Abstract
Asymmetric division of progenitor/stem cells generates both self-renewing and differentiating progeny and is fundamental to development and regeneration. How this process is regulated in the vertebrate brain remains incompletely understood. Here we use time-lapse imaging to track radial glia progenitor behavior in the developing zebrafish brain. We find that asymmetric division invariably generates a basal self-renewing daughter and an apical differentiating sibling. Gene expression and genetic mosaic analysis further show that the apical daughter is the source of Notch ligand that is essential to maintain higher Notch activity in the basal daughter. Notably, establishment of this intra-lineage and directional Notch signaling requires the intrinsic polarity regulator Partitioning defective protein-3 (Par-3), which segregates the fate determinant Mind bomb unequally to the apical daughter, thereby restricting the self-renewal potential to the basal daughter. These findings reveal with single-cell resolution how self-renewal and differentiation become precisely segregated within asymmetrically dividing neural progenitor/stem lineages.
Keywords: single cell imaging analysis in vivo, proliferation, cancer, dysplasia, neural stem cell, clonal analysis, in vivo lineage tracing, interkinetic nuclear migration (INM)
INTRODUCTION
Stem cells have the remarkable ability to continuously maintain a stem cell population (self-renew) while generating differentiating progeny. One important means to regulate such robust behavior of stem cells is through asymmetric cell division, which generates one daughter retaining the stem cell identity and the other committed to differentiation. Dys-regulation of this process has been implicated in human diseases ranging from dysplasia to cancer (Knoblich, 2010; Yong and Yan, 2011).
Asymmetric cell divisions of progenitor/stem cells have been extensively characterized in invertebrates. These studies have identified a set of intrinsic polarity regulators, which function to ensure proper segregation of cell fate determinants into two daughter cells (Doe, 2008; Guo and Kemphues, 1996; Knoblich, 2010; Lu et al., 2000). Compared to these advances, much less is understood about the regulation of asymmetric cell division and subsequent daughter cell fate choice in vertebrates. Despite that conserved counterparts to the invertebrate genes are found in vertebrates, the function of these proteins is only beginning to be elucidated (Doe, 2008; Gotz and Huttner, 2005; Knoblich, 2010; Williams et al., 2011). Available data suggest that vertebrates may deploy these factors in new and different ways that remain enigmatic.
Radial glia in the vertebrate developing central nervous system (CNS) have stem cell -like properties (Gotz and Huttner, 2005; Kriegstein and Alvarez-Buylla, 2009; Malatesta et al., 2000; Miyata et al., 2001; Noctor et al., 2001; Temple, 2001). Previous studies in mammals (Bultje et al., 2009; Cayouette et al., 2001; Chenn and McConnell, 1995; Miyata et al., 2001; Miyata et al., 2004; Noctor et al., 2004) and zebrafish (Alexandre et al., 2010; Baye and Link, 2007; Das et al., 2003) show that during the peak phase of neurogenesis, radial glia progenitors predominantly undergo asymmetric divisions, serving as an excellent model for understanding how asymmetric cell division, self-renewal, and differentiation are regulated in vertebrate stem cells.
An interesting behavior that vertebrate radial glia progenitors display is the interkinetic nuclear migration (INM) (Baye and Link, 2008; Miyata, 2008; Sauer, 1935), which refers to the movement of progenitor nuclei between the apical and basal surfaces of the neuroepithelium in phase with their cell cycle. Studies in the developing chick CNS (Murciano et al., 2002) and zebrafish retina (Baye and Link, 2007; Del Bene et al., 2008) suggest that proliferative (self-renewing) versus neurogenic (differentiating) potential of radial glia progenitors is largely determined by their pattern of INM. In particular, Del Bene et al proposes the presence of a Notch gradient between the apical and basal surfaces of the neuroepithelium, raising the possibility that extrinsic signals play a critical role in determining vertebrate progenitor self-renewal or differentiation in a location-dependent manner.
Here we carry out in vivo time-lapse imaging with single-cell resolution and perform clonal genetic mosaic analysis of individual radial glia lineages in the developing zebrafish brain. Our study uncovers a stereotyped pattern of asymmetric division that invariably generates a self-renewing daughter that migrates to a basal position and a differentiating sibling remaining at the apical position. We further reveal an asymmetry of Notch activity in paired daughters and show that Notch signaling between the daughters is critical for balancing self-renewal and differentiation. We also demonstrate that the ubiquitin E3 ligase Mind bomb (Mib), which promotes Notch signaling activity by modulating the endocytosis of Notch ligands (Itoh et al., 2003; Le Bras et al., 2011), is unequally segregated to the apical daughter. This Mib localization is critically dependent on Partitioning defective protein-3 (Par-3), an evolutionarily conserved polarity regulator (Alexandre et al., 2010; Etemad-Moghadam et al., 1995; Macara, 2004; von Trotha et al., 2006). Par-3 acts through Mib to restrict high Notch activity to the basal daughter thereby limiting self-renewal. Together, this study reveals with single-cell resolution that asymmetrically dividing vertebrate neural progenitors balance self-renewal and differentiation through directional intra-lineage Notch signaling that is established by intrinsic cell polarity.
RESULTS
In Vivo Time-Lapse Imaging Delineates Progenitor Division Pattern and Fate
To learn about the in vivo behavior of radial glia progenitors, we performed brain ventricle-targeted electroporation (Dong et al., 2011), which allowed for sparse labeling of individual progenitors in the developing zebrafish brain at ~26-somite stage (~22 hours post fertilization, hpf) (Figure 1A). Labeled embryos were subjected to time-lapse imaging for ~26 to 48 hours, during which the labeled progenitor undergoes INM and generally completes two successive rounds of divisions, yielding clonally related cells, which we termed mother, daughter, and granddaughter (Figures 1B and S1, Movie S1). The progenitor state was defined by distinct radial glia morphology and a lack of Elav/Hu, a marker for post-mitotic neurons (Kim et al., 1996; Mueller and Wullimann, 2002). The neuronal state was deduced from the lack of radial glia morphology, and further verified by positive expression of Elav/Hu (Figure 1B). These analyses allowed us to establish lineage relationships and the daughter cell fate choice (i.e. to self-renew or commit to differentiation). We did not discern whether divisions that produced two post-mitotic neurons were symmetric or asymmetric, given our focus on the fate choice between self-renewal and differentiation, and the lack of appropriate markers to follow neuronal subtype identity.
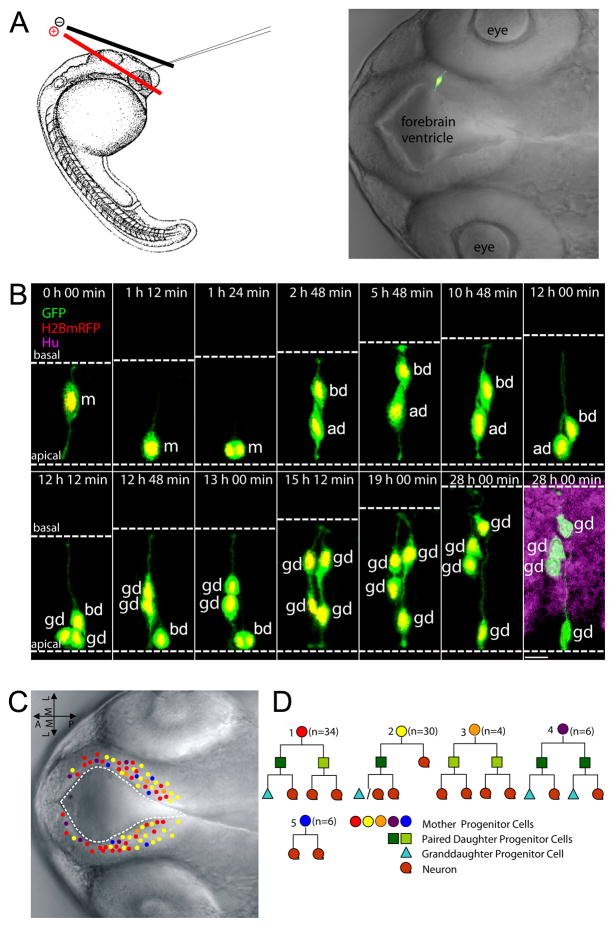
Figure 1. In vivo time-lapse imaging of radial glial progenitor cells in the developing zebrafish forebrain delineates self-renewal and differentiation divisions.
