with key elements of the EoE transcriptome, including cell-specific markers for key inflammatory cells (eosinophils and mast cells), as well as CCL26 (eotaxin-3), which is functionally involved in eosinophil recruitment,26 and POSTN (periostin), which is involved in tissue remodeling27 and eosinophilia28 and has recently been shown to be a key biomarker for anti–IL-13 responsiveness in human asthma.29 These data provide the first human evidence to substantiate the recent finding that miR-21 critically regulates the polarization of adaptive immunity in mice,8 supporting our previous finding that miR-21 regulates TH1 versus TH2 balance by targeting IL-12p35 expression.7 We also found downregulation of let-7c that could potentially regulate IL-13 levels.30 In addition, we found upregulation of miR-146a in patients with EoE. MiR-146a has recently been demonstrated to selectively regulate regulatory T cell–mediated suppression of TH1 cells.31 Therefore, upregulation of miR-146a could potentially suppress TH1 responses and promote TH2 responses. Together, these findings support a model whereby multiple miRNAs coordinate polarized TH responses in the pathogenesis of EoE. Indeed, recent human studies on 2 other TH2-associated diseases (atopic dermatitis and ulcerative colitis) have identified a role for miRNA in regulating T-cell proliferation and epithelium-derived chemokine production, as well as upregulating miR-21 in patients with ulcerative colitis and downregulating let-7 in patients with atopic dermatitis.32,33
One of the defining histologic features of EoE is intense eosinophil infiltration in the esophagus. We have found that a majority of the dysregulated miRNAs have significant correlation between the miRNA expression level and the esophageal eosinophil count, potentially reflecting disease severity. We performed functional enrichment analyses of the 2 miRNAs that most strongly correlated with eosinophil levels (miR-21 and miR-223); it is remarkable that this analysis empirically predicted that both miRNAs regulate levels of tissue eosinophilia, drawing further attention to the potential interplay between these 2 miRNAs in patients with allergic inflammation. Indeed, both miRNAs correlated significantly with IL-5, a key eosinophil growth factor shown to be contributory in murine models of EoE and human EoE.34,35 Another significant histologic finding in patients with EoE is epithelial basal layer hyperplasia. In particular, miR-203 is known to repress epithelial cell proliferation and promote epithelial cell differentiation.36 Thus, repression of miR-203 could in part explain the observed epithelial hyperplasia.
It is notable that several of the EoE-associated miRNAs have recently also been linked with esophageal squamous carcinoma or with Barrett esophagus, including let-7,37 miR-142-3p,38 miR-203,39 miR-210,40 miR-223,41 miR-375,42 and miR-21.43 Indeed, some miRNAs, such as miR-21, have already been shown to be oncomirs, tumor suppressors, or both.44,45 Although EoE is not considered a premalignant condition, it is notable that EoE involves marked epithelial cell hyperplasia.
We have identified miR-675 as the only disease remission–induced miRNA. Mir-675 is derived from the H19 gene, which is a paternally imprinted gene.46 The overexpression of H19 is commonly associated with various cancers.47 We have previously found that H19 is induced in glucocorticoid-responsive patients compared with patients with EoE or healthy control subjects; this induction is not seen in patients who did not respond to glucocorticoid therapy.20 Our current data indicate that the miR-675 expression pattern closely resembles that of H19. Because the exact roles of H19 and its miRNA product miR-675 are currently unknown in the disease remission process, elucidating their functions could potentially lead to a better understanding of the disease remission process in patients with EoE. On the basis of the likely role of H19 and miR-675 in DNA methylation responses and the unique overexpression of this miRNA specifically within patients in remission, it is tempting to speculate that miR-675 might be involved in epigenetic reprogramming in the esophageal cells of patients with EoE remission.
