Abstract
It has been suggested that xenoestrogens, a group of agents termed endocrine disruptors, may contribute to the development of hormone-dependent cancers such as breast and endometrial cancers. We previously demonstrated that the xenoestrogen bisphenol A (BPA) was able to induce transformation in vitro of human breast epithelial cells. The normal-like human breast epithelial cells MCF-10F form tubules in collagen (3-D cultures) although, after treatment with BPA (10-5M and 10-6M BPA), the cells produced less tubules (73% and 80%, respectively) and some spherical masses (27% and 20%, respectively). In the present work, expression and DNA methylation analyses were performed in these cells after being exposure to BPA. These cells showed an increased expression of BRCA1, BRCA2, BARD1, CtIP, RAD51, and BRCC3, all genes involved in DNA repair, and down-regulation of PDCD5 and BCL2L11 (BIM), both involved in apoptosis. Furthermore, DNA methylation analysis shown that BPA exposure induced hypermethylation of BCL2L11, PARD6G, FOXP1, and SFRS11, and hypomethylation of NUP98 and CtIP (RBBP8). Our results indicated that normal human breast epithelial cells exposed to BPA increased the expression of genes involved in DNA repair in order to overcome the DNA damage induced by this chemical. These results suggest that the breast tissue of women with BRCA1 or BRCA2 mutations could be more susceptible to be transformed by BPA.
Keywords: BPA, xenoestrogens, breast cancer, DNA-methylation, BRCA1, DNA repair
Introduction
Bisphenol A (BPA) is found as an environmental contaminant due to the fact it is a monomer that is polymerized to manufacture polycarbonate plastic and epoxy resins. Polycarbonate plastic is used to make baby and water bottles, dental fillings, and sealants; epoxy resins are used as coatings on the inside of almost all food and beverage's cans (1, 2). Thus, BPA leaches into food and beverages through the use of tin cans and polycarbonate plastic containers. The rate of leaching increases when polycarbonate is scratched and discolored (3-5). Decades of continuous release of free BPA into food, beverages, and the environment have resulted in a widespread human exposure to this chemical.
BPA is lipophilic and it has been detected in breast adipose tissue samples (6). It was also detected in human urine at concentrations ≥ 0.1 μg/L (3-5, 7-12). BPA was also found in maternal plasma (3.1 ng/ml), fetal plasma (2.3 ng/ml; ~10nM), placental tissues (1-104.9 ng/g), and amniotic fluid (8 ng/ml) indicating that there is a significant BPA exposure to pregnant women and their fetus (8, 11).
In vivo studies showed that early-life exposure in rodents to BPA results in persistent alterations in mammary gland morphogenesis and increased susceptibility to tumorigenesis (13, 14). In rats, maternal exposure to BPA during lactation decreased time to first tumor latency and increased the number of DMBA induced mammary tumors in their female offspring (15). However, there is less evidence for carcinogenic activity of BPA when administered to adult animals. Studies with human breast cancer cells showed inconsistent data regarding to the mitogenic, apoptotic, and transcriptional properties of BPA (16-19). This inconsistency is attributed to the lack of linear dose-dependence of BPA, which often shows a ‘U’ or an inverted ‘U’ shaped curve (20).
We had studied the effect of BPA on the normal-like human breast epithelial cells MCF-10F (21). These cells form tubules in collagen (3-D cultures) resembling the ducts of the normal mammary gland (22). We showed that treatment of MCF-10F with 10-5M or 10-6M of BPA was able to decrease the formation of tubules (73% and 80%, respectively) and increase the formation of spherical masses in collagen (27% and 20%, respectively), an indication of cell transformation (21). The objective of the present work was to study the expression and DNA-methylation changes in MCF-10F cells after BPA exposure.
Materials and Methods
Cells and treatments
MCF-10F is a normal-like human breast epithelial cell line that is estrogen receptor α (ERα) and progesterone receptor (PgR) negative. MCF-10F was maintained in DMEM: F12 media (1:1 Gibco/BRL, Gaithersburg, MD) supplemented with 5% horse serum (Gibco), 100 ng/ml cholera toxin (ICN Biomedicals, Cleveland, OH), 10μg/ml insulin (Sigma, St. Luis, MO), 0.5 μg/ml hydrocortisone (Sigma), 20 ng/ml epidermal growth factor (Gibco), 1.05mM CaCl2, and antibiotics (penicillin, 100 U/ml; streptomycin, 100 μg/ml; amphotericin, 0.25 μg/ml; Sigma). Bisphenol A (BPA; Sigma-Aldrich) was dissolved in dimethyl sulfoxide (DMSO) and the cells were treated with 10-5M or 10-6M BPA continuously for two weeks as described previously (21). As controls, MCF-10F cells were not treated and maintained in the regular media or treated with 0.284% DMSO (21). After BPA treatment, the cells were expanded and RNA or DNA were isolated for expression and DNA-methylation studies, respectively.
Expression arrays
RNA was isolated from the cells using RiboPure™ kit (Life Technologies, CA). The genome-wide gene expressions were performed using the Human Genome U133 Plus 2.0 arrays (Affymetrix, CA). The arrays were made in duplicate for MCF-10F treated with 10-6M BPA, 10-5M BPA, and control (cells without BPA treatment). After hybridization, the chips were scanned using GeneChip Scanner 3000.
MeDIP-on-chip
MeDIP-on chip (methylated DNA immuno-precipitated-on-chip) consists of an immuno-capturing approach for enriching methylated DNA in combination with detection by DNA microarray. DNA was isolated from cells treated with 10-5M, 10-6M BPA, and control MCF-10F cells (without BPA treatment) using DNeasy Blood and Tissue kit (Quiagen, CA). The DNA was fragmented (150-500 bp) by sonication and methylated DNA was immuno-precipitated with a monoclonal antibody against 5-methylcytidine (Eurogentec, San Diego, CA) (23). Methylated fragments were amplified using the GenomePlex Whole Genome Amplification kit (WGA, Sigma) (23). Double-stranded DNA was treated by a combination of UDG and APE1 that specifically recognizes the dUTP residues and breaks the DNA into fragments. Targets were then labeled with Affymetrix labeling reagent and terminal deoxynucleotidyl transferase (TdT) for 1 hour. The mixtures were hybridized to the Human promoter 1.0R Array (Affymetrix, CA) that comprises more than 4.6 million probes tiled to interrogate more than 25,500 genes; it interrogates 7.5kb upstream and 2.5kb downstream of transcription start sites. The methylation arrays were made in duplicate for cells treated with 10-6M BPA and in triplicate for cells treated with 10-5M BPA.