(A) Left: A schematic of electroporation. Right: a representative image of a labeled individual radial glia cell in 28 hpf zebrafish forebrain. (B) Representative montage of selected images from time-lapse in vivo imaging of a single fluorescently-labeled mother cell. The daughter cell on the left undergoes a differentiation division (generating two neurons), whereas the daughter cell on the right undergoes a self-renewal division (generating one progenitor and one neuron). Dashed white lines indicate the apical (bottom) and basal (top) surfaces. Time is shown on the top of each panel. 0 h 00 min equals the onset of time-lapse in vivo imaging (28 hpf). m: mother cell, ad: apical daughter cell, which maintains a more apical position, bd: basal daughter cell which migrates to and maintains a more basal position, gd: granddaughter cells. Scale bar, 10 μm. (C) Nomarski images of zebrafish forebrain depicting the location of the mother cells (A–P, anterior posterior; M–L, medial lateral). Colors represent different cell fate lineages as shown in (D). (D) Different clone types observed by time-lapse in vivo imaging. See also Figure S1 and Movie S1.
After conducting more than 50 independent experiments and following over 400 progenitor cells, we reconstructed 80 lineage trees. The analyzed mother cells were distributed around the forebrain ventricle, spreading along the dorsoventral and anteroposterior axes (Figure 1C). Of note, all progenitor divisions were observed at the apical surface, unlike the occurrence of divisions at both the apical surface and in the subventricular zone (SVZ) of the developing mammalian forebrain (Noctor et al., 2004). Among the 80 mother cells analyzed, 30 cells divided in an asymmetric manner sensu stricto, giving rise to one progenitor and one neuron (Figure 1D2). Remarkably, another 34 mother cells generate two differentially fated progenitor cells (Figure 1D1). The rest (16 out of 80) divided symmetrically with respect to self-renewal and differentiation, generating two differentiating progenitors (n=6, Figure 1D3), two self-renewing progenitors (n=4, Figure 1D4), or two neurons (n=6, Figure 1D5). This in vivo lineage analysis indicates that during active neurogenesis in the developing zebrafish forebrain, a majority of radial glia progenitors divide asymmetrically to produce both self-renewing and differentiating progeny, while a small proportion of radial glia divide to either self-renew or differentiate.
Clonal Analysis of Progenitor Behavior Reveals that the Self-Renewing Daughter Maintains a Basal Position Shortly after Birth and throughout INM
To identify distinguishing features of the self-renewing versus differentiating progenitors, we analyzed multiple parameters of progenitor behavior, including their cell cycle period, division orientation, apical to basal migration period, basal pause time, basal to apical migration period, and relative maximum basal migration (proportionate to the size of the germinal zone at a given location; see Experimental Procedures for details). We found that most of these parameters were highly heterogeneous spanning a broad range (Figure S2), in agreement with a previous study in the retina (Baye and Link, 2007). In addition to the heterogeneity of each parameter measured, a statistical correlation analysis did not detect any parameters that co-varied with one another.
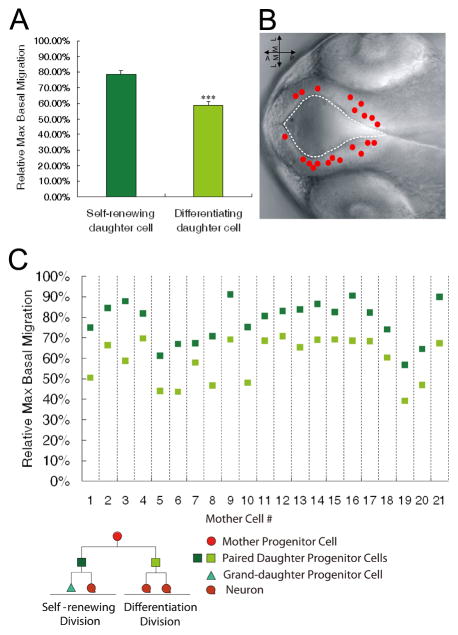
We then analyzed each parameter in two bins, one consisting of the self-renewing daughters and the other consisting of the differentiating daughters. While most of the parameters did not differ significantly between the two bins, interestingly, the self-renewing daughters migrated to and maintained a more basal position (hence termed the basal daughter in this study, see Figure 1B) than their differentiating siblings (termed the apical daughter in this study, see Figure 1B) when the max basal migration was assessed (Figure 2A). Since our imaging analysis tracked clonally related cells with single-cell resolution, we were able to further examine the maximum basal migration in paired daughter progenitors derived from asymmetric divisions (n=21; the maximum basal migration was not tracked in all lineages analyzed, see Experimental Procedures for details). The mother cells giving rise to these daughters were more or less randomly distributed around the forebrain ventricle (Figure 2B). This analysis revealed a striking correlation: In all 21 pairs of daughter progenitors, the self-renewing one always displayed more basal migration than the differentiating sibling (Figure 2C).
Figure 2. In vivo time-lapse imaging coupled with clonal analyses reveals that the self-renewing daughter cell migrates more basally than the differentiating sibling.
(A) Quantification of the relative maximum basal migration of the self-renewing and differentiating siblings in paired daughter cells. *** p < 0.001, t-test. Data are shown as the mean ± SEM. (B) Nomarski image of zebrafish forebrain depicting the location of the mother cells giving rise to the 21 paired daughter cells that show different cell fates. (C) Relative maximum basal migration of the self-renewing daughter cell (dark green) and the differentiating daughter cell (light green). See also Figure S2.
When we examined the cell positioning throughout the entire INM, we further noted that, shortly after the asymmetric division with a cleavage plane largely parallel to the apical-basal axis (See Figures S1 and S2), the two daughter cells assumed differential positions along the apical-basal axis. By carefully comparing each frame at 12-minute imaging intervals, we were able to deduce that the distinct positioning of the two daughter cells was maintained throughout INM (n=21) (Figure S1). These results show that after asymmetric divisions, daughter cells assume differential positions along the apical-basal axis, and this position predicts the self-renewing versus differentiating fates: the basal daughter is the one that retains the ability to self-renew.
The Basal Daughter Displays Higher Notch Activity than Its Apical Sibling
To determine why the basal daughter self-renews whereas the apical sibling embarks on a differentiation path, we considered the Notch signaling pathway, the activation of which inhibits neurogenesis and maintains progenitor characteristics(Artavanis-Tsakonas et al., 1999; Gaiano et al., 2000; Louvi and Artavanis-Tsakonas, 2006; Mizutani et al., 2007; Yoon and Gaiano, 2005; Yoon et al., 2008) Components of the Notch pathway,. including the Notch ligands DeltaA (Dla) and DeltaD (Dld), the Notch receptors, and the primary target of activated Notch, Hairy related 4.1 (Her4.1, orthologous to mammalian hes5), are expressed in the developing brain (Thisse and Thisse, 2005) (Figure 3). Notably, our expression analysis did not reveal a gradient pattern of Notch signaling in the developing brain, as what has been previously reported in the retina (Del Bene et al., 2008). Instead, the expression of her4.1, as well as that of Notch receptor and ligands, displayed interspersed patterns in the germinal zone (Figure 3).
Figure 3. Expression of Notch signaling components in the developing zebrafish brain.

Fluorescent in situ hybridization (FISH) shows the expression of dla (green; A), notch 1b (green; C) and her 4.1 (green; D) in 36 hpf embryos. Fluorescent immunohistochemistry shows the expression of Dld protein (red; B) in 36 hpf embryos. Hu (red) labels post-mitotic neurons, and β-catenin (blue) depicts the outline of the embryo. fb: forebrain, v: ventricle, mb: midbrain.
