Abstract
Almost a decade after the publication of the complete sequence of the genome of the human malaria parasite Plasmodium falciparum, the mechanisms involved in gene regulation remain poorly understood. Like other eukaryotic organisms, P. falciparum’s genomic DNA organizes into nucleosomes. Nucleosomes are the basic structural units of eukaryotic chromatin and their regulation is known to play a key role in regulation of gene expression. Despite its importance, the relationship between nucleosome positioning and gene regulation in the malaria parasite has only been investigated recently. Using two independent and complementary techniques followed by next-generation high-throughput sequencing, our laboratory recently generated a dynamic atlas of nucleosome-bound and nucleosome-free regions (NFRs) at single-nucleotide resolution throughout the parasite erythrocytic cycle. We have found evidences that genome-wide changes in nucleosome occupancy play a critical role in controlling the rigorous parasite replication in infected red blood cells. However, the role of nucleosome positioning at remarkable locations such as transcriptional start sites (TSS) was not investigated. Here we show that a study of NFR in experimentally determined TSS and in silico-predicted promoters can provide deeper insights of how a transcriptionally permissive organization of chromatin can control the parasite’s progression through its life cycle. We find that NFRs found at TSS and core promoters are strongly associated with high levels of gene expression in asexual erythrocytic stages, whereas nucleosome-bound TSSs and promoters are associated with silent genes preferentially expressed in sexual stages. The implications in terms of regulatory evolution, adaptation of gene expression and their impact in the design of antimalarial strategies are discussed.
Keywords: Nucleosome, Malaria, TSS, Promoter, Transcription, Virulence, Evolution, MAINE-seq, FAIRE-seq
1. Introduction
With up to one million deaths per year, malaria remains one of the deadliest infectious diseases in the world. Malaria is a mosquito-borne infectious disease caused by a eukaryotic protist of the genus Plasmodium. Plasmodium can parasitize a wide array of organisms including birds (e.g. Plasmodium gallinaceum), rodents (e.g. Plasmodium berghei, Plasmodium chabaudi), and monkeys (e.g. Plasmodium reichenowi, Plasmodium knowlesi). A specialization into mosquitoes of the genus Anopheles corresponds with the expansion of Plasmodium parasites into mammals including humans (Martinsen et al., 2008). Four different species of Plasmodia can infect human: Plasmodium malariae, Plasmodium ovale, Plasmodium vivax and Plasmodium falciparum. In recent years, human cases of lethal or febrile malaria have also occurred with Plasmodium knowlesi, a parasite usually found in macaques, in certain forested areas of South-East Asia (Galinski and Barnwell, 2009; Jongwutiwes et al., 2004). P. falciparum and P. vivax are the most common strains infecting humans, P. falciparum being responsible for the most severe malignant malaria leading to coma and death. At the molecular level, P. falciparum is most closely related to P. reichenowi (Escalante and Ayala, 1994) and may have directly originated from P. reichenowi of chimpanzees (Rich et al., 2009) or bonobos (Krief et al., 2010) by a single host transfer that occurred as early as 2–3 million or as recently as 10,000–70,000 years ago (Rich et al., 2009). Phenotypic differences between each of these species find their bases at the genomic and genetic level, and the evolution of gene expression leading to host specificity.
Almost a decade after the publication of the complete sequence of the human malaria parasite P. falciparum (Gardner et al., 2002), the mechanisms involved in gene regulation in the malaria parasite remain poorly understood. Whilst microarray analyses have revealed a cascade of mRNA accumulation suggesting that genes are transcribed in a “just in time” manner throughout P. falciparum’s asexual developmental cycle (Bozdech et al., 2003; Le Roch et al., 2003), no specific transcriptional response to stress (e.g. environmental perturbations and small molecule inhibitors) has been observed besides a cell cycle arrest and strong activation of sexual differentiation (Ganesan et al., 2008a; Le Roch et al., 2008). These data are consistent with the relative paucity of transcription factors and regulatory elements observed in the parasite genome (Coulson et al., 2004; Young et al., 2008). All together these data highlight an unusual mechanism of transcriptional regulation in the parasite with a relative transcriptional rigidity during the Plasmodium asexual cycle progression.
Despite the importance of epigenetic mechanisms to regulate transcriptional regulation in eukaryotic cells, the role of epigenetics in regulating the malaria parasite’s gene expression has been investigated only recently (Issar et al., 2008; Lopez-Rubio et al., 2009; Pérez-Toledo et al., 2009; Ponts et al., 2010). Like other eukaryotic organisms, P. falciparum’s genomic DNA organizes into nucleosomes (Ponts et al., 2010), i.e. ~147 bp of double-stranded DNA wrapped around a histone core consisting of the assembly of four histone homodimers, one from each type of the histone proteins H2A, H2B, H3 and H4. No clear homolog of the linker histone H1, responsible for a higher order of chromosome compaction known as the transcriptionally inactive “30 nm fiber”, has been identified in Plasmodia genomes. This absence may indicate that Plasmodium genome could exhibit a lower level of chromosome packaging when compared to other eukaryotes. A genuinely lower degree of compaction is consistent with our recent observation of a general relaxed and transcriptionally active status of P. falciparum’s chromatin throughout its asexual red blood cell cycle (Ponts et al., 2010). These recent data are also consistent with a constitutive binding of the transcription pre-initiation complex on both active and inactive promoters (Gopalakrishnan et al., 2009). Moreover, recent studies identified very high levels of histone H3 acetylated on its lysine 9 (Trelle et al., 2009), an epigenetic mark of active transcription, distributed on the entire genome with the exception of the subtelomeric regions that contain the inactive mark trimethylated histone H3 (Salcedo-Amaya et al., 2009). These subtelomeric regions contain the parasite’s virulence genes that are silenced during the erythrocytic cycle and seem to be regulated by specific mechanisms, notably those involving intronic regulatory elements (Ponts et al., 2010; Dzikowski and Deitsch, 2009). All together, these data suggest a chromatin-driven model of gene regulation in the human malaria parasite asexual stages rather than transcription factor-centered regulation of gene expression. The study of chromatin structure at remarkable genomic positions, such as transcriptional start sites, can therefore provide new insights on to how a transcriptionally permissive organization of chromatin can control the parasite’s progression through its life cycle within multiple sexual/asexual, host/vector and tissue transitions.