Data analyses
The mRNA expression arrays were analyzed using Bioconductor “limma” package. Briefly, the raw data were normalized using “rma” method (24). Differentially expressed genes were identified using empirical beyes methods implemented in the limma package (25). The criteria for significance are determined by fold change > 2 and Benjamini-Hochberg false discover rate < 5%. The functional significance of up- or down-regulated genes in the BPA treated cells were analyzed using Ingenuity Pathways Analysis software (IPA) version 5.0 (Ingenuity Systems, Redwood City, CA). The differentially expressed genes were uploaded into IPA to identify significantly enriched canonical pathways. The significance of a canonical pathway is controlled by the p-value calculated using the right-tailed Fisher Exact Test for 2×2 contingency tables.
The MeDIP-on Chip data was analyzed using CisGenome Software, an integrated tool package for tiling array and ChIP-seq analysis (26). Two-sample analyses were done using BPA treated cells versus MCF-10F control (without BPA treatment) precipitated using the antibody against 5-methylcytidines. Data were quantile normalized before the comparison. The ChIP-chip peak calling were detected with the Model-based Analysis of Tiling-array (MAT) algorithm (27) integrated in CisGenome. MAT was run with the following parameters to capture regions of increased signal intensity: bandwidth=300, maximum gap=300, max run of insignificant probes within a region =5, minimum region length =100, minimal number of significant probes within a region =5, window P-value cutoff =0.0001. Hyper- or hypo-methylated regions were determined by the signals of BPA treated cells significantly higher or lower than that of MCF-10F control cells. The MAT library and mapping files based on the March 2006 Human Genome Assembly (HG18) was used to link ChIP Hits to RefSeq Genes. Briefly, the files containing TSS (transcription start site) and end point were linked to RefSeq table with accession numbers. Chromosomal positions were then used to associate ChIP hits with RefSeq gene IDs. Specifically, hits falling within a window of −0.5 kb to +2.5 kb of a given RefSeq TSS were annotated as being associated with that gene. To illustrate the impact of methylation on gene expression, the hyperor hypo-methylated genes were compared to the down- or up-regulated genes determined by expression array. The raw data were submitted to NCBI GEO data base with accession number GSE26884 and GSE27865 for the expression and methylation arrays, respectively.
Results
Expression studies of the human breast epithelial cells after BPA exposure
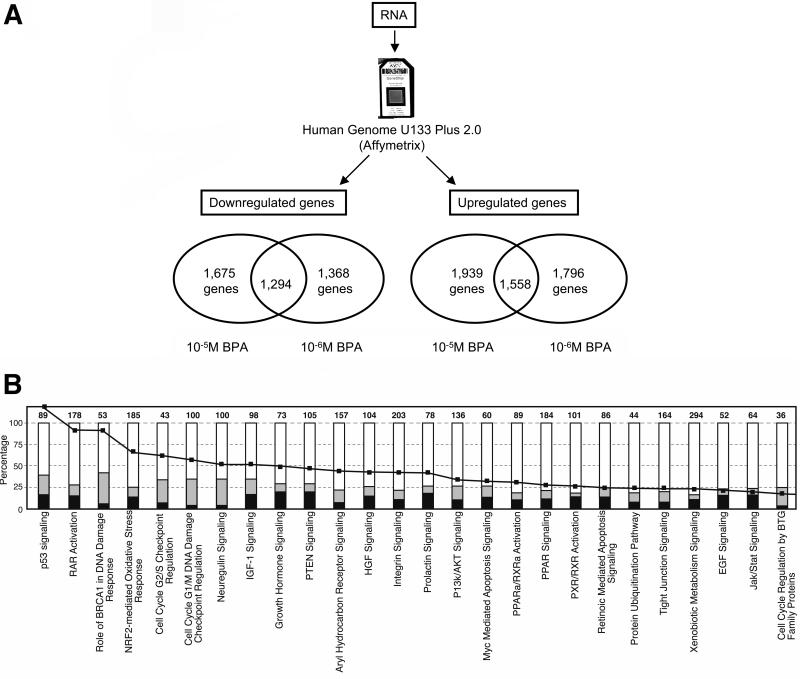
We characterized the cells exposed to 10-5M or 10-6M BPA by expression studies using microarrays. MCF-10F cells exposed to 10-5M BPA or 10-6M BPA showed 3,614 and 3,164 deregulated genes (up- or down-regulated compared to MCF-10F control), respectively (Figure 1A). We found a total of 1,675 genes down-regulated in the cells exposed to 10-5M BPA and 1,368 genes down-regulated in the cells exposed to 10-6M BPA; from these genes 1,294 genes were found to be down-regulated with both BPA concentrations (Figure 1A). A total of 1,939 genes were up-regulated in the cells treated with 10-5M BPA and 1,796 genes were up-regulated in the cells exposed to 10-6M BPA; from these genes, 1,558 genes were up-regulated in cells treated with both concentrations (Figure 1A).
Figure 1. Expression studies of the MCF-10F cells exposed to BPA.
A) RNA was isolated from the cells treated with 10-5M and 10-6M BPA and expression studies were performed using the Human Genome U133 Plus 2.0 arrays (Affymetrix); B) Canonical pathways enriched with deregulated genes in MCF-10F cells exposed to 10-6M BPA. In black, number of down-regulated genes; in gray, number of up-regulated genes; black line, log (p-value).
The functional enrichment of up- or down-regulated genes in the BPA exposed cells were analyzed using Ingenuity Pathways Analysis software (IPA). Gene networks and canonical pathways representing key genes were identified. The canonical pathways more affected in the cells exposed to BPA were the DNA damage response, p53 signaling (activate by genotoxic or non-genotoxic stress), the retinoic acid receptor activation, and the neuregulin signaling (Figure 1B). Cells treated with 10-5M or 10-6M BPA showed decreased expression of PDCD5 and BCL2L11 (also known as BIM), both involved in apoptosis, and an increased expression of BRCA1, BARD1, CtIP (also called RBBP8), RAD51, and BRCC3, all involved in DNA repair.
Table I shows genes which were at least two times up- or down-regulated in the cells after being exposure to 10-6M BPA. Several genes involved in DNA damage response were up-regulated such as BRCA1 (24.25-fold induction), BRCA2 (6.82-fold), BRCC3 (5.43-fold), BARD1 (4.5-fold), CtIP (2.06- fold), and RAD51 (36.25-fold). Other genes down-regulated after exposure to 10-6M BPA were: JAG1 (0.35-fold induction), JAG2 (0.26-fold), SMAD5 (0.19-fold), TWIST1 (0.09-fold), VIM (0.02-fold), TSPAN5 (0.02-fold), CD44 (0.18-fold), and HDAC5 (0.02-fold) (Table I). Exposure to 10-6M BPA induced the over-expression of RARRES1 (1016.92-fold) and RARRES3 (3.97-fold), both involved in the retinoic acid receptor pathway (Table I). Some genes up-regulated were CEACAM1 (317.37-fold), ALDH1A3 (20.82-fold), AURKA (23.43-fold), ID2 (17.03-fold), FN1 (11.96-fold induction), and CD24 (9.25-fold) (Table I).
Table I. Expression values of some genes in MCF-10F cells treated with 10-6M BPA.