To closely examine Notch activity in paired daughter cells, we sparsely labeled radial glia progenitors by brain ventricle-targeted electroporation of GFP constructs at ~22 hpf, and performed fluorescent in situ hybridization (FISH) for her4.1 coupled with immunostaining for GFP. Various developmental stages were examined, which covered different phases of the cell cycle and INM of the paired daughters. Quantitative analyses using Metamorph software showed that majority of paired daughter cells (83%, n=127) exhibited asymmetric her4.1 expression: it was always the basal daughter that exhibited higher her4.1 expression than its apical sibling (Figures 4A–E). Scatter plot analysis showed that the remaining 17% paired daughter cells had approximately equal level of her4.1 expression between siblings (Figure 4F). The percentage of paired daughters with asymmetric her4.1 expression (83%) matched well with that of radial glia progenitors undergoing asymmetric divisions [Clone types 1 and 2, 64/80, see Figure 1D], suggesting that asymmetrically dividing radial glia progenitors generate daughter cells with asymmetric her4.1 expression. Additionally, another Notch target gene her15.1 (previously also called hes5) (Thisse and Thisse, 2005) also showed asymmetric expression in paired daughter cells (Figure S3A–C).
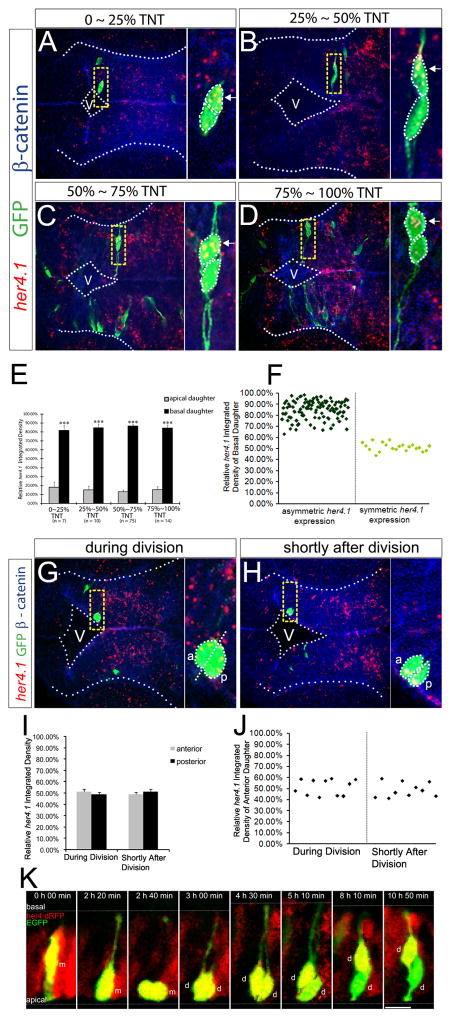
Figure 4. The basal daughter cell expresses a higher level of her4.1.
(A to D) FISH of her4.1 (red) coupled with immunohistochemistry of GFP (green) and β-catenin (blue) in forebrain paired daughter cells. Images are assembled according to the distance of the basal daughter cell to the ventricular surface. TNT: total neuroepithelium thickness. Enlargement of the yellow-boxed area is shown on the right of each panel. V: ventricle. (E–F) Quantification of the FISH signal of her4.1 in paired daughter cells shown in a bar graph (E) or scatter plot (F). *** p < 0.001 vs apical, t-test. (G–H) her4.1 expression in forebrain progenitor cells during division (G) or shortly after division (H). V: ventricle, a: anterior, p: posterior. (I–J) Quantification for (G–H) in bar graph (I) or scatter plot (J). (K) Representative montage of selected images from time-lapse in vivo imaging of a single EGFP-labeled mother cell in the hindbrain of her4:dRFP transgenic embryo. m: mother cell, d: daughter cells. Scale bar, 10 μm. See also Figure S3 and Movie S2.
To address whether the asymmetry of her4.1 mRNA arose before, during, or after cell division, we performed FISH analysis on progenitors around the time of division and found her4.1 expression to be symmetric (Figure 4G–J, n=21). We further carried out time-lapse imaging using the Notch activity reporter line her4.1:dRFP (Yeo et al., 2007). We observed that the Notch activity was high and uniformly distributed in the mother progenitor before, during, and shortly after division. As the two daughter cells began to adopt a differential positioning along the apical-basal neural axis, Notch activity started to decrease in the apical daughter but remained high in the basal daughter (Figure 4K, Figure S3D, and Movie S2, n=10). We did observe that some daughter cells of labeled progenitors (n=3) had extremely low level of Notch activity that did not change over time, likely corresponding to symmetrically dividing progenitors. Together, these results reveal an asymmetric Notch activity in paired siblings and indicate that such asymmetry is not due to differential inheritance of her4.1 mRNA, bur arises after asymmetric division and during the time when the two daughter cells assume differential positioning along the apical-basal neural axis.
The Apical Daughter Expresses Higher Notch Ligand than Its Basal Sibling
Notch activity in a given cell is maintained through contact with ligand-expressing neighboring cells. Four genes in zebrafish encode Delta ligands, among which dla and dld are prominently expressed in the developing brain (Thisse and Thisse, 2005) (Figure 3). After performing clonal analysis of dla expression in paired daughters, using the method similar to that implemented above to assess her4.1 expression, we found that, strikingly, in daughter cells with differential dla expression (81% of all dla-expressing paired daughters examined, n=124), the apical daughter always expressed a higher level of dla than its basal sibling (Figure 5A–F). Dla expression around the time of division showed no asymmetry, indicating that the asymmetric dla expression is not due to differential inheritance of dla mRNA by the two daughter cells (Figure 5G–J). Dld also exhibited asymmetric expression in paired daughter cells (Figure S4). Together, these results demonstrate an asymmetric distribution of Notch ligands in paired siblings that is not due to differential mRNA inheritance.
Figure 5. The apical daughter cell expresses a higher level of dla.
(A–D) FISH of dla (red) coupled with immunohistochemistry of GFP (green) and β-catenin (blue) in forebrain paired daughter cells. TNT: total neuroepithelium thickness; V: ventricle. (E) Quantification for (A–D) in bar graph (E) and scatter plot (F). *** p < 0.001 vs apical, t-test. (G and H) dla expression during (G) or shortly after division (H). V: ventricle, a: anterior, p: posterior. (I) Quantification for (G–H) in bar graph (I) and scatter plot (J). See also Figure S4.