The role of nucleosome positioning changes in regulatory variation and evolution has been intensively studied in various organisms such as yeast, flies, and humans (see Choi and Kim, 2009 for a review). Promoter architecture itself is believed to play a role in gene expression evolution driven by particular features like the presence of nucleosome-free regions at certain positions (see Tirosh et al., 2009 for a summary). So far none of these features have been described at the chromatin level in the human malaria parasite, P. falciparum. Using next-generation high-throughput sequencing, we recently generated dynamic genome-wide maps of both nucleosome-bound and nucleosome-free regions (NFRs) at single-nucleotide resolution (Ponts et al., 2010). Here we use these maps to examine the chromatin structure (NFR vs. nucleosome content) at remarkable genomic positions using experimentally proposed transcription start sites (TSS, Watanabe et al., 2001, 2004) and predicted promoters (Brick et al., 2008). The present analysis compares chromatin architecture at TSS and predicted core promoters to gene expression in asexual stages of P. falciparum. We find that promoters and TSS are associated with NFRs at every asexual stages of the erythrocytic cycle. On the contrary, genes exclusively expressed during sexual stages are associated with nucleosome-bound TSS and core promoters. The implications in terms of regulatory evolution and adaptation of gene expression are discussed.
2. Materials and methods
2.1. Dataset source
The datasets used in the present study were generated by formaldehyde-assisted isolation of regulatory elements and MNase-assisted isolation of nucleosomal elements coupled to deep sequencing (respectively FAIRE-seq and MAINE-seq, Ponts et al., 2010). Briefly, FAIRE-seq isolates genomic regions that are free of nucleosomes whereas MAINE-seq isolates genomic regions that are bound to nucleosomes (see Ponts et al., 2010 for experimental details on sample preparation and sequencing). Seven time points along the asexual erythrocytic developmental cycle of P. falciparum were considered: 0 h post-invasion (hpi), 6 hpi, 12 hpi, 18 hpi, 24 hpi, 30 hpi and 36 hpi (respectively early ring, mid ring, late ring, early trophozoite, late trophozoite, early schizont, and late schizont stages). The integrality of the data has been deposited on PlasmoDB (www.plasmodb.org) and can be displayed in the genome browser. In addition, all sequenced reads can be downloaded from the NCBI short read archive under the accession numbers SRA010122 and SRA010123.
2.2. Mapping of short sequenced reads
Short sequenced reads (36 bp) were mapped to the P. falciparum genome v6.3 (www.plasmoDB.org) according to a previously described procedure (Ponts et al., 2010), i.e. allowing up to two mismatches and rejecting reads that could map at more than one position on the genome. Since the sizes of the sequenced DNA fragments were 200–225 bp for FAIRE-seq and ~150 bp for MAINE-seq, the length of each mapped read was extended post-mapping to 200 bp and 150 bp for FAIRE-seq and MAINE-seq samples, respectively. This mapping procedure reflects more accurately the composition of each sample based on the principle that if the read is present and aligns to a unique location (i.e. a single locus without any ambiguity) in the reference genome, then the whole fragment is present in the sample and can only align to this particular location.
2.3. Read count per base normalization
For each time point in each experiment, each chromosome was normalized in two steps, similarly to a normalization method previously proposed (Ponts et al., 2010). First, the total number of nucleotides in reads mapped to the reference genome was normalized per 10-fold coverage (the sum of the total read count per base along the chromosome divided by ten times the length of the chromosome). Since the datasets reflect differential occupancies of nucleosomes along chromosomes, a second normalization was performed to account for the differences in the percentage of genome covered and data spreading across time points (e.g. a coverage obtained with one million nucleotides spread on 5000 genomic bases will look twice less intense than one million nucleotides spread on 10,000 genomic bases).
2.4. Asexual nucleosome occupancy profiles of selected regions of the genome
Asexual nucleosome occupancy profiles that would reflect a general chromatin organization of genes during the asexual developmental cycle were prepared by computing the mean of the read counts for each position using our FAIRE-seq and MAINE-seq profiles. P. falciparum’s genome annotation v6.3 was downloaded from www.plasmoDB.org and the start and stop positions of each protein-coding gene was extracted (see complete list in Supplementary Table S1). Experimentally proposed TSS locations were downloaded from the Full-Malaria database (Watanabe et al., 2001, 2004). These TSS were determined by the 5′-end position of full-length cDNAs (from erythrocytic stages of the parasite) sequencing and mapping. Only those with an identified overlapping gene were retained in our analysis, i.e. 2855 TSS. The central positions of in silico-predicted core promoters were obtained from Brick et al. (2008). The predictions were made using the cDNA collection from the Full-Malaria database as a training set. FAIRE-seq and MAINE-seq asexual read count profiles of the regions [−500; +500] centered on the position of interest were retrieved, scaled (zscore) and averaged at every position.
2.5. Nucleosome occupancy profiles across seven timepoints of the NFRs upstream of the TSS
TSSs for which overlapping genes were not identified were filtered out from the Full-Malaria database list (Watanabe et al., 2001, 2004). A total of 2855 TSSs (see complete list in Supplementary Table S2) were then used to analyze the changes of chromatin availability to the transcription machinery at the NFR immediately upstream of the TSS. For each region spanning [−150; 50] bases around the TSS (empirically determined), FAIRE-seq and MAINE-seq read count profiles at each of the seven time points were retrieved and integrated to determine the total area of enrichment. For each time point, the degree of chromatin availability of a given locus was measured by the enrichment value obtained with FAIRE divided by the enrichment value obtained with MAINE (Ponts et al., 2010). Results are expressed in a log2 scale.
2.6. K-means clustering and comparison with mRNA levels expression data
K-means clustering was performed on the log2(FAIRE/MAINE) time-series (seven time points) using the Euclidean distance method (replicates = 5000, confidence measurement = silhouette value, K = 2). Outliers within clusters were filtered out (confidence value lower than 0.7). For each cluster, a general profile of chromatin availability at TSS across P. falciparum’s asexual stages (log2(FAIRE/MAINE) considered at the seven time points) was calculated as the average of the profile of chromatin availability at TSS of genes belonging to the considered cluster (scaled using the fold difference to the mean). These profiles were compared to average expression profiles computed from previously published mRNA levels measurements (Le Roch et al., 2003).