Some genes that are at least two times up- or down-regulated are indicated.
| Gene Symbol | GenBank | Fold change |
|---|---|---|
| ALDH1A3 | NM_000693 | 20.82 |
| AURKA | NM_003600 | 23.43 |
| AURKB | AB011446 | 4.99 |
| BARD1 | NM_000465 | 4.50 |
| BCL11A | AF080216 | 0.13 |
| BCL2L11 (BIM) | AA629050 | 0.12 |
| BCL2L13 | AA156605 | 0.42 |
| BOLA3 | AI380704 | 3.58 |
| BRCA1 | NM_007295 | 24.25 |
| BRCA2 | NM_000059 | 6.82 |
| BRCC3 | X64643 | 5.43 |
| CD24 | X69397 | 9.25 |
| CD44 | AL552534 | 0.18 |
| CEACAM1 | X16354 | 317.37 |
| CtIP (RBBP8) | NM 002894 | 2.06 |
| ERBB3 | NM_001982 | 6.73 |
| FN1 | AK026737 | 11.96 |
| HDAC5 | NM_005474 | 0.02 |
| HDAC8 | AF212246 | 6.06 |
| HDAC9 | NM_014707 | 0.11 |
| ID2 | AI819238 | 17.03 |
| IL18R1 | NM_003855 | 0.04 |
| JAG1 | U73936 | 0.35 |
| JAG2 | Y14330 | 0.26 |
| LOX | NM_002317 | 0.29 |
| MTSS1 | NM_014751 | 0.02 |
| MUC1 | AF348143 | 3.81 |
| MUC16 | NM_024690 | 124.50 |
| MUC20 | AW084511 | 21.26 |
| MYCBP | D50692 | 15.78 |
| PARD6G | AI817448 | 0.06 |
| PDCD5 | AI817145 | 0.04 |
| RAD51 | NM_002875 | 36.25 |
| RARRES1 (TIG1) | NM_002888 | 1016.92 |
| RARRES3 (TIG3) | NM_004585 | 3.97 |
| SFRP1 | AF017987 | 0.34 |
| SLIT2 | AF055585 | 0.22 |
| SMAD5 | AF010601 | 0.19 |
| TSPAN5 | AA059445 | 0.02 |
| TWIST1 | X99268 | 0.09 |
| VIM | AI922599 | 0.02 |
DNA-methylation studies of MCF-10F cells after BPA exposure
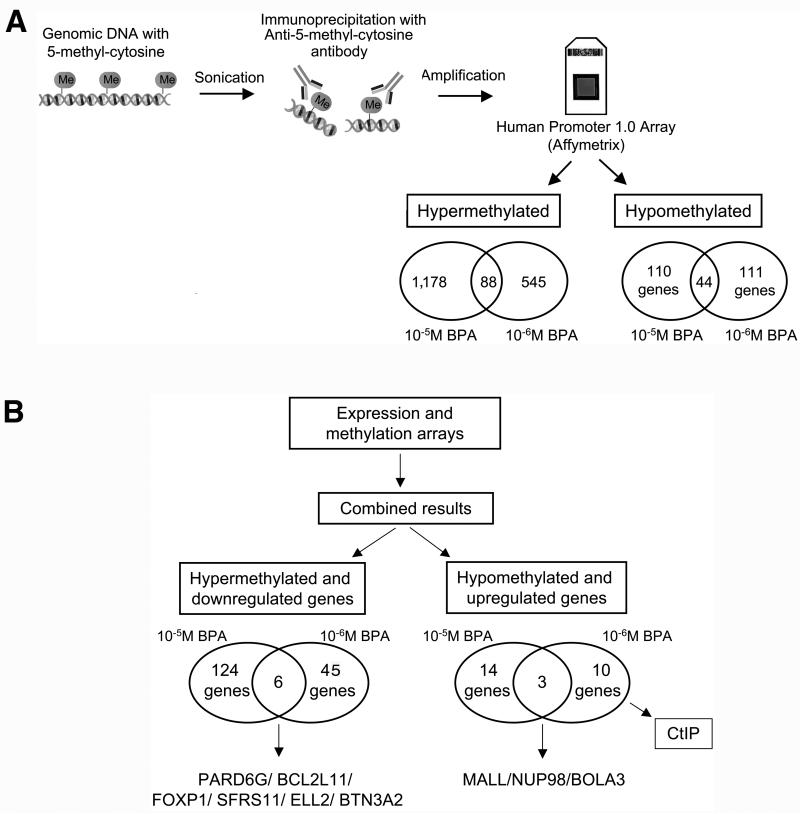
We studied DNA-methylation changes in the human breast epithelial cells MCF-10F after being exposure to BPA using MeDIP-on-Chip (23). Gene regulatory regions that were hypo- or hypermethylated were identified. Hypermethylated targets were sequences with significantly increased signals in the cells exposed to BPA relative to the control (MCF-10F growth without treatment), both DNA's immunoprecipitated with the antibody against 5-methyl-cytosines. The cells treated with 10-5M BPA showed 1,178 genes hypermethylated and cells treated with 10-6M BPA showed 545 hypermethylated genes; from these genes, 88 genes were hypermethylated at both concentrations (Figure 2A). In difference, hypomethylated targets are sequences that are significantly increased in the control relative to the cells treated with BPA. Using these criteria, we identified 110 genes that were hypomethylated in cells treated with 10-5M BPA and 111 hypomethylated genes in the cells treated with 10-6M BPA; from these genes, 44 genes were hypomethylated at both concentrations (Figure 2A).
Figure 2. DNA methylation studies in MCF-10F cells exposed to BPA.
A) DNA was isolated from the cells and fragmented by sonication. Methylated DNA was isolated using an antibody against 5-methylcytidine and amplified, followed by hybridization using a promoter microarray to identify regions with altered methylation in the promoters. The Human Promoter 1.0R Arrays were used and hyper- and hypomethylated promoters were identified. Hypermethylated targets were sequences with significantly increased signals in the cells exposed to BPA relative to the control without treatment immunoprecipitated using the antibody against 5-methylcytidine; B) Combined results from the expression and DNA methylation arrays: The list of genes that were found hypermethylated by MeDIP-on Chip and the list of genes found down-regulated by expression arrays were compared. At the left, the number of genes hypermethylated and down-regulated is indicated for the cells exposed to 10-5M and 10-6M BPA. The same was done for genes hypomethylated and up-regulated; the number of genes hypomethylated and up-regulated is indicated for the cells treated with 10-5M and 10-6M BPA at the right.
As hypermethylation is related with gene down-regulation and hypomethylation is related with increased gene expression (28), the expression and DNA methylation data were superimposed. The down-regulated and hypermethylated genes by BPA exposure were identified (Figure 2B). In the cells treated with 10-5M BPA, 124 genes were found hypermethylated and down-regulated (Figure 2B). In the cells treated with 10-6M BPA, 45 genes were found hypermethylated and down-regulated (Figure 2B). As it is indicated in Figure 2B, six genes were found hypermethylated and down-regulated in cells treated with both 10-5M or 10-6M BPA: PARD6G, BCL2L11 (or BIM), FOXP1, SFRS11, ELL2, and BTN3A2.