Intra-lineage Notch Signaling Is Essential for Maintaining Self-Renewal in the Basal Daughter
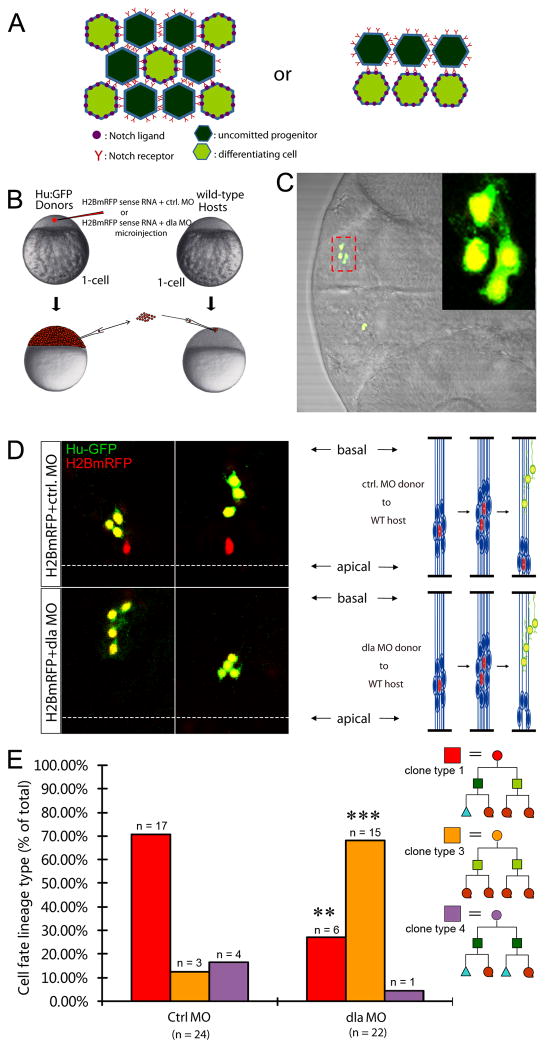
Our observation that asymmetric division generates a basal self-renewing daughter with higher Notch activity and apical differentiating daughter with higher Notch ligand expression prompted us to investigate whether Notch signaling operates within lineage to regulate daughter cells’ decision to self-renew or differentiate. While the classical mode of Notch signaling is lateral inhibition (Figure 6A, left), which selects one cell from a group of equivalent precursors, Notch also plays a role in lineage decisions that make two daughter cells adopt different fates (Figure 6A, right). Progenitors resided in the vertebrate neural tube are thought to signal via lateral inhibition (Pierfelice et al., 2011), but a careful evaluation of literature finds little experimental evidence. Therefore, to explore the mode of Notch signaling in the self-renewal and differentiation of daughter cells derived from asymmetric division, we performed genetic mosaic experiments by transplanting cells deficient for dla activity into a wild-type host embryo at the blastula stage (Figure 6B) and analyzed 4-cell clones at the pharyngula stage (~56 hpf) (Figure 6C). A well-established morpholino antisense oligonucleotide targeting dla (Diks et al., 2008; Latimer et al., 2002) was used to knock down dla activity (Figure S5). The transplanted dla-deficient cells also expressed H2BmRFP (red, lineage tracer) and Hu:GFP (green, marking differentiated neurons)(Figure 6B–C). In the control group, most 4-cell clones (~71%, n=24) contained one progenitor and three nascent neurons (Figure 6D, top panels, two representative clones were shown), hence representing granddaughters that were derived from one self-renewing daughter and one differentiating daughter (Figure 6E, red bar). In contrast, most dla-deficient 4-cell clones (~68%, n=22) contained four neurons (Figure 6D, bottom panels, two representative clones were shown). This difference between the control and the dla-deficient clones was highly significant (Figure 6E), indicating that clonal inactivation of dla is sufficient to bias progenitors toward differentiation. If lateral inhibition were the mode of Notch signaling, one would have not expected a loss of self-renewing potential in dla-deficient clones, given the wild-type level of Notch ligands in the surrounding cells. Since Notch signaling failed to be rescued in the dla-deficient clones despite the presence of Notch ligands in the surrounding cells, we conclude that intra-lineage Notch signaling is the predominant if not the exclusive mode of action that maintains a balanced self-renewal and differentiation in daughter cells of asymmetric division during active neurogenesis in the zebrafish neural tube.
Figure 6. Clonal knockdown of dla reveals intra-lineage Notch signaling in daughter cells of asymmetric division.
(A) A schematic depicting two different modes of Notch signaling. Left: lateral inhibition. Right: Intra-lineage. (B) Overview of the transplantation strategy. (C) Representative image of a single 4-cell clone. Inset is the enlargement of the area highlighted by the red dashed box. (D) Representative images of two single clones in control morpholino group (top) and dla morpholino group (bottom). In control, two single 4-cell clones contain one progenitor and three neuronal granddaughter cells. In the dla morpholino group, two single 4-cell clones contain four neuronal granddaughter cells. (E) Quantification for D. ** p < 0.01, *** p < 0.001 vs Ctrl MO, z-test. See also Figure S5.
The Notch Signaling Activator Mind Bomb Is Unequally Segregated to the Apical Daughter in a Par-3-Dependent Manner
The results delineated above, together with the observed asymmetric expression of Notch signaling components in paired daughter cells, informed us that Notch signaling is not only intra-lineage but also directional. What is the mechanism that sets up the directionality of Notch signaling? While the classical experiments in Drosophila have established a critical role of Numb in antagonizing Notch during neuroblast self-renewal and differentiation (Guo et al., 1996; Spana and Doe, 1996), the relationship between Numb and Notch in vertebrates has not been resolved (Li et al., 2003; Petersen et al., 2002). To determine how the directionality of Notch signaling is established in our system, we turned to the Notch signaling regulator Mind bomb (Mib) as a potential candidate. Mib is an E3 ubiquitin ligase that promotes Notch signaling by modulating the endocytosis of Notch ligands, and consistent with its role in regulating Notch signaling, the loss of mib function dramatically increases neuronal differentiation at the expense of progenitor cells (Itoh et al., 2003; Koo et al., 2005; Yoon et al., 2008). Together, these findings support the notion that Mib is a cell fate determinant that promotes Notch signaling and self-renewal.
The localization of Mib during asymmetric division is not known in any experimental system. To address this question in the absence of a working Mib antibody, we used a GFP-tagged full length Mib (Mib-GFP), which allows examination of the in vivo dynamics of the Mib protein. Multiple tagged forms of Mib (including GST-, Myc-, and FLAG-tagged versions) have been previously shown to be functional (Itoh et al., 2003). Nevertheless, we first verified whether the Mib-GFP reflected the endogenous Mib distribution. When transiently expressed in zebrafish embryos through either DNA electroporation or mRNA microinjection, Mib-GFP displayed a punctate pattern that is located in the cytosol near the membrane as well as adjacent to the nucleus (Figure 7A–B), in agreement with its previously reported localization and function in endosomes (Itoh et al., 2003; Koo et al., 2005). In addition, we performed double labeling with antibodies against GFP and Dld at ~24 hpf. Dld is expressed in the developing brain (Figure 3), albeit less prominently than Dla, for which a workable antibody was not available despite much failed effort with the previously published antibody (Tallafuss et al., 2009). This analysis showed that the Mib-GFP signal was co-localized with Dld (Figure 7C), although an exact co-localization was not expected due to the transient nature of Mib-GFP expression and the presence of other Notch ligands in the brain. Together, these results suggest that Mib-GFP reflects the endogenous Mib distribution pattern.
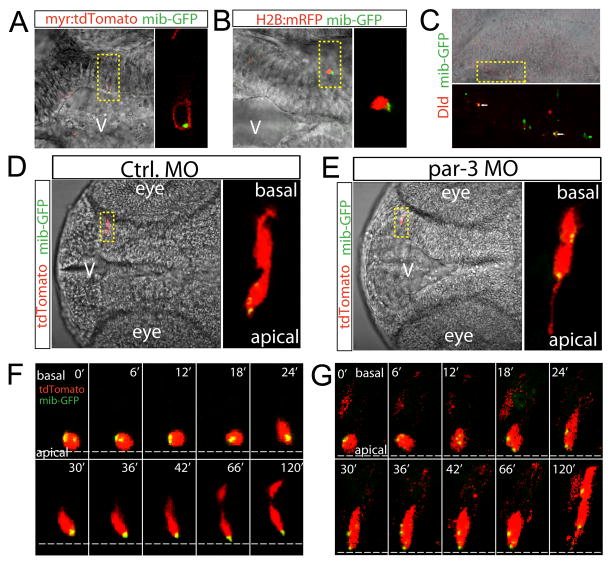
Figure 7. The fate determinant Mib is unequally segregated to the apical daughter in Par-3-dependent manner.
(A) Mib-GFP is detected in the cytosol within the membrane targeted myr:tdTomato. (B) Mib-GFP is detected in close proximity to the nucleus reporter H2B:mRFP. (C) Mib-GFP colocalizes with Dld. (D) Mib-GFP was unequally segregated into the apical daughter. (E) The unequal segregation of mib-GFP was eliminated by knockdown of par-3. (F) Selected frames of time-lapse live imaging shows unequal segregation of Mib-GFP to the apical daughter cell during division. (G) Selected frames of time-lapse live imaging shows equal distribution of Mib-GFP in both daughter cells during division in the par-3 morphant. See also Figure S6, Movie S3 and Movie S4.
Next, we analyzed the Mib-GFP distribution in paired daughter cells. Co-electroporation of a red fluorescent lineage tracer together with the Mib-GFP construct at ~22 hpf and analysis of paired daughters at ~37 hpf showed that Mib-GFP was exclusively detected in the apical daughter in 85% paired daughter cells analyzed (n=26)(Figure 7D). This observed percentage is consistent with the idea that Mib asymmetry is likely present in both Clone type 1 and 2 (as shown in Figure 1D). In addition, the Mib asymmetry appeared to be stably maintained during INM (Figure S6).