3. Results
3.1. Enhanced detection of nucleosome-bound and nucleosome-free regions
Using MNase-assisted isolation of nucleosomal elements and formaldehyde-assisted isolation of regulatory elements coupled to deep sequencing, alias MAINE-seq and FAIRE-seq, we recently identified the regions of the P. falciparum genome that are bound to or free of nucleosomes, respectively (our published results Ponts et al., 2010). Considering seven time points along the asexual erythrocytic developmental cycle, i.e. 0–36 h post-invasion (hpi) with 6-h increments, we observed that P. falciparum’s genome undergoes drastic changes of chromatin structure (Ponts et al., 2010). At ring stage (0–12 hpi) chromatin is partly euchromatic. Then, at trophozoite stage (18 hpi), chromatin is completely open and most genes are available to the transcription machinery. Finally, DNA progressively packs until the most condensed state of the cycle at schizont stage (36 hpi) before merozoite differentiation and re-invasion of fresh erythrocytes. We also found that nucleosomes are mostly localized within exons, consistent with other observations in human and yeast (Nahkuri et al., 2009; Schwartz et al., 2009; Tilgner et al., 2009; Andersson et al., 2009). Here, we use these nucleosome occupancy data to explore chromatin status of promoters and TSS with regards to gene expression in the intra-erythrocytic asexual stages of P. falciparum.
To locally minimize the experimental variations of the read counts per base due to different qualities of the sequenced reads, and based on the fact that the presence of a sequenced read demonstrates the presence of the corresponding full-length gDNA fragment, we extended the length of each mapped read to the length of the sequenced library (see Section 2). We compared the performances of this method, here called “extended mapping”, to the original “classical” mapping results obtained by Ponts et al. (2010). Results are shown in Table 1. The percentage of the genome covered by at least one read, the mean, and the median read coverage per base are considered (performances are measured before normalization is applied to the dataset). For both FAIRE-seq and MAINE-seq, the extended mapping procedure significantly increases and homogenizes the percentage of genome covered by at least one read. We further compared the profiles of coverage obtained with extended mapping to the ones previously obtained with classical mapping (see our example Fig. 1A vs. B). The positioning of the peaks is identical in both classical and extended mapping. The extended mapping, however, allows a denser coverage of the region, improves the shapes of the peaks, and reveals small peaks that are lost in the background of the classical mapping (e.g. position ~390,000). We also verified the complementarity of FAIRE-seq profiles (nucleosome-free regions) with MAINE-seq profiles (nucleosome-bound regions) that was described in the original experiment (Ponts et al., 2010). We visually compared the distributions of reads obtained with FAIRE-seq to the ones measured with MAINE-seq along chromosome 4 (see Fig. 1B for an example of our results) and found that regions highly covered with FAIRE-seq reads have very low coverage with MAINE-seq reads, which demonstrates that extended mapping preserves FAIRE-seq and MAINE-seq complementarities. The read counts per base obtained using this method were used to identify nucleosome-bound and nucleosome-free regions (NFRs) in the genome. Nucleosome-bound regions are characterized by a high density of MAINE-seq reads (and a low density of FAIRE-seq reads), and NFRs are defined by a high density of FAIRE-seq reads (and a low density of MAINE-seq reads).
Table 1.
Comparison of mapping performances between classical and extended mapping.
| Performance criteriaa | FAIRE-seq data
|
MAINE-seq data
|
||
|---|---|---|---|---|
| Classical mapping (Ponts et al., 2010) | Extended mapping | Classical mapping (Ponts et al., 2010) | Extended mapping | |
| Proportion of genome covered by at least one read | 0.64 ± 0.1 | 0.94 ± 0.04 | 0.44 ± 0.1 | 0.71 ± 0.1 |
| Mean read count per base | 5.8 ± 1.4 | 24.1 ± 6.7 | 8.4 ± 1.7 | 23.0 ± 5.3 |
| Median read count per base | 3.9 ± 1.0 | 16.9 ± 6.2 | 4.4 ± 1.0 | 11.4 ± 3.8 |
All values are the average measurements among all time points ± standard deviation.
Fig. 1.
Example of the read count profiles obtained with FAIRE-seq and MAINE-seq using extended mapping by comparison with classical mapping. An example of the correlation between (A) classical and (B) extended mapping, and the complementarity between FAIRE-seq and MAINE-seq, is given for the region [365,000; 395,000] on chromosome 4. The read count profiles obtained at 18 hpi with both FAIRE-seq (in black) and MAINE-seq (in red) are overlaid. Extended mapping allows a denser coverage of the region, improves the shapes of the peaks, and reveals small peaks that are lost in the background of the classical mapping (e.g. position ~390,000). Extended mapping also preserves the complementarity of FAIRE-seq and MAINE-seq. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of the article.)
3.2. Identification of nucleosome-bound and nucleosome-free region at remarkable sites in the genome of P. falciparum
To characterize the general nucleosome occupancy in P. falciparum’s genome asexual stages, we averaged the read counts for each position obtained at the seven different time points for both MAINE-seq and FAIRE-seq. Using these average profiles, we examined the chromatin status of regions surrounding the ends of protein-coding genes, their TSS, and their predicted core promoters.
3.3. Chromatin structure at the 5′ and 3′ ends of protein-coding genes
The average nucleosome occupancy at the 5′ and 3′ ends of protein-coding genes during P. falciparum’s asexual cycle is shown in Fig. 2 (the complete list of protein-coding genes was downloaded from PlasmoDB v6.3 and is provided in Supplementary Table S1). Consistent with our previous observations in P. falciparum, nucleosome content is higher within gene bodies than in their 5′ and 3′ flanking regions (Ponts et al., 2010; Westenberger et al., 2009). In addition, we observe the presence of a well-positioned nucleosome on both ends of the gene, at the sites of the ATG and the STOP codons. Furthermore, we observe marked NFRs flanking the gene bodies. The 5′ NFR is believed to contain the TSS and play a role in transcription initiation in yeast whereas the 3′ NFR could intervene with transcription termination (see Jiang and Pugh, 2009 for a review). The high nucleosome content of gene bodies is suspected to be involved in the regulation of transcription elongation (Choi et al., 2009; Vaillant et al., 2010). Moreover, observations in various eukaryotes suggested an additional role in exon recognition during RNA processing (see Tilgner and Guigó, 2010 for a review).
Fig. 2.