In Table II, the 124 genes down-regulated and hypermethylated in the cells after being exposed to 10-5M BPA are indicated; some of these genes are PARD6G (0.21-fold induction), BCL2L11 (0.2-fold), FOXP1 (0.43-fold), SFRS11 (0.38-fold), ELL2 (0.39-fold), and BTN3A2 (0.24-fold). Other genes down-regulated and hypermethylated were STAT5B (0.31-fold induction), WWOX (0.41-fold induction), and SULT1E1 (0.07-fold induction). Table III shows the 45 genes down-regulated and hypermethylated in the cells after being exposed to 10-6M BPA. Some of these genes are PARD6G (0.16-fold induction), BCL2L11 (0.18-fold), FOXP1 (0.38-fold), SFRS11 (0.22-fold), ELL2 (0.31-fold), and BTN3A2 (0.28-fold). Other genes down-regulated and hypermethylated were RHOU (0.02-fold induction), TWIST1 (0.11-fold induction), and SFRP1 (0.39-fold induction).
Table II. Downregulated and hypermethylated genes in MCF-10F cells treated with 10-5M BPA.
Hypermethylated genes and their expression values are indicated (fold change).
| Gene Symbol | Fold change | Gene Symbol | Fold change | Gene Symbol | Fold change | Gene Symbol | Fold change |
|---|---|---|---|---|---|---|---|
| SGSH | 0.5 | FADS3 | 0.42 | PURA | 0.34 | ZNF197 | 0.22 |
| OGFRL1 | 0.5 | ABCA12 | 0.42 | RBMS3 | 0.34 | MARK1 | 0.22 |
| IL13RA1 | 0.49 | AFF4 | 0.42 | STK16 | 0.33 | TOMM20 | 0.22 |
| ANKDD1A | 0.49 | MLL | 0.41 | PEX11A | 0.32 | PARD6G | 0.21 |
| UBE2Z | 0.49 | WWOX | 0.41 | SENP6 | 0.32 | C1orf21 | 0.21 |
| PRKAG2 | 0.49 | SLC7A8 | 0.40 | KIAA0182 | 0.32 | BCL2L11 (BIM) | 0.20 |
| ZNF488 | 0.48 | MCEE | 0.40 | PARD3 | 0.31 | PRKCH | 0.20 |
| ZC3H11A | 0.48 | HMHA1 | 0.40 | RNF135 | 0.31 | C7orf31 | 0.19 |
| KCNAB2 | 0.48 | RIOK3 | 0.39 | STAT5B | 0.31 | TMEM91 | 0.19 |
| RHOQ | 0.48 | ELL2 | 0.39 | ATF6 | 0.30 | MGST1 | 0.18 |
| DFFB | 0.47 | C7orf38 | 0.39 | ADARB1 | 0.30 | ANKRD28 | 0.18 |
| TBC1D8B | 0.47 | METTL9 | 0.39 | JAK1 | 0.29 | DHRS3 | 0.18 |
| DGAT2 | 0.47 | FBXO9 | 0.38 | CP | 0.29 | CCDC11 | 0.16 |
| ZNF219 | 0.47 | EFHC1 | 0.38 | TMEM67 | 0.29 | TMEM37 | 0.15 |
| LRCH3 | 0.47 | CD99L2 | 0.38 | FARP1 | 0.28 | HSD11B1 | 0.15 |
| SH3BP2 | 0.47 | CAPN1 | 0.38 | FAM119B | 0.28 | IL18R1 | 0.13 |
| RUFY1 | 0.47 | SFRS11 | 0.38 | VPS41 | 0.28 | LETMD1 | 0.12 |
| UBE2Q1 | 0.46 | ACVR2A | 0.38 | TMEM80 | 0.28 | FAM19A2 | 0.11 |
| SSR2 | 0.46 | ATP9A | 0.38 | TRIM69 | 0.27 | BBOX1 | 0.10 |
| CIR | 0.46 | DUSP1 | 0.38 | GJA3 | 0.27 | EBF1 | 0.10 |
| GRAMD2 | 0.46 | PPIL6 | 0.38 | IFNGR1 | 0.27 | GBP2 | 0.08 |
| DNAJC16 | 0.46 | MAP2K5 | 0.37 | GPR177 | 0.24 | SULT1E1 | 0.07 |
| HHLA3 | 0.45 | RAB13 | 0.37 | BTN3A2 | 0.24 | TSLP | 0.07 |
| SMYD3 | 0.45 | PKIA | 0.37 | PIK3R1 | 0.24 | PLEKHA6 | 0.07 |
| C5orf 25 | 0.45 | SFXN1 | 0.37 | ZBTB20 | 0.24 | KLHDC8B | 0.07 |
| TMLHE | 0.45 | FGFR1OP2 | 0.37 | TPPP | 0.23 | PDZK1IP1 | 0.06 |
| KIAA1244 | 0.44 | MSX2 | 0.37 | PIR | 0.23 | DCN | 0.05 |
| ITSN2 | 0.44 | LYPLAL1 | 0.36 | DUSP16 | 0.22 | SORL1 | 0.04 |
| MAPT | 0.44 | KLRK1 | 0.36 | PRKAG2 | 0.22 | ZBTB10 | 0.03 |
| RNPC3 | 0.44 | GNPTAB | 0.35 | DSC3 | 0.22 | BCL6 | 0.03 |
| FOXP1 | 0.43 | ADRBK2 | 0.34 | ITGA4 | 0.22 | GALNTL2 | 0.003 |
Table III. Downregulated and hypermethylated genes in MCF-10F cells treated with 10-6M BPA.
Hypermethylated genes and their expression values (fold change) are indicated.