We measured the expression levels of a selected set of EoE-associated miRNAs in plasma samples from patients with EoE and found that miR-146a, miR-146b, and miR-223 were upregulated in these plasma samples compared with those from healthy control subjects. This suggests that miRNAs could potentially serve as noninvasive biomarkers of EoE alone or in combination with other noninvasive biomarkers. Plasma miRNAs have been reported to exist both within exosomes and in protein-bound vesicle-free form.48,49 The circulating miRNAs could potentially be uptaken by cells through exosome uptake or pinocytosis.50,51 Mast cells have recently been found to release exosomes containing miRNA.50 It is particularly interesting that mast cells express high levels of both miR-146a and miR-146b.52,53 Notably, patients with EoE have concomitant esophageal mastocytosis,13,54,55 which could be a potential source for the increased miR-146a and miR-146b levels seen in the plasma of patients with EoE. Because miR-146a has been found to selectively promote regulatory T cell–mediated suppression of TH1 responses, it is possible that increased circulating levels of plasma miR-146a could further propagate or help maintain the TH2 responses seen in patients with EoE. We noted that although plasma miR-146a and miR-223 levels returned to baseline during EoE remission, miR-146b levels remain increased in patients with EoE remission. Although the specific role of miR-146b in regulating adaptive immune responses has not been investigated, miR-146a and miR-146b have an identical seed sequence that is critical for miRNA-mediated target gene expression. Thus, it is plausible that miR-146b could also suppress TH1 responses and promote TH2 responses. Patients with EoE in remission often relapse as time progresses.56 We speculate that the increased miR-146b level might predispose the patients with EoE in remission to a relapse.
In summary, we report an miRNA signature of patients with EoE. We demonstrated that the EoE miRNA signature is distinct from that of patients with chronic esophagitis and is largely reversible on disease remission. We showed that 3 of the differentially regulated miRNAs in the esophageal biopsy specimens, miR-146a, miR-146b, and miR-223, were also differentially regulated in plasma samples from patients with EoE. Our study supports a key role of miRNA in regulating TH2-associated diseases. Future studies will further refine the clinical utility of miRNAs as biomarkers during EoE diagnosis and as an indicator of disease activity during treatment, as well as their use in determining steroid susceptibility in patients with EoE. Further elucidating and understanding of the roles and regulations of the miRNAs differentially regulated in patients with EoE might lead to improved patient diagnosis and facilitate the development of miRNA mimics and inhibitors as novel therapies for patients with EoE.
Supplementary Material
Key messages.
EoE is associated with 32 differentially regulated miRNAs in the esophagus, and the miRNA expression panel can distinguish patients with EoE from healthy control subjects, including those with noneosinophilic forms of esophagitis (eg, reflux esophagitis).
Of the identified miRNAs, miR-21 and miR-223 levels strongly correlate with esophageal eosinophil levels, as well as a subset of the previously described EoE transcriptome.
The differentially expressed miRNAs are largely reversible in patients who respond to glucocorticoid treatment.
MiR-146a, miR-146b, and miR-223 are the most differentially expressed miRNAs in the plasma and could potentially be used as noninvasive biomarkers.
Acknowledgments
We thank the teams from the Cincinnati Center for Eosinophilic Disorders for their support with the collection of clinical samples.
Supported by the National Heart, Lung, and Blood Institute’s Ruth L. Kirschstein National Research Service Award for individual predoctoral MD/PhD fellows F30HL104892 (T.X.L), the Ryan Fellowship from the Albert J. Ryan Foundation (T.X.L.), and the Organogenesis Training Grant (National Institutes of Health [NIH] grant T32HD046387 supporting T.X.L.). Additionally, this work was supported by NIH grants R01DK076893 (M.E.R), U19 AI070235 (M.E.R), and P30 DK078392; the Campaign Urging Research for Eosinophilic Disease (CURED); the Buckeye Foundation; and the Food Allergy Initiative.
Disclosure of potential conflict of interest: J. P. Abonia has received research support from the National Institutes of Health (NIH), Ception Therapeutics, and the Children’s Digestive Health and Nutrition Foundation. L. J. Martin has received research support from the NIH and the Department of Defense (DOD). M. E. Rothenberg has equity interest in reslizumab through Cephalon; is a consultant for and Chief Scientific Officer of Immune Pharmaceuticals; has received research support from the NIH, the Food Allergy & Anaphylaxis Network, and the DOD; is on the American Partnership for Eosinophilic Disorders Medical Advisory Board; and is on the International Eosinophil Society’s Executive Council.
Abbreviations used
- EoE
Eosinophilic esophagitis
- miRNA
MicroRNA
Footnotes
The rest of the authors declare that they have no relevant conflicts of interest.