Nucleosome occupancy at gene boundaries. Average nucleosome occupancy profiles (zscored) were computed for regions spanning 1000 bp around the ATG or STOP codons of annotated protein-coding genes (see complete list in Supplementary Table S1) using FAIRE-seq profiles (nucleosome-free DNA content, in black) and MAINE-seq profiles (nucleosome-bound DNA content, in red). Protein-coding genes are flanked by NFRs during P. falciparum’s asexual erythrocytic stages. Well-positioned nucleosomes are found at both the 5′ and 3′ end of the coding region (nucleosome represented by a grey circle).
3.4. Chromatin structure at experimentally proposed TSS (Watanabe et al., 2001, 2004)
In P. falciparum, the mechanisms involved in the regulation of transcription initiation are poorly known. To determine the nucleosome occupancy at P. falciparum’s TSSs, we examined the average FAIRE-seq and MAINE-seq read count profiles of 2855 experimentally proposed transcription start sites (Watanabe et al., 2001, 2004) (see complete list in Supplementary Table S2). Our results are represented in Fig. 3. Genomic regions containing TSSs are relatively depleted in nucleosomes. Additionally, a clear ~150–200 bp-long NFR is present immediately upstream of the TSS, consistent with the common situation in other eukaryotes. Interestingly, the length of the NFR is about the combined size of a nucleosome and its linker DNA.
Fig. 3.
Nucleosome occupancy at experimentally proposed TSS. Average nucleosome occupancy profiles (zscored) were computed for regions spanning 1000 bp centered on experimentally proposed TSS (Watanabe et al., 2001, 2004) (see list of TSS in Supplementary Table S2). FAIRE-seq profile (nucleosome-free DNA content) is shown in black; MAINE-seq profile (nucleosome-bound DNA content) is shown in red. TSSs are preceded by NFRs during P. falciparum’s asexual erythrocytic stages. The NFR approximately ranges from −150 bp to +50 bp around the TSS.
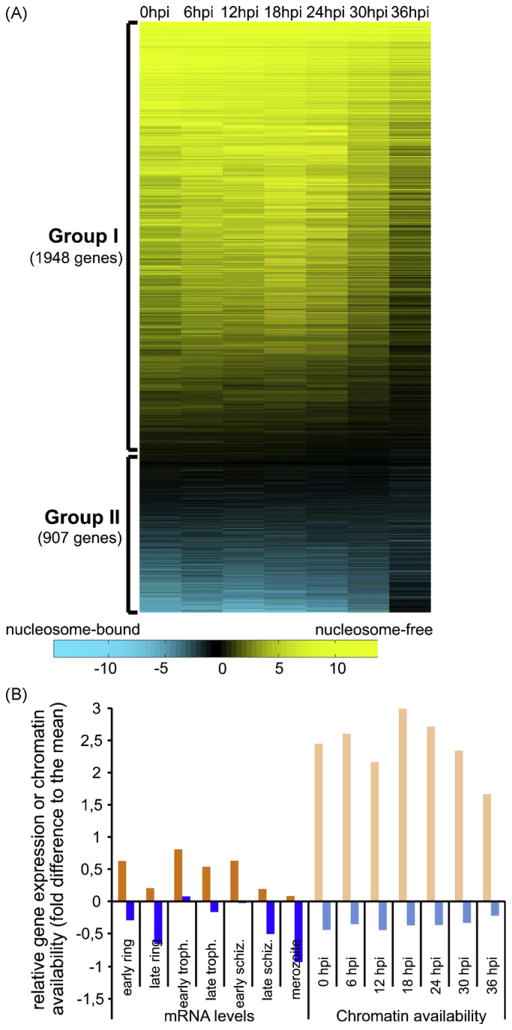
We investigated in more details the occurrence of these NFRs dynamically across the asexual erythrocytic cycle (using the data from the seven time points separately and the TSSs for which a corresponding gene has been proposed) and compared it to RNA transcript accumulation (data from Le Roch et al., 2003). For each region spanning [−150; 50] bases around the TSSs taken at each time point, we retrieved the level of enrichment in nucleosome-free (FAIRE) or nucleosome-bound (MAINE) DNA. The level of chromatin availability to the transcription machinery is expressed by the ratio between enrichment with FAIRE and enrichment with MAINE at a log2 scale (see Ponts et al., 2010 and Section 2 for details). Using K-means, we clustered the log2(FAIRE/MAINE) profiles (seven time points) of chromatin availability into two groups (Fig. 4A, see Supplementary Table S3 for the list of the genes). Group I consists of 1948 genes for which the NFR upstream of the TSS is always present (virtually never occupied by a nucleosome). Group II consists of 907 genes for which the NFR is very often occupied by a nucleosome. We compared the average expression profiles (mRNA levels from Le Roch et al., 2003) and the average profiles of chromatin availability (see Section 2) among the groups (Fig. 4B). Genes that belong to group I (orange bars) have higher mRNA levels at every asexual stage than the ones observed among group II (blue bars). In parallel, chromatin around TSS sites is more open for genes from group I (light orange blue bars) than genes from group II (light blue bars). These results indicate a strong association between gene expression and nucleosome landscape at the TSS. Open chromatin is indicative of higher mRNA levels. Conversely, the binding of a nucleosome at a TSS is associated to lower mRNA levels. Transcript levels, nonetheless, vary across the asexual cycle, which suggest the existence of other downstream regulations such as regulation of mRNA stability, binding of transcription factors (positive or negative), enhancers, the use of alternative TSS/promoters, etc. This analysis, however, cannot be applied to the parasite’s virulence genes that are located in the silenced subtelomeric regions of the chromosomes. Their regulation involves complex mechanisms of allelic exclusion in which regulatory elements located within their single intron play major roles (Ponts et al., 2010; Dzikowski and Deitsch, 2009). Regulation of virulence genes in the malaria parasite goes beyond chromatin structure and may require the intervention of specialized transcription factors. Recent work, indeed, found one of the members of the ApiAP2 family of transcription factors to be exclusively distributed at subtelomeric regions where it controls the formation of densely packed chromatin and thus could play a role in the silencing of virulence gene (Flueck et al., 2010).
Fig. 4.