| Gene Symbol | Fold change | Gene Symbol | Fold change |
|---|---|---|---|
| NIPA1 | 0.50 | UACA | 0.24 |
| EDA | 0.49 | RAB4A | 0.24 |
| RERE | 0.48 | SFRS11 | 0.22 |
| SLC25A28 | 0.47 | KIAA0564 | 0.21 |
| FAM19A1 | 0.45 | BCL2L11 (BIM) | 0.18 |
| MGEA5 | 0.44 | KLHL13 | 0.18 |
| TLR3 | 0.44 | IGF1R | 0.17 |
| ZHX2 | 0.41 | RPL37 | 0.16 |
| OSBPL6 | 0.40 | PARD6G | 0.16 |
| NUDT2 | 0.40 | GPC4 | 0.15 |
| SFRP1 | 0.39 | GAB2 | 0.15 |
| FOXP1 | 0.38 | MRPL39 | 0.14 |
| SYNE1 | 0.38 | KLF9 | 0.13 |
| NPL | 0.37 | TWIST1 | 0.11 |
| C17orf69 | 0.36 | RNF128 | 0.10 |
| TPCN2 | 0.35 | DST | 0.10 |
| TOP1MT | 0.35 | TOX | 0.07 |
| TMEM47 | 0.33 | TTC7B | 0.04 |
| ELL2 | 0.31 | NXN | 0.04 |
| PCDHB14 | 0.29 | RHOU (WRCH1) | 0.02 |
| BTN3A2 | 0.28 | FHL1 | 0.01 |
| TARSL2 | 0.27 | SOD2 | 0.01 |
| LONRF1 | 0.27 |
Similarly, hypomethylated and up-regulated genes by BPA were identified (Figure 2B). In the cells exposed to 10-5M BPA, 14 genes were found hypomethylated and up-regulated; in the cells treated with 10-6M BPA, 10 genes were found hypomethylated and up-regulated (Figure 2B). From these genes, 3 were hypomethylated and up-regulated in both cells: MALL, NUP98, and BOLA3 (Figure 2B and Table IV). In cells treated with 10-5M BPA, MLL, NUP98, and BOLA3 shown: 67.65, 9.65 and 5.43-fold induction, respectively (Table IV). In cells treated with 10-6M BPA, MLL, NUP98, and BOLA3 shown 72, 5.39 and 3.10-fold induction, respectively (Table III); also these cells shown up-regulation and hypomethylation of CtIP (2.06-fold induction).
Table IV. Hypomethylated and upregulated genes in MCF-10F cells treated BPA.
Hypomethylated genes and their expression values (fold change) are indicated.
| Cells treated with 10-5M BPA | ||
|---|---|---|
| Gene Symbol | Gene Title | Fold change |
| MALL | mal, T-cell differentiation protein-like | 67.65 |
| NUP98 | nucleoporin 98kDa | 9.65 |
| ARHGAP11A | Rho GTPase activating protein 11A | 7.67 |
| BOLA3 | bolA homolog 3 (E. coli) | 5.43 |
| CA2 | carbonic anhydrase II | 5.10 |
| GPR172A | G protein-coupled receptor 172A | 4.20 |
| CCDC80 | coiled-coil domain containing 80 | 3.76 |
| BID | BH3 interacting domain death agonist | 3.53 |
| NT5E | 5′-nucleotidase, ecto (CD73) | 3.23 |
| SHC4 | SHC (Src homology 2 domain containing) family, member 4 | 2.73 |
| C12orf30 | chromosome 12 open reading frame 30 | 2.62 |
| CCDC90A | Coiled-coil domain containing 90A | 2.31 |
| FAM86A | family with sequence similarity 86, member A | 2.31 |
| SYNCRIP | synaptotagmin binding, cytoplasmic RNA interacting protein | 2.25 |
| Cells treated with 10-6M BPA | ||
| MALL | mal, T-cell differentiation protein-like | 72.00 |
| RPL27A | ribosomal protein L27a | 13.93 |
| GDA | guanine deaminase | 7.46 |
| NUP98 | nucleoporin 98kDa | 5.39 |
| TIAM1 | T-cell lymphoma invasion and metastasis 1 | 4.44 |
| BCMO1 | beta-carotene 15,15′-monooxygenase 1 | 4.35 |
| BOLA3 | bolA homolog 3 (E. coli) | 3.10 |
| DDX52 | DEAD (Asp-Glu-Ala-Asp) box polypeptide 52 | 3.01 |
| CtIP (RBBP8) | retinoblastoma binding protein 8 | 2.06 |
| PPME1 | protein phosphatase methylesterase 1 | 2.04 |
Discussion
The normal-like human breast epithelial cells MCF-10F showed increased expression of genes involved in DNA repair (BRCA1, BARD1, CtIP, RAD51, and BRCC3) and decreased expression of genes involved in apoptosis (PDCD5 and BCL2L11) after being exposed to BPA for two weeks. Also, these cells showed hypermethylation of different genes such as BCL2L11, PARD6G, FOXP1, SFRS11, and hypomethylation of CtIP (or RBBP8) and NUP98.
Epigenetic changes derived from exposure to endocrine disruptors have been described in several tissues and organisms (29, 30) although, this is the first demonstration that BPA induces DNA methylation changes in genes related to apoptosis and DNA repair in human breast epithelial cells. BCL2L11 (or BIM) has an ability to trigger apoptosis in various cells, such as epithelia and neuronal cells (31); the fact that this gene is hypermethylated after BPA treatment suggest that apoptosis was inhibited in the cells after being exposed to BPA. CtIP was hypomethylated and up-regulated after BPA treatment; CtIP is involve in double strand brake (DSB) repair and plays a role in DNA-damage-induced cell cycle checkpoint control at the G2/M transition (32). MCF-10F cells treated with BPA also showed changes in the DNA methylation pattern of PARD6G (partitioning defective 6 homolog gamma) which is an adapter-protein involved in asymmetrical cell division and cell polarization processes.
It has been suggested that BPA is a weak carcinogen (33). Evidence of the estrogenic effects of BPA has been reported in several studies showing that it activates estrogen receptors (ER) alpha and beta (34, 35) although, BPA's affinity is at least 10,000-fold less than estrogen for both receptors (36). It has been proposed that BPA, similar to certain estrogen metabolites, can react with the DNA to cause mutations that can lead to cancer initiation (33, 37-41). One mechanism by which estrogen and BPA can initiates breast cancer is by generating adducts that can produce a variety of DNA modifications that, if not countered by DNA repair, can lead to cell transformation. It was shown that BPA is able to form DNA adducts in vitro and in vivo (42-45); BPA can be converted to bisphenol O-quinone (46) and, the BPA semiquinone and/or quinone intermediates may be the ultimate DNA binding metabolites. In the present study, the ERα-negative breast epithelial cells MCF-10F were used indicating that in these cells other mechanisms independent of the ER were responsible for the biological effect of BPA.