The unequal segregation of Mib-GFP into the apical daughter made us wonder whether it is dependent on the conserved intrinsic polarity regulator Par-3, since Par-3 has been found asymmetrically localized to the apical domain of dividing neural progenitors in zebrafish (Alexandre et al., 2010; von Trotha et al., 2006). We analyzed paired daughters in the embryos injected with a well-established morpholino antisense oligonucleotide targeting par-3 (referred to as the par-3 morphant)(Alexandre et al., 2010; Tawk et al., 2007). As expected, the par-3 morphants in our experiments displayed a loss of apico-basal cell polarity and suffered a mild defect in brain morphology at 37 hpf (Figure S7A–I). In the par-3 morphant, Mib-GFP was detected in both daughter cells (91%, n=23 pairs of daughter cells analyzed) (Figure 7E and Figure S6).
To determine the onset of Mib-GFP localization in paired daughters, we carried out time-lapse imaging. The segregation of Mib-GFP into the apical daughter was apparent at the time of birth (Figure 7F and Movie S3, ~24 min). However, in the par-3 morphant, Mib-GFP was present in both the apical and basal daughter at the time of their birth (Figure 7G and Movie S4, ~18 min). Together, these results suggest that Mib is unequally segregated into the apical daughter upon asymmetric division in a Par-3 – dependent manner and such asymmetry is maintained in the daughter cells.
Par-3 Is Essential to Restrict Notch Activity and Self-renewal to the Basal Daughter through Mib
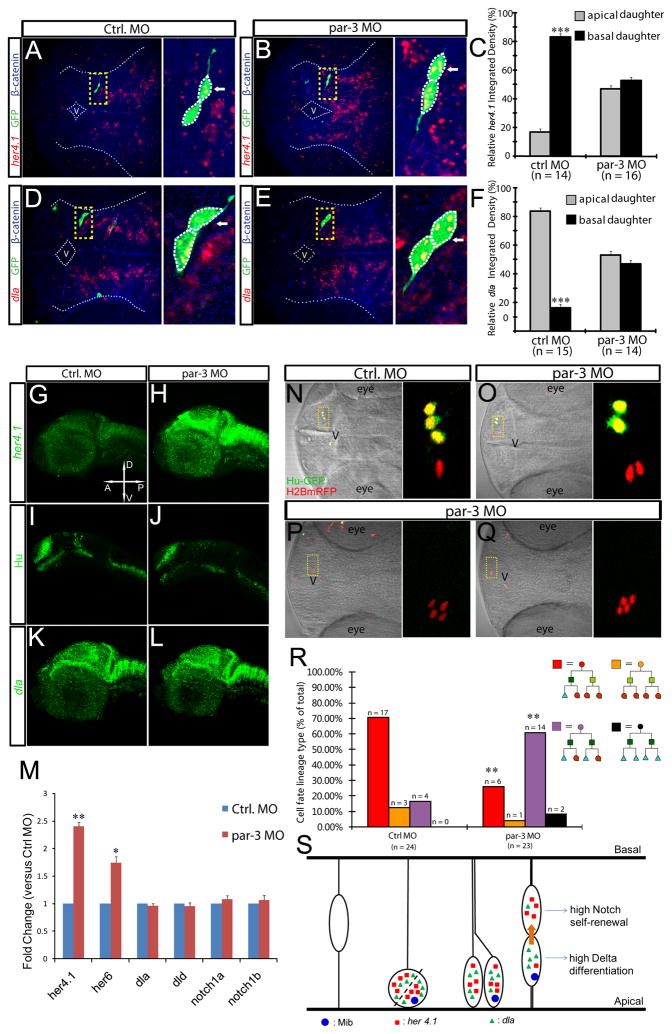
In agreement with the disrupted Mib localization in the par-3 morphant, we found that the asymmetry of both her4.1 (Figure 8A–C) and dla (Figure 8D–F) expression was lost in par-3 -deficient embryos, demonstrating that Par-3 is essential for establishing Notch asymmetry in paired siblings.
Figure 8. Par-3 is essential for restricting Notch activity to the basal daughter thereby limiting self-renewal.
(A–F)her4.1 (A–B) or dla(D–E) expression in control (A,D) and par-3 (B,E) morphants. (C) and (F) are quantifications. *** p < 0.001 vs apical, t-test. (G–L) Expression of her4.1(G–H), Hu (I–J), and dla(K–L) in control (G,I,K) or par-3 morphants (H,J,L). (M) Quantitative RT-PCR shows relative fold change of her4.1, her6, dla, dld, notch1a and notch1b in control versus par-3 morphants. * p < 0.05, ** p < 0.01, vs Ctrl MO. (N-R) Transplantation similar to that described in Figure 5B was carried out. Representative image of 4-cell clones derived from control (N) or par-3 morphants (O–Q) in otherwise wild-type brains. In control, the single clone is composed of 1 progenitor (red) and 3 neurons (green), whereas par-3 –deficient clones contained fewer neurons (green) and more progenitors (red). (R) Quantification for N–Q. ** p < 0.01, vs Ctrl MO, Z-test. (S) A model for regulated self-renewal and differentiation in asymmetrically dividing radial glia progenitors. See Discussion. See also Figure S7.
Mib mis-localization and lost asymmetry of Notch signaling components in par-3 morphants could result in either increased or diminished Notch activity in both daughter cells, which would in turn impact progenitor fate choice differently. To determine how Notch activity and cell fate might be affected in par-3-deficient embryos, we first analyzed the overall expression level of her4.1, dla, and the pan-neuronal marker Hu. These analyses showed that her4.1 expression (Figure 8G–H, 88%, n=16) was increased while neuronal numbers (Figure 8I–J, 83%, n=18) were decreased in the par-3 morphant. In contrast, the expression of dla was not changed significantly (Figure 8K–L, 100%, n=15). This is surprising, given the increase of her4.1 expression and the known negative feedback regulation of Notch ligands by hes/her genes. Quantitative RT-PCR analysis further confirmed the significant up-regulation of her4.1 (and her6) mRNA expression, whereas the mRNA levels of dla, dld, notch1a, and notch1b were unchanged (Figure 8M)(Supplementary Table S1). Thus, par-3 function is essential to restrict Notch activity, and is somehow also required for the feedback repression of Notch ligand expression.
To understand the nature of these par-3 functions, we asked whether they are dependent on mib. In the mib−/− mutant, consistent with the disruption of Notch signaling, her4.1 expression was significantly reduced (Figure S7J–K). The par-3 and mib – double deficient embryos also showed reduced her4.1 expression (Figure S7M) that was indistinguishable from the mib−/− single mutant (Figure S7K). This result indicates that Par-3 restricts Notch activity through Mib.
While the diminished Notch activation in the mib−/− mutant is expected to up-regulate Notch ligand expression via the negative feedback loop, this was not what we observed. Instead, the dla mRNA level was significantly reduced in the mib−/− mutant (Figure S7N–O) as well as in the par-3 and mib – double deficient embryos (Figure S7Q). This finding indicates that intact Mib activity is critical for the manifestation of negative feedback regulation of Notch ligand expression, possibly due to the effect of Mib on regulating Notch ligand protein turnover that also impacts transcription, albeit in an opposing way.
To further determine whether par-3 acts to limit self-renewal, we performed genetic mosaic experiments by transplanting par-3-deficient cells into wild-type host embryos, as we have done previously with the analysis of dla (Figure 6B). This clonal analysis of par-3 function showed that par-3-deficient 4-cell clones had a greater propensity (~61%, n=23) to contain 2 progenitors and 2 neurons (Figure 8O, 8R-purple bar). Moreover, some par-3-deficient 4-cell clones contained all four progenitors (8.7%, n=23), which were never observed in the control group (Figure 8P, 8Q, 8R, black bar). This clonal analysis indicates that par-3 is essential to limit self-renewal within asymmetrically dividing radial glia lineages.