Nucleosome occupancy at NFRs preceding TSS and gene expression. (A) K-means clustering of genes according to their asexual profiles of chromatin availability within the NFR preceding the TSS. The degree of nucleosome occupancy (log2(FAIRE/MAINE)) within the NFR is color coded from cyan (nucleosome-bound) to yellow (nucleosome-free). Two groups are delineated: group I with high chromatin availability, and group II with low/no chromatin availability. (B) Comparison of gene expression levels and chromatin availability at NFRs preceding TSS. Average asexual expression profiles (data from Le Roch et al., 2003) were calculated for genes in group I (dark orange) and group II (dark blue). Similarly, average asexual chromatin availability profiles (log2(FAIRE/MAINE)) were calculated for genes among group I (light orange) and group II (light blue). Genes within group I are generally more expressed and have open NFR upstream of their TSS. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of the article.)
We expanded our investigation and performed a similar analysis considering in silico-predicted core promoters (Brick et al., 2008). The algorithm that Brick et al. developed is based on the physicochemical properties of the DNA sequence surrounding TSS sites (the authors trained their promoter predicting tool on the sequences surrounding TSS obtained using the cDNA collection from the Full-Malaria Database; their following predictions correlate extremely well with the TSS from Full Malaria (Brick et al., 2008). They predicted more than 200,000 promoters that could be assigned to downstream annotated genes, with various levels of confidence (from moderately to highly confident, with EGASP values ranging from 0.4 to 1). To determine the nucleosome occupancy at these predicted core promoters during P. falciparum’s asexual cycle, we examined the average FAIRE-seq and MAINE-seq read count profiles surrounding the predicted positions. Fig. 5A shows our results for 3477 highly confident promoters (EGASP = 1) proposed by Brick et al. (2008) for a full evaluation of their prediction and the EGASP score. Genomic regions containing putative core promoters are clearly nucleosome-free, and the length of the NFR is about 300–350 bp. We assigned 3397 of these putative promoters to their downstream genes (see Supplementary Table S4 for the list of promoter positions and the corresponding downstream gene) and calculated their profiles of chromatin availability across time (log2(FAIRE/MAINE) for the regions spanning 300 bp centered on MAPP predicted promoter positions). Results are displayed in Fig. 5B. We immediately observed that the very large majority of the promoters are nucleosome-free, regardless their expression profile (data not shown). This observation is consistent with Gopalakrishnan and colleagues’ results showing that the P. falciparum transcription pre-initiation complex assembles on both active and inactive promoters, regardless of transcriptional activity (Gopalakrishnan et al., 2009). Interestingly, the average profile of chromatin availability peaks around 18–24 hpi. This “opening” of chromatin coincides with DNA replication (S phase) during the P. falciparum’s erythrocytic cell cycle. Only a subset of putative promoters appears to be densely packed. We investigated the nature of these genes for which putative promoter regions are packed.
Fig. 5.
Nucleosome occupancy at predicted promoters and gene function. (A) Nucleosome occupancy at in silico-predicted promoter regions. Average nucleosome occupancy profiles (zscored) were computed for regions spanning 1000 bp centered on predicted promoters (Brick et al., 2008) (see Supplementary Table S3 for the list of genes in each group). FAIRE-seq profile (nucleosome-free DNA content) is shown in black; MAINE-seq profile (nucleosome-bound DNA content) is shown in red. Predicted promoters are free of nucleosomes during P. falciparum’s asexual erythrocytic stages. The NFR approximately spans about 300 bp around the predicted promoter position. (B) Comparison of gene expression levels and chromatin availability at predicted core promoters. Genes with asexual expression percentiles higher (or equal) than 90% and lower (or equal) than 10% were downloaded from PlasmoDB (expression based on data from Le Roch et al., 2003). Genes for which no confident core promoter was predicted were filtered out. The group of highly expressed genes consisted of 491 members, and the group with low/moderate expression contained 350 genes. Average asexual chromatin availability profiles (log2(FAIRE/MAINE)) were calculated for highly (orange curve) and moderately (blue curve) expressed genes. Chromatin status for both groups of genes follows similar dynamics. Nonetheless, chromatin tends to be more open in core promoters of highly expressed that in the ones of modestly expressed genes. (C) Heatmap representation of general chromatin availability within predicted promoters during the asexual cycle. The regions spanning 300 bp centered on MAPP predicted promoter positions are considered. The degree of nucleosome occupancy (log2(FAIRE/MAINE)) within the NFR is color coded from cyan (nucleosome-bound) to yellow (nucleosome-free). The very large majority of the promoters are strikingly open. Only a subset of them appears to be densely packed, subset significantly enriched in sexual genes (more than a 10-fold over-representation). (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of the article.)
Subsets of genes were prepared according to their expression levels (data from Le Roch et al., 2003): genes with the maximum of expression during the asexual cycle at the 90th percentile (and higher) and genes with the minimum of expression during the asexual cycle at the 10th percentile (and lower), with a predicted core promoter at EGAP = 1.0. A total of 491 highly expressed genes and 350 moderately expressed ones were retrieved (see Supplementary Table S5 for the list of these genes). We then examined the average asexual chromatin availability profiles (log2(FAIRE/MAINE)) of highly (orange curve) and moderately (blue curve) expressed genes (Fig. 5B). Chromatin status for both groups of genes follows similar dynamics. Chromatin, however, tends to be more open in core promoters of highly expressed than in the ones of modestly expressed genes. These observations are consistent with low expression genes likely to be silenced whereas high expression genes are more likely to be expressed.
Finally, we sampled the 5% most densely packed genes (at their promoter region) and compared their functions to the ones carried on by the 5% most open genes (i.e. ~170 genes for each group, see Supporting Table S6). To perform our task we used the classification of genes into functional clusters proposed by Le Roch et al. (2003). We found that the group containing genes with densely packed promoters is significantly enriched (p < 0.05, measured using a hypergeometric test) from cluster 2 and cluster 9 that contain genes that are highly expressed during the parasite’s sexual stage (Fig. 5C). These findings suggest that promoters of genes involved in sexual stage function are constituted of closed chromatin during the erythrocytic development, consistent with silencing during asexual stages. The activation of these genes during gametocytogenesis is then an active process that involves the intervention of one specific transcription factor at least. Indeed, a recent work identified a member of the ApiAP2 family of transcription factor as the positive regulator of all the genes that are required for mosquito midgut infection at the ookinete stage (Yuda et al., 2009).
All together, these observations support a model of a genuine transcriptionally permissive state of promoter chromatin during P. falciparum’s development throughout its asexual erythrocytic cycle. Nucleosome positioning within promoters might silence the expression of sexual stage-specific genes in a regulated manner.