REFERENCES
- 1.Sayed D, Abdellatif M. MicroRNAs in development and disease. Physiol Rev. 2011;91:827–887. doi: 10.1152/physrev.00006.2010. [DOI] [PubMed] [Google Scholar]
- 2.Smith CM, Watson DI, Michael MZ, Hussey DJ. MicroRNAs, development of Barrett’s esophagus, and progression to esophageal adenocarcinoma. World J Gastroenterol. 2010;16:531–537. doi: 10.3748/wjg.v16.i5.531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mattes J, Collison A, Plank M, Phipps S, Foster PS. Antagonism of microRNA-126 suppresses the effector function of TH2 cells and the development of allergic airways disease. Proc Natl Acad Sci U S A. 2009;106:18704–18709. doi: 10.1073/pnas.0905063106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kumar M, Ahmad T, Sharma A, Mabalirajan U, Kulshreshtha A, Agrawal A, et al. Let-7 microRNA-mediated regulation of IL-13 and allergic airway inflammation. J Allergy Clin Immunol. 2011;128:1077–1085. doi: 10.1016/j.jaci.2011.04.034. [DOI] [PubMed] [Google Scholar]
- 5.Collison A, Mattes J, Plank M, Foster PS. Inhibition of house dust mite-induced allergic airways disease by antagonism of microRNA-145 is comparable to glucocorticoid treatment. J Allergy Clin Immunol. 2011;128:160–167. doi: 10.1016/j.jaci.2011.04.005. [DOI] [PubMed] [Google Scholar]
- 6.Jiang X. The emerging role of microRNAs in asthma. Molecular and cellular biochemistry. 2011;353:35–40. doi: 10.1007/s11010-011-0771-z. [DOI] [PubMed] [Google Scholar]
- 7.Lu TX, Munitz A, Rothenberg ME. MicroRNA-21 is up-regulated in allergic airway inflammation and regulates IL-12p35 expression. J Immunol. 2009;182:4994–5002. doi: 10.4049/jimmunol.0803560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lu TX, Hartner J, Lim EJ, Fabry V, Mingler MK, Cole ET, et al. MicroRNA-21 limits in vivo immune response-mediated activation of the IL-12/IFN-γ pathway, Th1 polarization, and the severity of delayed-type hypersensitivity. J Immunol. 2011;187:3362–3373. doi: 10.4049/jimmunol.1101235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liacouras CA, Furuta GT, Hirano I, Atkins D, Attwood SE, Bonis PA, et al. Eosinophilic esophagitis: updated consensus recommendations for children and adults. J Allergy Clin Immunol. 2011;128:3–20. doi: 10.1016/j.jaci.2011.02.040. [DOI] [PubMed] [Google Scholar]
- 10.DeBrosse CW, Franciosi JP, King EC, Butz BK, Greenberg AB, Collins MH, et al. Long-term outcomes in pediatric-onset esophageal eosinophilia. J Allergy Clin Immunol. 2011;128:132–138. doi: 10.1016/j.jaci.2011.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Blanchard C, Wang N, Stringer KF, Mishra A, Fulkerson PC, Abonia JP, et al. Eotaxin-3 and a uniquely conserved gene-expression profile in eosinophilic esophagitis. J Clin Invest. 2006;116:536–547. doi: 10.1172/JCI26679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Blanchard C, Mingler MK, Vicario M, Abonia JP, Wu YY, Lu TX, et al. IL-13 involvement in eosinophilic esophagitis: transcriptome analysis and reversibility with glucocorticoids. J Allergy Clin Immunol. 2007;120:1292–1300. doi: 10.1016/j.jaci.2007.10.024. [DOI] [PubMed] [Google Scholar]