DISCUSSION
In the present study, we have carried out in vivo time-lapse imaging and genetic mosaic analysis, both at single-cell resolution in an intact vertebrate brain. We show that radial glia progenitors divide predominantly in an asymmetric fashion in the developing zebrafish brain during active neurogenesis. Such asymmetric division invariably generates basal self-renewing and apical differentiating daughters. The basal daughter maintains higher Notch activity, whereas the apical sibling expresses higher Notch ligand. We further establish that intra-lineage Notch signaling is critical for maintaining self-renewal in the basal daughter. Finally, we demonstrate that the directionality of Notch signaling is established through Par-3 -dependent asymmetric localization of Mib to the apical daughter (Figure 8S).
Radial Glia Progenitor Behavior Revealed by in vivo Time-lapse Imaging
Direct observation of cellular behavior in its native environment is a powerful approach to gain new biological insights. Our work has extended previous time-lapse imaging studies in vitro in mammalian cultured cells (Temple, 1989) and cortical slices (Chenn and McConnell, 1995; Miyata et al., 2001; Noctor et al., 2004) as well as in vivo in zebrafish brains (Alexandre et al., 2010) in important ways. First, through imaging clonally labeled progenitors for two rounds of cell divisions, we are able to construct lineages spanning three generations (mother, daughter, and granddaughter) that have uncovered five clonal types. In agreement with the mammalian cortical slice study (Noctor et al., 2004), we find that a majority (~80%) of progenitors divide asymmetrically, half of which generate two differentially fated progenitors (Clone type 1) whereas the other half generates a progenitor and a neuron (Clone type 2). What mechanisms differentiate Clone type 1 versus 2 is an interesting and unresolved question. It is possible that the difference in the absolute Notch activity level may underlie the difference in these two lineages. Future experiments are needed to test this idea, together with determining what makes a cell decide to choose any one of the five lineages and whether other type of more rare lineages exist.
Secondly, analysis of INM and the relative positioning of daughter cells in conjunction with their fate has allowed us to discern that the paired daughters assume a differential positioning along the apical-basal neural axis shortly after asymmetric division. This differential position is maintained throughout INM, with the apical daughter taking on a differentiation path whereas the basal sibling remaining as a progenitor. In agreement with our results, a recent study in zebrafish, which has examined the asymmetric division that produces one progenitor and one neuron, also finds that the apical daughter inheriting the Par-3-expressing apical domain usually becomes a neuron whereas the basal daughter inheriting the basal process remains a progenitor (Alexandre et al., 2010). In contrast, previous studies in the mammalian brain show that the apical daughter remains a progenitor while the daughter inheriting the basal process becomes a neuron (Chenn and McConnell, 1995; Miyata et al., 2001). What accounts for these opposite observations are not entirely clear, but possibilities include differences in timing, tissue region under study, or species. Nevertheless, results from zebrafish (Alexandre et al 2010 and the present study) indicate that the notion of the presence of “stemness” factors in the apical domain (Gotz and Huttner, 2005; Kosodo et al., 2004) is not universally true.
Notch Asymmetry in Daughter Cells of Asymmetric Division
The apical domain and the basal process have been used as convenient morphological marks for correlating with self-renewing or differentiating fates (Gotz and Huttner, 2005). How they might actually determine progenitor fate choice is not clear. We show that Notch signaling components are expressed asymmetrically in daughters of asymmetric division, with the apical daughter expressing higher level of Notch ligands and the basal daughter exhibiting higher Notch activity. The time-lapse imaging using the Notch activity reporter further reveals that such asymmetry is not due to asymmetric inheritance of mRNAs but arises after asymmetric division, concurrently with the appearance of differential daughter cell positioning along the apical basal neural axis. During INM, the two daughter cells appear to maintain a direct contact, raising the possibility that they interact through Notch signaling at their interface. It will be interesting to determine whether the Notch ligand or the receptor is concentrated at this interface.
Asymmetric inheritance of Notch1 immunoreactivity by the basal daughter (albeit a neuron) is previously reported in the developing ferret cortex (Chenn and McConnell, 1995). Additionally, at population levels, it has been observed that neural stem cells have higher Notch reporter activity than intermediate progenitors of the developing mouse telencephalon (Mizutani et al., 2007). Collectively, the Notch asymmetry in daughter cells of asymmetric division observed in the present study may be a conserved phenomenon in vertebrates.
Intra-lineage Notch Signaling Regulates Self-renewal and Differentiation in Daughter Cells of Asymmetric Division
The mode of Notch signaling has been studied in many cellular contexts (Bray, 1998). The classical lateral inhibition is demonstrated in Drosophila neuroblast delamination (Bourouis et al., 1989) and vertebrate primary neurogenesis at the neural plate stage (Chitnis et al., 1995). In both cases, cells of distinct fates are selected from a field of equi-potent cells. In addition to lateral inhibition, Notch signaling can also act in a binary mode to influence lineage decisions. Studies in Drosophila have established an important role of Numb in antagonizing Notch signaling during neuroblast lineage decisions; however, the source of Notch ligand (i.e. whether it is from intra-lineage or elsewhere) is not known.
The mode of Notch signaling during active neurogenesis in the vertebrate neural tube has not been resolved. The present study, to our knowledge, is the first to combine in vivo time lapse imaging and lineage-restricted genetic mosaic analysis to show that asymmetrically dividing radial glial progenitors in the developing zebrafish brain segregate self-renewal and differentiation through intra-lineage Notch signaling.
It is worth pointing out that our present study is focused on neural progenitor cells that undergo asymmetric divisions. It remains to be determined whether and how Notch signaling operates in lineages that undergo symmetric divisions or at different stages of neural tube development, and whether intra-lineage Notch signaling occurs in asymmetrically dividing progenitors of other vertebrate systems. Interestingly, a recent study (Shitamukai et al., 2011) reveals that clonal Notch signaling is essential for the outer VZ progenitors to self-renew in the developing mouse neocortex, which indicates that intra-lineage Notch signaling may be a shared mechanism for maintaining neural progenitor self-renewal in vertebrates.
Role of Par-3 and Mib in Establishing the Directionality of Notch Signaling in Daughter cells of Asymmetric Division
In Drosophila neural progenitors, multiple cell fate determinants including Brat (Betschinger et al., 2006), Neuralized (Le Borgne and Schweisguth, 2003), Numb (Rhyu et al., 1994), and Prospero (Hirata et al., 1995; Knoblich et al., 1995; Spana and Doe, 1995) are asymmetrically localized in mitotic progenitors and unequally inherited by the two daughter cells. Importantly, the asymmetric inheritance of Numb biases Notch in Drosophila neuroblast lineages (Guo et al., 1996). However, it is not known whether Numb has a role in regulating Notch signaling in the vertebrate brain. Studies have shown polarized distribution of Numb in the basolateral domain of mitotic neural progenitors in both zebrafish and mice and at the adherens junctions of mammalian interphase radial glia (Rasin et al., 2007; Reugels et al., 2006).
Our results establish Mib as a cell fate determinant that is unequally inherited by the apical daughter of asymmetric division. We further show that the intrinsic polarity regulator Par-3 is required to segregate Mib to the apical daughter. How might this occur? While it is possible that Par-3 may directly interact with Mib, a previous study has reported that Par-1, a conserved protein kinase that regulates asymmetric division and is localized in a complementary manner to Par-3 (Guo and Kemphues, 1995), can phosphorylate Mib, leading to its degradation (Ossipova et al., 2009). Based on these findings, it is possible that Par-1 protein or activity is enriched in the basal daughter, where it acts to phosphorylate Mib and cause its degradation.
Our loss-of-function studies at both population and clonal levels reveal that Par-3 is required to restrict Notch activity to the basal daughter, thereby limiting progenitor self-renewal. A repressive role of Par-3 on self-renewal is in agreement with previous studies in the developing zebrafish (Alexandre et al., 2010) and the mammalian mammary gland (McCaffrey and Macara, 2009). However, in the developing mammalian cortex, Par-3 is found to promote radial glia self-renewal by promoting Notch activity (Bultje et al., 2009; Costa et al., 2008). Tissue-, species-, or temporally- specific functions of these factors may account for these different observations.