4. Discussion
In P. falciparum, like in many other eukaryotes, nucleosomes distribute preferentially within exons (Ponts et al., 2010; Westenberger et al., 2009). This preference might have strong consequences on gene expression by regulating transcription elongation and splicing for example (Choi et al., 2009). In addition, and this is probably the most well-known case of gene regulation, nucleosome occupancy of TSS and core promoters impedes the binding of the transcription machinery to the template DNA and inhibits transcription (repressive state). On the contrary, the presence of a NFR on the TSS would be a sign of possible transcriptional activity (permissive state). In yeast, gene bodies are flanked on both ends by NFRs (see Jiang and Pugh, 2009 for a review). The 5′ NFR is believed to contain the TSS whereas the 3′ NFR could intervene with transcription termination. We found that P. falciparum is not different from other eukaryotes in the way that its genes are flanked by NFRs on both ends. These NFRs, however, seem to be hardly ever occupied by nucleosomes.
We examined in more details the 5′ NFR and found that, on the whole, TSS and core promoters are mostly nucleosome-free during the asexual erythrocytic cycle (transcriptionally permissive chromatin). A transcriptionally permissive status however does not necessarily mean that transcription actually occurs. Transcript levels are not equal among genes with “open” TSS, and they are not necessarily constant across the cell cycle (illustrated in Fig. 4). A fundamental reason is the important fact that mRNA levels do not accurately reflect transcriptional activity. They are the combined result of production (transcription) and degradation (mRNA stability). After transcription of a gene in an “open” state, regulation of mRNA stability can increase (stable RNAs accumulate in the cell) or decrease (rapid degradation of the transcripts) gene expression. Regulation of transcripts half-lives is one of the many regulatory events that can occur downstream of the modulation of chromatin availability. Gene expression is indeed the result of the interplay between nucleosome occupancy, the action of general regulatory factors, the affinity of the transcription machinery for DNA template, post-transcriptional regulations, etc. In addition, we observe that the average chromatin availability profile reaches a maximum during the S phase of the parasite’s erythrocytic cycle (18–24 hpi), i.e. when the genome is replicated. In eukaryotes, DNA replication origins are usually enriched in open chromatin, and nucleosome removal or displacement is required for the replication machinery to access the DNA. In that sense, the large opening that we observe at 18 hpi in Fig. 5B is consistent with active DNA replication. In that sense, DNA replication and transcriptional activity at genes loci on or in the vicinity of DNA replication origins are related events. Transcriptionally active promoters are indeed constituted of open chromatin, which is a permissive state for DNA replication. In Saccharomyces cerevisiae for example, the histone acetyltransferase GCN5 is associated with transcriptional activity (Rodríguez-Navarro, 2009) and increases DNA replication when bound to a replication origin (Vogelauer et al., 2002). Active transcription, DNA replication, and chromatin architecture are therefore interplaying events in eukaryotes in general, and very likely in P. falciparum in particular.
We also identified small subsets of genes that are more likely to contain a nucleosome within the TSS or core promoter area. This subset of genes is enriched in those that play a role during the sexual development of the parasite, which suggests that chromatin architecture may be of major importance for the silencing of genes involved in sexual development. This apparent dichotomy in the chromatin architecture, i.e. low nucleosome occupancy vs. higher nucleosome occupancy at the locus of the NFR surrounding the TSS, is consistent with recent observations in 12 species of yeast (Tsankov et al., 2010). Tsankov and colleagues found a conserved dual status of chromatin structure at NFR loci. Yeast genes are either in “growth” mode, characterized by low nucleosome occupancy, or in “stress” mode, characterized by higher nucleo-some occupancy. Evolution and adaptation in yeast would be the result of a gene switch from “growth” mode to “stress” mode and vice versa. By analogy, sexual differentiation in P. falciparum would correspond to a switch of the sexual genes from “stress” mode into “growth” mode. Sexual differentiation in the parasite is indeed a common response to environmental perturbations and small molecule inhibitors in the parasite (Le Roch et al., 2008; Ganesan et al., 2008b). Interestingly, the genes involved in sexual differentiation are largely represented in the dataset describing inter-species and environmental transcriptional variations in Plasmodium (Hu et al., 2010; van Brummelen et al., 2009; Gonzales et al., 2008; Daily et al., 2007) (recent analyses further showed that part of the variations described by Daily et al., 2007) could be due to gametocytes (Lemieux et al., 2009).
All data considered together, a broad model of gene regulation in P. falciparum starts to emerge. During the asexual cycle of development, the parasite’s environment is well maintained (homeostasis of the red blood cell). The need for adaptation to environmental changes is minimum; gene regulation could be hard-wired and turned on by default, with a general active status of chromatin always accessible to general transcription factors and the rest of the transcriptional machinery. This hypothesis finds supports in the absence of specific transcriptional response to stress (e.g. environmental perturbations and small molecule inhibitors), besides only a general cell cycle arrest and strong induction of sexual differentiation (Le Roch et al., 2003; Ganesan et al., 2008a). Moreover, the nucleosome-depleted character of P. falciparum’s promoters (and non-exonic regions in general, Ponts et al., 2010) might be related to the transcriptional rigidity of the parasite since highly responsive genes in yeast are usually covered with nucleosomes (see Tirosh et al., 2009 for a review). Indeed, the removal of a nucleosome represents a quick way to adapt gene expression to a changing environment. If regulatory regions are nucleosome-free by default, regulation of transcription becomes rigid, transcriptional response to stress becomes limited, and adaptation and evolution become more difficult (see Choi and Kim, 2009 for a review). In the case of P. falciparum, non-coding regions are highly depleted in nucleosomes (Ponts et al., 2010; Westenberger et al., 2009). As a consequence, its virulence could be the result of a reduced capacity to evolve during its asexual cycle combined with a fairly recent jump from monkey to human (and thus a shorter history of co-evolution), and having minimal environmental pressure during the intra-erythrocytic asexual cycle (well-maintained intra-erythrocytic homeostasis).