- 13.Abonia JP, Blanchard C, Butz BB, Rainey HF, Collins MH, Stringer K, et al. Involvement of mast cells in eosinophilic esophagitis. J Allergy Clin Immunol. 2010;126:140–149. doi: 10.1016/j.jaci.2010.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sherrill JD, Rothenberg ME. Genetic dissection of eosinophilic esophagitis provides insight into disease pathogenesis and treatment strategies. J Allergy Clin Immunol. 2011;128:23–32. doi: 10.1016/j.jaci.2011.03.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lim EJ, Lu TX, Blanchard C, Rothenberg ME. Epigenetic regulation of the IL-13-induced human eotaxin-3 gene by CREB-binding protein-mediated histone 3 acetylation. J Biol Chem. 2011;286:13193–13204. doi: 10.1074/jbc.M110.210724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Blanchard C, Stucke EM, Burwinkel K, Caldwell JM, Collins MH, Ahrens A, et al. Coordinate interaction between IL-13 and epithelial differentiation cluster genes in eosinophilic esophagitis. J Immunol. 2010;184:4033–4041. doi: 10.4049/jimmunol.0903069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sato F, Tsuchiya S, Meltzer SJ, Shimizu K. MicroRNAs and epigenetics. FEBS J. 2011;278:1598–1609. doi: 10.1111/j.1742-4658.2011.08089.x. [DOI] [PubMed] [Google Scholar]
- 18.Blanchard C, Stucke EM, Rodriguez-Jimenez B, Burwinkel K, Collins MH, Ahrens A, et al. A striking local esophageal cytokine expression profile in eosinophilic esophagitis. J Allergy Clin Immunol. 2011;127:208–217. doi: 10.1016/j.jaci.2010.10.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Straumann A, Bauer M, Fischer B, Blaser K, Simon HU. Idiopathic eosinophilic esophagitis is associated with a T(H)2-type allergic inflammatory response. J Allergy Clin Immunol. 2001;108:954–961. doi: 10.1067/mai.2001.119917. [DOI] [PubMed] [Google Scholar]
- 20.Caldwell JM, Blanchard C, Collins MH, Putnam PE, Kaul A, Aceves SS, et al. Glucocorticoid-regulated genes in eosinophilic esophagitis: a role for FKBP51. J Allergy Clin Immunol. 2010;125:879–888. doi: 10.1016/j.jaci.2010.01.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Griffiths-Jones S, Saini HK, van Dongen S, Enright AJ. miRBase: tools for micro-RNA genomics. Nucleic Acids Res. 2008;36:D154–D158. doi: 10.1093/nar/gkm952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xiang Y, Tan YR, Zhang JS, Qin XQ, Hu BB, Wang Y, et al. Wound repair and proliferation of bronchial epithelial cells regulated by CTNNAL1. J Cell Biochem. 2008;103:920–930. doi: 10.1002/jcb.21461. [DOI] [PubMed] [Google Scholar]
- 23.Mitchell PS, Parkin RK, Kroh EM, Fritz BR, Wyman SK, Pogosova-Agadjanyan EL, et al. Circulating microRNAs as stable blood-based markers for cancer detection. Proc Natl Acad Sci U S A. 2008;105:10513–10518. doi: 10.1073/pnas.0804549105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zahm AM, Thayu M, Hand NJ, Horner A, Leonard MB, Friedman JR. Circulating MicroRNA is a biomarker of pediatric Crohn disease. J Pediatr Gastroenterol Nutr. 2011;53:26–33. doi: 10.1097/MPG.0b013e31822200cc. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shen J, Todd NW, Zhang H, Yu L, Lingxiao X, Mei Y, et al. Plasma microRNAs as potential biomarkers for non-small-cell lung cancer. Lab Invest. 2011;91:579–587. doi: 10.1038/labinvest.2010.194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yuan Q, Campanella GS, Colvin RA, Hamilos DL, Jones KJ, Mathew A, et al. Membrane-bound eotaxin-3 mediates eosinophil transepithelial migration in IL-4-stimulated epithelial cells. Eur J Immunol. 2006;36:2700–2714. doi: 10.1002/eji.200636112. [DOI] [PubMed] [Google Scholar]
- 27.Stansfield WE, Andersen NM, Tang RH, Selzman CH. Periostin is a novel factor in cardiac remodeling after experimental and clinical unloading of the failing heart. Ann Thorac Surg. 2009;88:1916–1921. doi: 10.1016/j.athoracsur.2009.07.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Blanchard C, Mingler MK, McBride M, Putnam PE, Collins MH, Chang G, et al. Periostin facilitates eosinophil tissue infiltration in allergic lung and esophageal responses. Mucosal Immunol. 2008;1:289–296. doi: 10.1038/mi.2008.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Corren J, Lemanske RF, Hanania NA, Korenblat PE, Parsey MV, Arron JR, et al. Lebrikizumab treatment in adults with asthma. N Engl J Med. 2011;365:1088–1098. doi: 10.1056/NEJMoa1106469. [DOI] [PubMed] [Google Scholar]