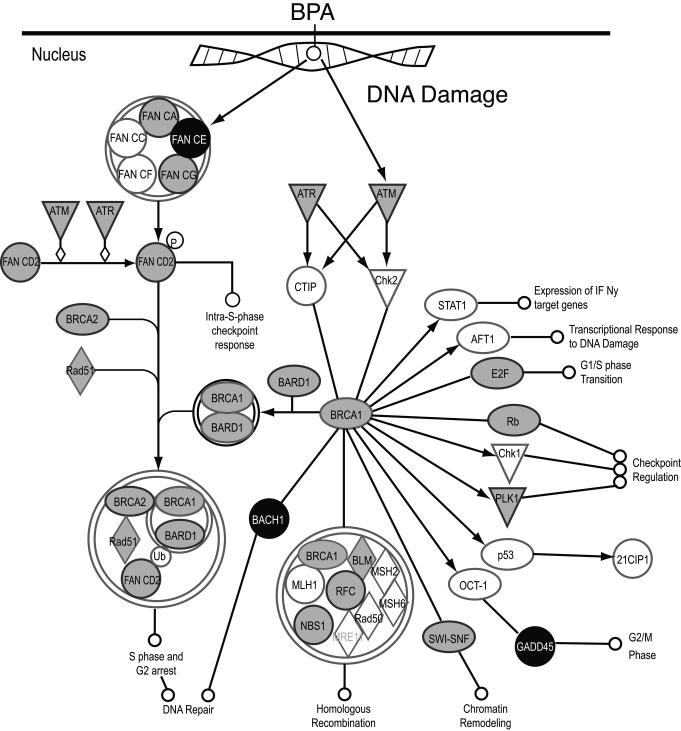
In addition to DNA adduct formation, oxidative stress could be another reason for the alterations produced by BPA in the DNA (47). Oxidative DNA lesions include the oxidation of nucleotidic bases, modifications to the sugar moiety of DNA which may result in base-loss abasic (apurinic/apyrimidinic) sites and/or strand breakage (single and double strand breaks), DNA intra-strand adducts, and DNA-protein crosslinks, all of which are cytotoxic and some can be mutagenic (48, 49). Our studies showed that after BPA treatment, the cells showed up-regulation of genes involved in DNA repair suggesting that BPA could have produced DNA double strand breaks and, the normal breast epithelial cells increased the expression of DNA repair genes to overcome the damage (Figure 3). The normal-like human breast epithelial cells MCF-10F showed increased expression of BRCA1, BARD1, CtIP, RAD51, and BRCC3, all genes involved in DNA repair, after being exposed to BPA.
Figure 3. DNA repair genes were induced in the normal breast epithelial cells after being exposed to BPA.
The MCF-10F cells showed increase expression of BRCA1, BARD1, BRCA2, and RAD51 after being exposed to 10-6M BPA. Up-regulated genes are shown in gray. The down-regulated genes are shown in black.
The human BRCA1 is a nuclear polypeptide consisting of 1,863 amino acids and it contains several functional domains that interact directly or indirectly with a variety of molecules, including tumor suppressor, oncogenes, DNA damage repair proteins, cell cycle regulators, and transcriptional activators and repressors (Figure 3) (50). BRCA1 exists as a heterodimer with BARD1 and most of the functions of BRCA1 have been attributed to occur in association with BARD1 (51, 52). Disruption of the BRCA1-BARD1 interaction would impair the cell cycle checkpoint control as well as DNA repair functions of BRCA1 which could lead to tumorigenesis. BRCA1 ubiquitinates its phosphorylation –dependent partner CtIP (RBBP8) and this reaction plays a role in the G2/M checkpoint control upon DNA damage (53). RAD51, also involved in DNA-damage repair, interacted with BRCA1 (54). Our results showed that BRCA2 was down-regulated in the cells treated with 10-6M BPA; the BRCA2 protein, which has a function similar to that of BRCA1, also interacted with RAD51. By influencing DNA damage repair, BRCA1, BRCA2, and RAD51 play a role in maintaining the stability of the human genome. BRCC3 encodes a subunit of BRCA1-BRCA2 containing complex (BRCC), which is an E3 ubiquitin ligase; this protein is also thought to be involved in the cellular response to progression through the G2/M checkpoint. Our results demonstrated that normal breast epithelial cells treated with BPA showed an increased expression of BRCA1, BARD1, CtIP, RAD51, and BRCC3; all involved in DNA repair. This supports the hypothesis that BPA can initiate breast cancer by generating adducts or reactive oxidative species (ROS) that can produce a variety of DNA modifications that, if not countered by DNA repair, can lead to cell transformation.
Our results suggest that loss of BRCA1 could lead to an increased sensitivity to BPA as it was shown by Jones et al (55). We had isolated primary breast epithelial cells from a BRCA1 carrier; these cells were treated continuously during one week with media containing 10-5M or 10-6M BPA and, at the end of the treatment the ductulogenic and invasion assays were performed. These BRCA1 cells treated with BPA formed an increased number of spherical masses in collagen and showed increased invasion (data not shown). Our results suggest that women that carry BRCA1 mutations could be more susceptible to the effects of BPA.
Human exposure to BPA is widespread and studies have shown detectable levels of BPA ranging from 0.2 to 10ng/ml (~0.5-40 nM) in adult and fetal human serum (56). Although, the doses of 10-5M and 10-6 M BPA that were used in our studies are higher compared to the concentrations found in serum samples, the cells were exposed for 2 weeks in contrast to humans that are exposure to low doses for a longer period of time.
In conclusion, our results showed that BPA induced the expression of genes related to DNA repair in normal human breast epithelial cells. Up-regulation of BRCA1, BRCA2, RAD51, BARD1, and BRCC3 expression was induced after BPA exposure in MCF-10F cells. This suggests that in BRCA1 carriers BPA exposure could lead to increased frequency of DNA mutations. Our results suggest that loss of BRCA1 could lead to an increased sensitivity to BPA. Furthermore, this is the first demonstration that BPA induced DNA methylation changes in human breast epithelial cells.
Acknowledgements
This work was supported by the grants R21 ES015894 and U01 ES/CA 12771 from the NIH.