On the whole, the basis of P. falciparum’s mechanisms of asexual gene regulation and adaptation to environmental stress are fairly simple. The parasite maintains a globally active transcriptional state during its asexual cycle that resembles the hyperactive state of embryonic cells. After each cycle, up to 32 daughter cells are derived from a single parasite. Then, under environmental stress (e.g. higher temperature i.e. fever), this cycle arrests and silenced genes involved in sexual differentiation are activated by specific mechanisms involving at least one ApiAP2 transcription factor (Yuda et al., 2009). The parasite differentiates and can spread further via a mosquito vector. In this scenario, P. falciparum would have achieved a high fitness: infection is maximized by its high division rate (permitted by a hyperactive transcription) and transmissibility is maximized by the systematic sexual differentiation under environmental stress. These considerations are of major importance whilst the research for a vaccine is intensive. Improving host immunity will have drastic consequences on the parasite’s fitness. Vaccination does not block the parasite’s progression but controls the parasite’s population within the vaccinated host. Recent studies on rodent malaria and human field data predict that reducing asexual density is expected to lead to evolution of higher virulence by selecting more virulent pathogen (see Mackinnon et al., 2008 for a review). On the other hand, strategies designed to block infection (e.g. prevention, drugs) are expected to lead to lower virulence. In the search of new antimalarial, understanding the parasite’s biology is of major importance.
Supplementary Material
Acknowledgments
We thank Glenn Hicks, Barbara Walter, Thomas Girke, Tyler Backman, and Rebecca Sun (IIGB, University of California, Riverside), and Gary Hardiman, Colleen Eckhardt-Ludka, and Ivan Wick (BIOGEM, University of California, San Diego) for assistance with Illumina® sequencing and Pipeline analysis. We also thank Jacques Prudhomme for assistance with P. falciparum cultures. 3D7 parasites were obtained through the MR4 (MRA-102) deposited by D.J. Carucci. This work was supported by the University of California, Riverside starting funds.
Appendix A. Supplementary data
Supplementary data associated with this article can be found, in the online version, at doi:10.1016/j.meegid.2010.08.002.
References
- Andersson R, Enroth S, Rada-Iglesias A, Wadelius C, Komorowski J. Genome Res. 2009;19:1732–1741. doi: 10.1101/gr.092353.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bozdech Z, Llinás M, Pulliam BL, Wong ED, Zhu J, DeRisi JL. PLoS Biol. 2003;1:E5. doi: 10.1371/journal.pbio.0000005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brick K, Watanabe J, Pizzi E. Genome Biol. 2008;9:R178. doi: 10.1186/gb-2008-9-12-r178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi JK, Kim Y. Epigenetics. 2009;4:291–295. doi: 10.4161/epi.4.5.9281. [DOI] [PubMed] [Google Scholar]
- Choi JK, Bae J, Lyu J, Kim T, Kim Y. Genome Biol. 2009;10:R89. doi: 10.1186/gb-2009-10-9-r89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coulson RMR, Hall N, Ouzounis CA. Genome Res. 2004;14:1548–1554. doi: 10.1101/gr.2218604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daily JP, Scanfeld D, Pochet N, Le Roch K, Plouffe D, Kamal M, Sarr O, Mboup S, Ndir O, Wypij D, Levasseur K, Thomas E, Tamayo P, Dong C, Zhou Y, Lander ES, Ndiaye D, Wirth D, Winzeler EA, Mesirov JP, Regev A. Nature. 2007;450:1091–1095. doi: 10.1038/nature06311. [DOI] [PubMed] [Google Scholar]
- Dzikowski R, Deitsch KW. Curr Genet. 2009;55:103–110. doi: 10.1007/s00294-009-0233-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escalante AA, Ayala FJ. Proc Natl Acad Sci USA. 1994;91:11373–11377. doi: 10.1073/pnas.91.24.11373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flueck C, Bartfai R, Niederwieser I, Witmer K, Alako BTF, Moes S, Bozdech Z, Jenoe P, Stunnenberg HG, Voss TS. PLoS Pathog. 2010;6:e1000784. doi: 10.1371/journal.ppat.1000784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galinski MR, Barnwell JW. Trends Parasitol. 2009;25:200–204. doi: 10.1016/j.pt.2009.02.002. [DOI] [PubMed] [Google Scholar]
- Ganesan K, Ponmee N, Jiang L, Fowble JW, White J, Kamchonwongpaisan S, Yuthavong Y, Wilairat P, Rathod PK. PLoS Pathog. 2008a;4:e1000214. doi: 10.1371/journal.ppat.1000214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganesan K, Ponmee N, Jiang L, Fowble J, White J, Kamchonwongpaisan S, Yuthavong Y, Wilairat P, Rathod P. PLoS Pathog. 2008b;4:e1000214. doi: 10.1371/journal.ppat.1000214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner MJ, Hall N, Fung E, White O, Berriman M, Hyman RW, Carlton JM, Pain A, Nelson KE, Bowman S, Paulsen IT, James K, Eisen JA, Rutherford K, Salzberg SL, Craig A, Kyes S, Chan M, Nene V, Shallom SJ, Suh B, Peterson J, Angiuoli S, Pertea M, Allen J, Selengut J, Haft D, Mather MW, Vaidya AB, Martin DMA, Fairlamb AH, Fraunholz MJ, Roos DS, Ralph SA, McFadden GI, Cummings LM, Subramanian GM, Mungall C, Venter JC, Carucci DJ, Hoffman SL, Newbold C, Davis RW, Fraser CM, Barrell B. Nature. 2002;419:498–511. doi: 10.1038/nature01097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzales JM, Patel JJ, Ponmee N, Jiang L, Tan A, Maher SP, Wuchty S, Rathod PK, Ferdig MT. PLoS Biol. 2008;6:e238. doi: 10.1371/journal.pbio.0060238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gopalakrishnan AM, Nyindodo LA, Ross Fergus M, López-Estraño C. Exp Parasitol. 2009;121:46–54. doi: 10.1016/j.exppara.2008.09.016. [DOI] [PubMed] [Google Scholar]