- 30.Polikepahad S, Knight JM, Naghavi AO, Oplt T, Creighton CJ, Shaw C, et al. Proinflammatory role for let-7 microRNAS in experimental asthma. J Biol Chem. 2010;285:30139–30149. doi: 10.1074/jbc.M110.145698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lu LF, Boldin MP, Chaudhry A, Lin LL, Taganov KD, Hanada T, et al. Function of miR-146a in controlling Treg cell-mediated regulation of Th1 responses. Cell. 2010;142:914–929. doi: 10.1016/j.cell.2010.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wu F, Zikusoka M, Trindade A, Dassopoulos T, Harris ML, Bayless TM, et al. MicroRNAs are differentially expressed in ulcerative colitis and alter expression of macrophage inflammatory peptide-2 alpha. Gastroenterology. 2008;135:1624–1635. doi: 10.1053/j.gastro.2008.07.068. [DOI] [PubMed] [Google Scholar]
- 33.Sonkoly E, Janson P, Majuri ML, Savinko T, Fyhrquist N, Eidsmo L, et al. MiR-155 is overexpressed in patients with atopic dermatitis and modulates T-cell proliferative responses by targeting cytotoxic T lymphocyte-associated antigen 4. J Allergy Clin Immunol. 2010;126:581–589. doi: 10.1016/j.jaci.2010.05.045. [DOI] [PubMed] [Google Scholar]
- 34.Mishra A, Wang M, Pemmaraju VR, Collins MH, Fulkerson PC, Abonia JP, et al. Esophageal remodeling develops as a consequence of tissue specific IL-5-induced eosinophilia. Gastroenterology. 2008;134:204–214. doi: 10.1053/j.gastro.2007.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Assa’ad AH, Gupta SK, Collins MH, Thomson M, Heath AT, Smith DA, et al. An antibody against IL-5 reduces numbers of esophageal intraepithelial eosinophils in children with eosinophilic esophagitis. Gastroenterology. 2011;141:1593–1604. doi: 10.1053/j.gastro.2011.07.044. [DOI] [PubMed] [Google Scholar]
- 36.Yi R, Poy MN, Stoffel M, Fuchs E. A skin microRNA promotes differentiation by repressing “stemness.”. Nature. 2008;452:225–229. doi: 10.1038/nature06642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu Q, Lv GD, Qin X, Gen YH, Zheng ST, Liu T, et al. Role of microRNA let-7 and effect to HMGA2 in esophageal squamous cell carcinoma. Mol Biol Rep. 2012;39:1239–1246. doi: 10.1007/s11033-011-0854-7. [DOI] [PubMed] [Google Scholar]
- 38.Lin RJ, Xiao DW, Liao LD, Chen T, Xie ZF, Huang WZ, et al. MiR-142-3p as a potential prognostic biomarker for esophageal squamous cell carcinoma. J Surg Oncol. 2012;105:175–182. doi: 10.1002/jso.22066. [DOI] [PubMed] [Google Scholar]
- 39.Yuan Y, Zeng ZY, Liu XH, Gong DJ, Tao J, Cheng HZ, et al. MicroRNA-203 inhibits cell proliferation by repressing DeltaNp63 expression in human esophageal squamous cell carcinoma. BMC Cancer. 2011;11:57. doi: 10.1186/1471-2407-11-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tsuchiya S, Fujiwara T, Sato F, Shimada Y, Tanaka E, Sakai Y, et al. MicroRNA-210 regulates cancer cell proliferation through targeting fibroblast growth factor receptor-like 1 (FGFRL1) J Biol Chem. 2011;286:420–428. doi: 10.1074/jbc.M110.170852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li S, Li Z, Guo F, Qin X, Liu B, Lei Z, et al. miR-223 regulates migration and invasion by targeting Artemin in human esophageal carcinoma. J Biomed Sci. 2011;18:24. doi: 10.1186/1423-0127-18-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li X, Lin R, Li J. Epigenetic silencing of microRNA-375 regulates PDK1 expression in esophageal cancer. Dig Dis Sci. 2011;56:2849–2856. doi: 10.1007/s10620-011-1711-1. [DOI] [PubMed] [Google Scholar]