References
- 1.Brotons JA, Olea-Serrano MF, Villalobos M, Pedraza V, Olea N. Xenoestrogens released from lacquer coatings in food cans. Environ Health Perspect. 1995;103:608–612. doi: 10.1289/ehp.95103608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Olea N, Pulgar R, Perez P, et al. Estrogenicity of resin-based composites and sealants used in dentistry. Environ Health Perspect. 1996;104:298–305. doi: 10.1289/ehp.96104298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brede C, Fjeldal P, Skjevrak I, Herikstad H. Increased migration levels of bisphenol A from polycarbonate baby bottles after dishwashing, boiling and brushing. Food Addit Contam. 2003;20:684–689. doi: 10.1080/0265203031000119061. [DOI] [PubMed] [Google Scholar]
- 4.Burridge E. Bisphenol A: product profile. Eur. Chem. News. 2003;17 [Google Scholar]
- 5.Howdeshell KL, Peterman PH, Judy BM, et al. Bisphenol A is released from used polycarbonate animal cages into water at room temperature. Environ Health Perspect. 2003;111:1180–1187. doi: 10.1289/ehp.5993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fernandez MF, Arrebola JP, Taoufiki J, et al. Bisphenol-A and chlorinated derivatives in adipose tissue of women. Reprod Toxicol. 2007;24:259–264. doi: 10.1016/j.reprotox.2007.06.007. [DOI] [PubMed] [Google Scholar]
- 7.Calafat AM, Kuklenyik Z, Reidy JA, Caudill SP, Ekong J, Needham LL. Urinary concentrations of bisphenol A and 4-nonylphenol in a human reference population. Environ Health Perspect. 2005;113:391–395. doi: 10.1289/ehp.7534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ikezuki Y, Tsutsumi O, Takai Y, Kamei Y, Taketani Y. Determination of bisphenol A concentrations in human biological fluids reveals significant early prenatal exposure. Hum Reprod. 2002;17:2839–2841. doi: 10.1093/humrep/17.11.2839. [DOI] [PubMed] [Google Scholar]
- 9.Matsumoto A, Kunugita N, Kitagawa K, et al. Bisphenol A levels in human urine. Environ Health Perspect. 2003;111:101–104. doi: 10.1289/ehp.5512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ouchi K, Watanabe S. Measurement of bisphenol A in human urine using liquid chromatography with multi-channel coulometric electrochemical detection. J Chromatogr B Analyt Technol Biomed Life Sci. 2002;780:365–370. doi: 10.1016/s1570-0232(02)00547-0. [DOI] [PubMed] [Google Scholar]
- 11.Schonfelder G, Wittfoht W, Hopp H, Talsness CE, Paul M, Chahoud I. Parent bisphenol A accumulation in the human maternal-fetal-placental unit. Environ Health Perspect. 2002;110:A703–707. doi: 10.1289/ehp.110-1241091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yang M, Kim SY, Lee SM, et al. Biological monitoring of bisphenol a in a Korean population. Arch Environ Contam Toxicol. 2003;44:546–551. doi: 10.1007/s00244-002-2124-0. [DOI] [PubMed] [Google Scholar]
- 13.Keri RA, Ho SM, Hunt PA, Knudsen KE, Soto AM, Prins GS. An evaluation of evidence for the carcinogenic activity of bisphenol A. Reprod Toxicol. 2007;24:240–252. doi: 10.1016/j.reprotox.2007.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Soto AM, Maffini MV, Sonnenschein C. Neoplasia as development gone awry: the role of endocrine disruptors. Int J Androl. 2008;31:288–293. doi: 10.1111/j.1365-2605.2007.00834.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jenkins S, Raghuraman N, Eltoum I, Carpenter M, Russo J, Lamartiniere CA. Oral exposure to bisphenol a increases dimethylbenzanthracene-induced mammary cancer in rats. Environ Health Perspect. 2009;117:910–915. doi: 10.1289/ehp.11751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dairkee SH, Seok J, Champion S, et al. Bisphenol A induces a profile of tumor aggressiveness in high-risk cells from breast cancer patients. Cancer Res. 2008;68:2076–2080. doi: 10.1158/0008-5472.CAN-07-6526. [DOI] [PubMed] [Google Scholar]
- 17.Diel P, Olff S, Schmidt S, Michna H. Effects of the environmental estrogens bisphenol A, o,p'-DDT, p-tert-octylphenol and coumestrol on apoptosis induction, cell proliferation and the expression of estrogen sensitive molecular parameters in the human breast cancer cell line MCF-7. J Steroid Biochem Mol Biol. 2002;80:61–70. doi: 10.1016/s0960-0760(01)00173-x. [DOI] [PubMed] [Google Scholar]
- 18.Singleton DW, Feng Y, Yang J, Puga A, Lee AV, Khan SA. Gene expression profiling reveals novel regulation by bisphenol-A in estrogen receptor-alpha-positive human cells. Environ Res. 2006;100:86–92. doi: 10.1016/j.envres.2005.05.004. [DOI] [PubMed] [Google Scholar]
- 19.Soto AM, Sonnenschein C, Chung KL, Fernandez MF, Olea N, Serrano FO. The E-SCREEN assay as a tool to identify estrogens: an update on estrogenic environmental pollutants. Environ Health Perspect. 1995;103(Suppl 7):113–122. doi: 10.1289/ehp.95103s7113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hugo ER, Brandebourg TD, Woo JG, Loftus J, Alexander JW, Ben-Jonathan N. Bisphenol A at environmentally relevant doses inhibits adiponectin release from human adipose tissue explants and adipocytes. Environ Health Perspect. 2008;116:1642–1647. doi: 10.1289/ehp.11537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fernandez SV, Russo J. Estrogen and xenoestrogens in breast cancer. Toxicol Pathol. 2010;38:110–122. doi: 10.1177/0192623309354108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kocdor H, Kocdor MA, Russo J, et al. Human chorionic gonadotropin (hCG) prevents the transformed phenotypes induced by 17 beta-estradiol in human breast epithelial cells. Cell Biol Int. 2009;33:1135–1143. doi: 10.1016/j.cellbi.2009.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Weber M, Davies JJ, Wittig D, et al. Chromosome-wide and promoter-specific analyses identify sites of differential DNA methylation in normal and transformed human cells. Nat Genet. 2005;37:853–862. doi: 10.1038/ng1598. [DOI] [PubMed] [Google Scholar]
- 24.Irizarry R, Hobbs B, Collin F, et al. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4:249–264. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- 25.Smyth GK. Linear models and empirical Bayes methods for assessing diferential expression in microarray experiments. Statistical Applications in Genetics and Molecular Biology. 2004;3 doi: 10.2202/1544-6115.1027. Article 3. [DOI] [PubMed] [Google Scholar]
- 26.Ji H, Jiang H, Ma W, Johnson DS, Myers RM, Wong WH. An integrated software system for analyzing ChIP-chip and ChIP-seq data. Nat Biotechnol. 2008;26:1293–1300. doi: 10.1038/nbt.1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Johnson WE, Li W, Meyer CA, et al. Model-based analysis of tiling-arrays for ChIP-chip. Proc Natl Acad Sci U S A. 2006;103:12457–12462. doi: 10.1073/pnas.0601180103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jaenisch R, Bird A. Epigenetic regulation of gene expression: how the genome integrates intrinsic and environmental signals. Nat Genet. 2003;33(Suppl):245–254. doi: 10.1038/ng1089. [DOI] [PubMed] [Google Scholar]