- Hu G, Cabrera A, Kono M, Mok S, Chaal BK, Haase S, Engelberg K, Cheemadan S, Spielmann T, Preiser PR, Gilberger T, Bozdech Z. Nat Biotechnol. 2010;28:91–98. doi: 10.1038/nbt.1597. [DOI] [PubMed] [Google Scholar]
- Issar N, Roux E, Mattei D, Scherf A. Cell Microbiol. 2008;10:1999–2011. doi: 10.1111/j.1462-5822.2008.01183.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang C, Pugh BF. Nat Rev Genet. 2009;10:161–172. doi: 10.1038/nrg2522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jongwutiwes S, Putaporntip C, Iwasaki T, Sata T, Kanbara H. Emerg Infect Dis. 2004;10:2211–2213. doi: 10.3201/eid1012.040293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krief S, Escalante AA, Pacheco MA, Mugisha L, André C, Halbwax M, Fischer A, Krief J, Kasenene JM, Crandfield M, Cornejo OE, Chavatte J, Lin C, Letourneur F, Grüner AC, McCutchan TF, Rénia L, Snounou G. PLoS Pathog. 2010;6:e1000765. doi: 10.1371/journal.ppat.1000765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Roch KG, Zhou Y, Blair PL, Grainger M, Moch JK, Haynes JD, De La Vega P, Holder AA, Batalov S, Carucci DJ, Winzeler EA. Science. 2003;301:1503–1508. doi: 10.1126/science.1087025. [DOI] [PubMed] [Google Scholar]
- Le Roch K, Johnson J, Ahiboh H, Chung D, Prudhomme J, Plouffe D, Henson K, Zhou Y, Witola W, Yates J, Mamoun C, Winzeler E, Vial H. BMC Genomics. 2008;9:513. doi: 10.1186/1471-2164-9-513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemieux JE, Gomez-Escobar N, Feller A, Carret C, Amambua-Ngwa A, Pinches R, Day F, Kyes SA, Conway DJ, Holmes CC, Newbold CI. Proc Natl Acad Sci USA. 2009;106:7559–7564. doi: 10.1073/pnas.0811829106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Rubio J, Mancio-Silva L, Scherf A. Cell Host Microb. 2009;5:179–190. doi: 10.1016/j.chom.2008.12.012. [DOI] [PubMed] [Google Scholar]
- Mackinnon MJ, Gandon S, Read AF. Vaccine. 2008;26(Suppl 3):C42–C52. doi: 10.1016/j.vaccine.2008.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinsen ES, Perkins SL, Schall JJ. Mol Phylogenet Evol. 2008;47:261–273. doi: 10.1016/j.ympev.2007.11.012. [DOI] [PubMed] [Google Scholar]
- Nahkuri S, Taft RJ, Mattick JS. Cell Cycle. 2009;8:3420–3424. doi: 10.4161/cc.8.20.9916. [DOI] [PubMed] [Google Scholar]
- Pérez-Toledo K, Rojas-Meza AP, Mancio-Silva L, Hernández-Cuevas NA, Delgadillo DM, Vargas M, Martínez-Calvillo S, Scherf A, Hernandez-Rivas R. Nucleic Acids Res. 2009;37:2596–2606. doi: 10.1093/nar/gkp115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponts N, Harris EY, Prudhomme J, Wick I, Eckhardt-Ludka C, Hicks GR, Hardiman G, Lonardi S, Le Roch KG. Genome Res. 2010;20:228–238. doi: 10.1101/gr.101063.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rich SM, Leendertz FH, Xu G, LeBreton M, Djoko CF, Aminake MN, Takang EE, Diffo JLD, Pike BL, Rosenthal BM, Formenty P, Boesch C, Ayala FJ, Wolfe ND. Proc Natl Acad Sci USA. 2009;106:14902–14907. doi: 10.1073/pnas.0907740106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodríguez-Navarro S. EMBO Rep. 2009;10:843–850. doi: 10.1038/embor.2009.168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salcedo-Amaya AM, van Driel MA, Alako BT, Trelle MB, van den Elzen AMG, Cohen AM, Janssen-Megens EM, van de Vegte-Bolmer M, Selzer RR, Iniguez AL, Green RD, Sauerwein RW, Jensen ON, Stunnenberg HG. Proc Natl Acad Sci USA. 2009;106:9655–9660. doi: 10.1073/pnas.0902515106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz S, Meshorer E, Ast G. Nat Struct Mol Biol. 2009;16:990–995. doi: 10.1038/nsmb.1659. [DOI] [PubMed] [Google Scholar]
- Tilgner H, Guigó R. Epigenetics. 2010;5 doi: 10.4161/epi.5.3.11319. [DOI] [PubMed] [Google Scholar]
- Tilgner H, Nikolaou C, Althammer S, Sammeth M, Beato M, Valcárcel J, Guigó R. Nat Struct Mol Biol. 2009;16:996–1001. doi: 10.1038/nsmb.1658. [DOI] [PubMed] [Google Scholar]
- Tirosh I, Barkai N, Verstrepen KJ. J Biol. 2009;8:95. doi: 10.1186/jbiol204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trelle MB, Salcedo-Amaya AM, Cohen AM, Stunnenberg HG, Jensen ON. J Proteome Res. 2009;8:3439–3450. doi: 10.1021/pr9000898. [DOI] [PubMed] [Google Scholar]
- Tsankov AM, Thompson DA, Socha A, Regev A, Rando OJ. PLoS Biol. 2010;8:e1000414. doi: 10.1371/journal.pbio.1000414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaillant C, Palmeira L, Chevereau G, Audit B, d’Aubenton-Carafa Y, Thermes C, Arneodo A. Genome Res. 2010;20:59–67. doi: 10.1101/gr.096644.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Brummelen AC, Olszewski KL, Wilinski D, Llinás M, Louw AI, Birkholtz L. J Biol Chem. 2009;284:4635–4646. doi: 10.1074/jbc.M807085200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogelauer M, Rubbi L, Lucas I, Brewer BJ, Grunstein M. Mol Cell. 2002;10:1223–1233. doi: 10.1016/s1097-2765(02)00702-5. [DOI] [PubMed] [Google Scholar]
- Watanabe J, Sasaki M, Suzuki Y, Sugano S. Nucleic Acids Res. 2001;29:70–71. doi: 10.1093/nar/29.1.70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe J, Suzuki Y, Sasaki M, Sugano S. Nucleic Acids Res. 2004;32:D334–D338. doi: 10.1093/nar/gkh115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westenberger S, Cui L, Dharia N, Winzeler E, Cui L. BMC Genomics. 2009;10:610. doi: 10.1186/1471-2164-10-610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young J, Johnson J, Benner C, Yan SF, Chen K, Le Roch K, Zhou Y, Winzeler E. BMC Genomics. 2008;9:70. doi: 10.1186/1471-2164-9-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuda M, Iwanaga S, Shigenobu S, Mair GR, Janse CJ, Waters AP, Kato T, Kaneko I. Mol Microbiol. 2009;71:1402–1414. doi: 10.1111/j.1365-2958.2009.06609.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.