- 43.Matsushima K, Isomoto H, Kohno S, Nakao K. MicroRNAs and esophageal squamous cell carcinoma. Digestion. 2010;82:138–144. doi: 10.1159/000310918. [DOI] [PubMed] [Google Scholar]
- 44.Medina PP, Nolde M, Slack FJ. OncomiR addiction in an in vivo model of micro-RNA-21-induced pre-B-cell lymphoma. Nature. 2010;467:86–90. doi: 10.1038/nature09284. [DOI] [PubMed] [Google Scholar]
- 45.Hatley ME, Patrick DM, Garcia MR, Richardson JA, Bassel-Duby R, van Rooij E, et al. Modulation of K-Ras-dependent lung tumorigenesis by MicroRNA-21. Cancer Cell. 2010;18:282–293. doi: 10.1016/j.ccr.2010.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cai X, Cullen BR. The imprinted H19 noncoding RNA is a primary microRNA precursor. RNA. 2007;13:313–316. doi: 10.1261/rna.351707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tsang WP, Ng EK, Ng SS, Jin H, Yu J, Sung JJ, et al. Oncofetal H19-derived miR-675 regulates tumor suppressor RB in human colorectal cancer. Carcinogenesis. 2010;31:350–358. doi: 10.1093/carcin/bgp181. [DOI] [PubMed] [Google Scholar]
- 48.Arroyo JD, Chevillet JR, Kroh EM, Ruf IK, Pritchard CC, Gibson DF, et al. Argonaute2 complexes carry a population of circulating microRNAs independent of vesicles in human plasma. Proc Natl Acad Sci U S A. 2011;108:5003–5008. doi: 10.1073/pnas.1019055108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rabinowits G, Gercel-Taylor C, Day JM, Taylor DD, Kloecker GH. Exosomal microRNA: a diagnostic marker for lung cancer. Clin Lung Cancer. 2009;10:42–46. doi: 10.3816/CLC.2009.n.006. [DOI] [PubMed] [Google Scholar]
- 50.Valadi H, Ekstrom K, Bossios A, Sjostrand M, Lee JJ, Lotvall JO. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol. 2007;9:654–659. doi: 10.1038/ncb1596. [DOI] [PubMed] [Google Scholar]
- 51.Tian T, Wang Y, Wang H, Zhu Z, Xiao Z. Visualizing of the cellular uptake and intracellular trafficking of exosomes by live-cell microscopy. J Cell Biochem. 2010;111:488–496. doi: 10.1002/jcb.22733. [DOI] [PubMed] [Google Scholar]
- 52.Sonkoly E, Wei T, Janson PC, Saaf A, Lundeberg L, Tengvall-Linder M, et al. MicroRNAs: novel regulators involved in the pathogenesis of psoriasis? PLoS One. 2007;2:e610. doi: 10.1371/journal.pone.0000610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mayoral RJ, Pipkin ME, Pachkov M, van Nimwegen E, Rao A, Monticelli S. MicroRNA-221–222 regulate the cell cycle in mast cells. J Immunol. 2009;182:433–445. doi: 10.4049/jimmunol.182.1.433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Aceves SS, Chen D, Newbury RO, Dohil R, Bastian JF, Broide DH. Mast cells infiltrate the esophageal smooth muscle in patients with eosinophilic esophagitis, express TGF-beta1, and increase esophageal smooth muscle contraction. J Allergy Clin Immunol. 2010;126:1198–1204. doi: 10.1016/j.jaci.2010.08.050. [DOI] [PubMed] [Google Scholar]
- 55.Dellon ES, Chen X, Miller CR, Fritchie KJ, Rubinas TC, Woosley JT, et al. Tryptase staining of mast cells may differentiate eosinophilic esophagitis from gastroesophageal reflux disease. Am J Gastroenterol. 2011;106:264–271. doi: 10.1038/ajg.2010.412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Assa’ad AH, Putnam PE, Collins MH, Akers RM, Jameson SC, Kirby CL, et al. Pediatric patients with eosinophilic esophagitis: an 8-year follow-up. J Allergy Clin Immunol. 2007;119:731–738. doi: 10.1016/j.jaci.2006.10.044. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.