In conclusion, the present findings exemplify the importance of single-cell imaging analysis in a native environment for understanding how self-renewal and differentiation are regulated in vertebrate neural development. While our findings elucidate the significance of intrinsic polarity-established directional intra-lineage Notch signaling in balancing self-renewal and differentiation, extrinsic regulation may play roles in establishing and maintaining the intrinsic polarity, as well as to coordinate different cell lineages in order to generate appropriate neuronal types in a spatially and temporally regulated manner.
EXPERIMENTAL PROCEDURES
Zebrafish Strains
Wild-type embryos were obtained from natural spawning of AB adults, and raised according to Kimmel et al (Kimmel et al., 1995). The following zebrafish mutants and transgenic lines were used: mibta52b (Itoh et al., 2003), Hu:GFP (Park et al., 2000).
DNA Plasmid Cloning
The Cla I-BamH I fragment of mib and BamH I-Xba I fragment of gfp were isolated, and inserted between the Cla I-Xba I sites of the pCS2 to create pCS2-mib-GFP. The Xho I-Not I fragment of H2B-mRFP was isolated from plasmid pCS-H2B-mRFP (Megason and Fraser, 2003) and inserted between the EcoR I-Not I sites of the Puas-E1b-EGFP to create Puas-E1b-H2B-mRFP.
Electroporation
Electroporation and sparse labeling of neural progenitor cells in zebrafish embryos was previously described (Dong et al., 2011). Plasmid DNAs (e.g. Pef1a-gal4; Puas-E1b-EGFP; Puas-E1b-H2b:mRFP) were mixed and microinjected into the forebrain or hindbrain ventricles at a final concentration of 500 ng/μl for each plasmid. Electroporated embryos were then released from the agarose and transferred to a fresh dish of embryonic medium containing 0.003% PTU and incubated at 28.5 °C.
Time-lapse in vivo Imaging
Embryos with sparse labeling of radial glia progenitors were imaged on the temperature controlled (28.5 °C) stage of a confocal microscope (Nikon C1 spectral confocal microscope with up-right objectives). One group was imaged every 8 hours for 48 hours to examine cell fate and lineage. The second group was imaged for 26–32 hours with a fixed 12-minute interval. For the second group, the parameters of confocal imaging were determined to be sufficient to capture the INM for each cell, while reducing photobleaching during the extended imaging period. Data from both groups contributed to Figure 1C and Figure 1D, whereas only data from the second group contributed to Figure 2. For the analysis of Mib-GFP segregation in paired daughter cells, electroporated embryos are embedded and imaged using the same method as described above except the interval of time-lapse is 6 minutes. For the analysis of Notch activity in paired daughter cells using her4.1:dRFP transgenic fish, we electroporated the GFP reporter plasmid into the hindbrain region to label individual radial glia progenitors, since this transgenic line is reported to better recapitulate Notch activity in the hindbrain than in the forebrain (Yeo et al., 2007). Electroporated embryos are embedded and imaged using the same method as described above except that the interval of time-lapse is 10 minutes.
Cell Transplantation
Blastomere transplantation was performed as previously described (Ho and Kane, 1990). The Hu:GFP+ donor embryos were injected at the 1-cell stage with the morpholino antisense oligonucleotides against dla (or par-3) and the H2BmRFP sense RNA serving as a lineage tracer. At 3–4 hpf stage (1-k cell to sphere), 10~20 donor cells were transplanted to the animal-pole region of similarly staged wild-type hosts.
Morpholino Oligonucleotides and mRNA Injection
Morpholino and mRNA injections were performed at the 1- cell stage. The following gene specific morpholinos were used in this study: dla MO (5′-CTTCTCTTTTCGCCGACTGATTCAT-3′) (Latimer et al., 2002), par-3 MO(5′ - TCAAAGGCTCCCGTGCTCTGGTGTC -3′) (Echeverri and Oates, 2007). Approximately 1 pmol of dla morpholino or 0.35 pmol of par3MO was injected at the one cell stage per embryo. H2BmRFP 5′-capped sense mRNA was synthesized by SP6 transcription from NotI-linearized plasmid by using the mMESSAGE mMACHINE kit (Ambion). Approximately 4 nl mRNA at 100 ng/ml was injected per embryo.
In situ Hybridization and Immunohistochemistry
In situ hybridization and immunohistochemistry were performed on whole mount embryos as described (Guo et al., 1999) and imaged with a Nikon C1 confocal. The following antibodies were used in immunohistochemistry: chicken anti-GFP (Abcam), rabbit anti-β-catenin (Invitrogen), mouse anti-Hu (Molecular Probes), mouse anti-Dlc and mouse anti-Dld (Leslie et al., 2007), rabbit anti-aPKC (Santa Cruz).
Measurement and Analysis of Gene Expression Level
Expression levels of her4.1, her15.1, dla, and dld were examined by fluorescent in situ hybridization, followed by quantitative analysis using MetaMorph Imaging software (Universal Imaging, Philadelphia, PA). The relative integrated density is calculated as the ratio of the integrated density in each daughter cell to the sum of the integrated density of both daughter cells. Quantitative RT-PCR analysis was carried out as previously described (Chen et al., 2009).
Statistical Analysis
Statistical significance was confirmed by t test or z test comparison of mean values obtained from each experimental condition. All data are presented as mean ± SEM. * P< 0.05, ** P < 0.01, *** P < 0.001.
Supplementary Material
Acknowledgments
We thank Dr. J. Lewis for zebrafish Dla and Dld antibodies, Drs. P. Soba and Y.N. Jan for the UAS-TdTomato plasmids, Drs. A. Kriegstein, J. Rubenstein, S. Wilson, and the Guo lab members for discussions; Dr. B. Lu and Guo lab members for critically reading the manuscript; Dr. K. Thorn and the UCSF Nikon imaging center for assistance with confocal microscopy; and M. Munchua for fish husbandry. This work was supported by the NIH grant NS042626. S.G. was a Searle Scholar and a Science and Engineering Fellow of the David and Lucile Packard foundation.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Alexandre P, Reugels AM, Barker D, Blanc E, Clarke JD. Neurons derive from the more apical daughter in asymmetric divisions in the zebrafish neural tube. Nat Neurosci. 2010;13:673–679. doi: 10.1038/nn.2547. [DOI] [PubMed] [Google Scholar]
- Artavanis-Tsakonas S, Rand M, Lake R. Notch signaling: cell fate control and signal integration in development. Science. 1999;284:770. doi: 10.1126/science.284.5415.770. [DOI] [PubMed] [Google Scholar]
- Baye LM, Link BA. Interkinetic nuclear migration and the selection of neurogenic cell divisions during vertebrate retinogenesis. J Neurosci. 2007;27:10143–10152. doi: 10.1523/JNEUROSCI.2754-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baye LM, Link BA. Nuclear migration during retinal development. Brain Res. 2008;1192:29–36. doi: 10.1016/j.brainres.2007.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betschinger J, Mechtler K, Knoblich JA. Asymmetric segregation of the tumor suppressor brat regulates self-renewal in Drosophila neural stem cells. Cell. 2006;124:1241–1253. doi: 10.1016/j.cell.2006.01.038. [DOI] [PubMed] [Google Scholar]
- Bourouis M, Heitzler P, el Messal M, Simpson P. Mutant Drosophila embryos in which all cells adopt a neural fate. Nature. 1989;341:442–444. doi: 10.1038/341442a0. [DOI] [PubMed] [Google Scholar]