- 29.Ho SM, Tang WY, Belmonte de Frausto J, Prins GS. Developmental exposure to estradiol and bisphenol A increases susceptibility to prostate carcinogenesis and epigenetically regulates phosphodiesterase type 4 variant 4. Cancer Res. 2006;66:5624–5632. doi: 10.1158/0008-5472.CAN-06-0516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dolinoy DC, Huang D, Jirtle RL. Maternal nutrient supplementation counteracts bisphenol A-induced DNA hypomethylation in early development. Proc Natl Acad Sci U S A. 2007;104:13056–13061. doi: 10.1073/pnas.0703739104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.O'Connor L, Strasser A, O'Reilly LA, et al. Bim: a novel member of the Bcl-2 family that promotes apoptosis. Embo J. 1998;17:384–395. doi: 10.1093/emboj/17.2.384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Varma AK, Brown RS, Birrane G, Ladias JA. Structural basis for cell cycle checkpoint control by the BRCA1-CtIP complex. Biochemistry. 2005;44:10941–10946. doi: 10.1021/bi0509651. [DOI] [PubMed] [Google Scholar]
- 33.Cavalieri EL, Rogan EG. Is bisphenol A a weak carcinogen like the natural estrogens and diethylstilbestrol? IUBMB Life. 2010;62:746–751. doi: 10.1002/iub.376. [DOI] [PubMed] [Google Scholar]
- 34.Matthews JB, Twomey K, Zacharewski TR. In vitro and in vivo interactions of bisphenol A and its metabolite, bisphenol A glucuronide, with estrogen receptors alpha and beta. Chem Res Toxicol. 2001;14:149–157. doi: 10.1021/tx0001833. [DOI] [PubMed] [Google Scholar]
- 35.Routledge EJ, White R, Parker MG, Sumpter JP. Differential effects of xenoestrogens on coactivator recruitment by estrogen receptor (ER) alpha and ERbeta. J Biol Chem. 2000;275:35986–35993. doi: 10.1074/jbc.M006777200. [DOI] [PubMed] [Google Scholar]
- 36.Maruyama S, Fujimoto N, Yin H, Ito A. Growth stimulation of a rat pituitary cell line MtT/E-2 by environmental estrogens in vitro and in vivo. Endocr J. 1999;46:513–520. doi: 10.1507/endocrj.46.513. [DOI] [PubMed] [Google Scholar]
- 37.Cavalieri E, Chakravarti D, Guttenplan J, et al. Catechol estrogen quinones as initiators of breast and other human cancers: implications for biomarkers of susceptibility and cancer prevention. Biochim Biophys Acta. 2006;1766:63–78. doi: 10.1016/j.bbcan.2006.03.001. [DOI] [PubMed] [Google Scholar]
- 38.Cavalieri E, Frenkel K, Liehr JG, Rogan E, Roy D. Estrogens as endogenous genotoxic agents--DNA adducts and mutations. J Natl Cancer Inst Monogr. 2000:75–93. doi: 10.1093/oxfordjournals.jncimonographs.a024247. [DOI] [PubMed] [Google Scholar]
- 39.Cavalieri E, Kohli E, Zahid M, Rogan E. Greater reactivity of estradiol-3,4-quinone vs estradiol-2,3-quinone with DNA in the formation of depurinating DNA adducts. Proc. Amer. Assoc. Cancer Res. 2003;44:180. doi: 10.1021/tx050229y. [DOI] [PubMed] [Google Scholar]
- 40.Liehr JG. Is estradiol a genotoxic mutagenic carcinogen? Endocr Rev. 2000;21:40–54. doi: 10.1210/edrv.21.1.0386. [DOI] [PubMed] [Google Scholar]
- 41.Russo J, Fernandez SV, Russo PA, et al. 17-Beta-estradiol induces transformation and tumorigenesis in human breast epithelial cells. FASEB J. 2006;20:1622–1634. doi: 10.1096/fj.05-5399com. [DOI] [PubMed] [Google Scholar]
- 42.Izzotti A, Kanitz S, D'Agostini F, Camoirano A, De Flora S. Formation of adducts by bisphenol A, an endocrine disruptor, in DNA in vitro and in liver and mammary tissue of mice. Mutat Res. 2009;679:28–32. doi: 10.1016/j.mrgentox.2009.07.011. [DOI] [PubMed] [Google Scholar]
- 43.Bentley P, Bieri F, Kuster H, et al. Hydrolysis of bisphenol A diglycidylether by epoxide hydrolases in cytosolic and microsomal fractions of mouse liver and skin: inhibition by bis epoxycyclopentylether and the effects upon the covalent binding to mouse skin DNA. Carcinogenesis. 1989;10:321–327. doi: 10.1093/carcin/10.2.321. [DOI] [PubMed] [Google Scholar]
- 44.Steiner S, Honger G, Sagelsdorff P. Molecular dosimetry of DNA adducts in C3H mice treated with bisphenol A diglycidylether. Carcinogenesis. 1992;13:969–972. doi: 10.1093/carcin/13.6.969. [DOI] [PubMed] [Google Scholar]
- 45.Vanhoutte K, Van Dongen W, Hoes I, et al. Development of a nanoscale liquid chromatography/electrospray mass spectrometry methodology for the detection and identification of DNA adducts. Anal Chem. 1997;69:3161–3168. doi: 10.1021/ac970121q. [DOI] [PubMed] [Google Scholar]
- 46.Atkinson A, Roy D. In vitro conversion of environmental estrogenic chemical bisphenol A to DNA binding metabolite(s). Biochem Biophys Res Commun. 1995;210:424–433. doi: 10.1006/bbrc.1995.1678. [DOI] [PubMed] [Google Scholar]
- 47.Chitra KC, Latchoumycandane C, Mathur PP. Induction of oxidative stress by bisphenol A in the epididymal sperm of rats. Toxicology. 2003;185:119–127. doi: 10.1016/s0300-483x(02)00597-8. [DOI] [PubMed] [Google Scholar]
- 48.Powell CL, Swenberg JA, Rusyn I. Expression of base excision DNA repair genes as a biomarker of oxidative DNA damage. Cancer Lett. 2005;229:1–11. doi: 10.1016/j.canlet.2004.12.002. [DOI] [PubMed] [Google Scholar]
- 49.Hazra TK, Das A, Das S, Choudhury S, Kow YW, Roy R. Oxidative DNA damage repair in mammalian cells: a new perspective. DNA Repair (Amst) 2007;6:470–480. doi: 10.1016/j.dnarep.2006.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Deng CX, Brodie SG. Roles of BRCA1 and its interacting proteins. Bioessays. 2000;22:728–737. doi: 10.1002/1521-1878(200008)22:8<728::AID-BIES6>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- 51.Mallery DL, Vandenberg CJ, Hiom K. Activation of the E3 ligase function of the BRCA1/BARD1 complex by polyubiquitin chains. EMBO J. 2002;21:6755–6762. doi: 10.1093/emboj/cdf691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wu LC, Wang ZW, Tsan JT, et al. Identification of a RING protein that can interact in vivo with the BRCA1 gene product. Nat Genet. 1996;14:430–440. doi: 10.1038/ng1296-430. [DOI] [PubMed] [Google Scholar]
- 53.Yu X, Chen J. DNA damage-induced cell cycle checkpoint control requires CtIP, a phosphorylation-dependent binding partner of BRCA1 C-terminal domains. Mol Cell Biol. 2004;24:9478–9486. doi: 10.1128/MCB.24.21.9478-9486.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chen JJ, Silver D, Cantor S, Livingston DM, Scully R. BRCA1, BRCA2, and Rad51 operate in a common DNA damage response pathway. Cancer Res. 1999;59:1752s–1756s. [PubMed] [Google Scholar]
- 55.Jones LP, Sampson A, Kang HJ, et al. Loss of BRCA1 leads to an increased sensitivity to Bisphenol A. Toxicol Lett. 2010;199:261–268. doi: 10.1016/j.toxlet.2010.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Welshons WV, Nagel SC, vom Saal FS. Large effects from small exposures. III. Endocrine mechanisms mediating effects of bisphenol A at levels of human exposure. Endocrinology. 2006;147:S56–69. doi: 10.1210/en.2005-1159. [DOI] [PubMed] [Google Scholar]