- Bray S. Notch signaling in Drosophila: three ways to use a pathway. Sem Cell Dev Biol. 1998;9:591–597. doi: 10.1006/scdb.1998.0262. [DOI] [PubMed] [Google Scholar]
- Bultje RS, Castaneda-Castellanos DR, Jan LY, Jan YN, Kriegstein AR, Shi SH. Mammalian Par3 regulates progenitor cell asymmetric division via notch signaling in the developing neocortex. Neuron. 2009;63:189–202. doi: 10.1016/j.neuron.2009.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cayouette M, Whitmore AV, Jeffery G, Raff M. Asymmetric segregation of Numb in retinal development and the influence of the pigmented epithelium. J Neurosci. 2001;21:5643–5651. doi: 10.1523/JNEUROSCI.21-15-05643.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Contreras X, Yamaguchi Y, Handa H, Peterlin BM, Guo S. Repression of RNA polymerase II elongation in vivo is critically dependent on the C-terminus of Spt5. PLoS One. 2009;4:e6918. doi: 10.1371/journal.pone.0006918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chenn A, McConnell SK. Cleavage orientation and the asymmetric inheritance of Notch1 immunoreactivity in mammalian neurogenesis. Cell. 1995;82:631–641. doi: 10.1016/0092-8674(95)90035-7. [DOI] [PubMed] [Google Scholar]
- Chitnis A, Henrique D, Lewis J, Ish-Horowicz D, Kintner C. Primary neurogenesis in Xenopus embryos regulated by a homologue of the Drosophila neurogenic gene Delta. Nature. 1995;375:761–766. doi: 10.1038/375761a0. [DOI] [PubMed] [Google Scholar]
- Costa MR, Wen G, Lepier A, Schroeder T, Götz M. Par-complex proteins promote proliferative progenitor divisions in the developing mouse cerebral cortex. Development. 2008;135:11–22. doi: 10.1242/dev.009951. [DOI] [PubMed] [Google Scholar]
- Das T, Payer B, Cayouette M, Harris WA. In vivo time-lapse imaging of cell divisions during neurogenesis in the developing zebrafish retina. Neuron. 2003;37:597–609. doi: 10.1016/s0896-6273(03)00066-7. [DOI] [PubMed] [Google Scholar]
- Del Bene F, Wehman AM, Link BA, Baier H. Regulation of neurogenesis by interkinetic nuclear migration through an apical-basal notch gradient. Cell. 2008;134:1055–1065. doi: 10.1016/j.cell.2008.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diks SH, Sartori da Silva MA, Hillebrands JL, Bink RJ, Versteeg HH, van Rooijen C, Brouwers A, Chitnis AB, Peppelenbosch MP, Zivkovic D. d-Asb11 is an essential mediator of canonical Delta-Notch signalling. Nat Cell Biol. 2008;10:1190–1198. doi: 10.1038/ncb1779. [DOI] [PubMed] [Google Scholar]
- Doe CQ. Neural stem cells: balancing self-renewal with differentiation. Development. 2008;135:1575–1587. doi: 10.1242/dev.014977. [DOI] [PubMed] [Google Scholar]
- Dong Z, Wagle M, Guo S. Labeling and live imaging of clonally related progenitor cells in the developing zebrafish forebrain. J Vis Exp. 2011;50:pii:2594. doi: 10.3791/2594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Echeverri K, Oates AC. Coordination of symmetric cyclic gene expression during somitogenesis by Suppressor of Hairless involves regulation of retinoic acid catabolism. Dev Biol. 2007;301:388–403. doi: 10.1016/j.ydbio.2006.10.003. [DOI] [PubMed] [Google Scholar]
- Etemad-Moghadam B, Guo S, Kemphues KJ. Asymmetrically distributed PAR-3 protein contributes to cell polarity and spindle alignment in early C. elegans embryos. Cell. 1995;83:743–752. doi: 10.1016/0092-8674(95)90187-6. [DOI] [PubMed] [Google Scholar]
- Gaiano N, Nye JS, Fishell G. Radial glial identity is promoted by Notch1 signaling in the murine forebrain. Neuron. 2000;26:395–404. doi: 10.1016/s0896-6273(00)81172-1. [DOI] [PubMed] [Google Scholar]
- Gotz M, Huttner WB. The cell biology of neurogenesis. Nat Rev Mol Cell Biol. 2005;6:777–788. doi: 10.1038/nrm1739. [DOI] [PubMed] [Google Scholar]
- Guo M, Jan LY, Jan YN. Control of daughter cell fates during asymmetric division: interaction of Numb and Notch. Neuron. 1996;17:27–41. doi: 10.1016/s0896-6273(00)80278-0. [DOI] [PubMed] [Google Scholar]
- Guo S, Brush J, Teraoka H, Goddard A, Wilson SW, Mullins MC, Rosenthal A. Development of noradrenergic neurons in the zebrafish hindbrain requires BMP, FGF8, and the homeodomain protein Soulless/Phox2a. Neuron. 1999;24:555–566. doi: 10.1016/s0896-6273(00)81112-5. [DOI] [PubMed] [Google Scholar]
- Guo S, Kemphues KJ. par-1, a gene required for establishing polarity in C. elegans embryos, encodes a putative ser/thr kinase that is asymmetrically distributed. Cell. 1995;81:611–620. doi: 10.1016/0092-8674(95)90082-9. [DOI] [PubMed] [Google Scholar]
- Guo S, Kemphues KJ. Molecular genetics of asymmetric cleavage in the early Caenorhabditis elegans embryo. Curr Opin Genet Dev. 1996;6:408–415. doi: 10.1016/s0959-437x(96)80061-x. [DOI] [PubMed] [Google Scholar]
- Hirata J, Nakagoshi H, Nabeshima Y, Matsuzaki F. Asymmetric segregation of the homeodomain protein Prospero during Drosophila development. Nature. 1995;377:627–630. doi: 10.1038/377627a0. [DOI] [PubMed] [Google Scholar]
- Ho RK, Kane DA. Cell-autonomous action of zebrafish spt-1 mutation in specific mesodermal precursors. Nature. 1990;348:728–730. doi: 10.1038/348728a0. [DOI] [PubMed] [Google Scholar]
- Itoh M, Kim C, Palardy G, Oda T, Jiang Y, Maust D, Yeo S, Lorick K, Wright G, Ariza-McNaughton L. Mind bomb is a ubiquitin ligase that is essential for efficient activation of Notch signaling by Delta. Dev cell. 2003;4:67–82. doi: 10.1016/s1534-5807(02)00409-4. [DOI] [PubMed] [Google Scholar]
- Kim CH, Ueshima E, Muraoka O, Tanaka H, Yeo SY, Huh TL, Miki N. Zebrafish elav/HuC homologue as a very early neuronal marker. Neurosci Lett. 1996;216:109–112. doi: 10.1016/0304-3940(96)13021-4. [DOI] [PubMed] [Google Scholar]
- Kimmel CB, Ballard WW, Kimmel SR, Ullmann B, Schilling TF. Stages of embryonic development of the zebrafish. Dev Dyn. 1995;203:253–310. doi: 10.1002/aja.1002030302. [DOI] [PubMed] [Google Scholar]
- Knoblich JA. Asymmetric cell division: recent developments and their implications for tumour biology. Nat Rev Mol Cell Biol. 2010;11:849–860. doi: 10.1038/nrm3010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knoblich JA, Jan LY, Jan YN. Asymmetric segregation of Numb and Prospero during cell division. Nature. 1995;377:624–627. doi: 10.1038/377624a0. [DOI] [PubMed] [Google Scholar]
- Koo BK, Lim HS, Song R, Yoon MJ, Yoon KJ, Moon JS, Kim YW, Kwon MC, Yoo KW, Kong MP, et al. Mind bomb 1 is essential for generating functional Notch ligands to activate Notch. Development. 2005;132:3459–3470. doi: 10.1242/dev.01922. [DOI] [PubMed] [Google Scholar]
- Kosodo Y, Röper K, Haubensak W, Marzesco AM, Corbeil D, Huttner WB. Asymmetric distribution of the apical plasma membrane during neurogenic divisions of mammalian neuroepithelial cells. EMBO J. 2004;23:2314–2324. doi: 10.1038/sj.emboj.7600223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kriegstein A, Alvarez-Buylla A. The glial nature of embryonic and adult neural stem cells. Annu Rev Neurosci. 2009;32:49–84. doi: 10.1146/annurev.neuro.051508.135600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latimer AJ, Dong X, Markov Y, Appel B. Delta-Notch signaling induces hypochord development in zebrafish. Development. 2002;129:2555–2563. doi: 10.1242/dev.129.11.2555. [DOI] [PubMed] [Google Scholar]
- Le Borgne R, Schweisguth F. Unequal segregation of Neuralized biases Notch activation during asymmetric cell division. Dev Cell. 2003;5:139–148. doi: 10.1016/s1534-5807(03)00187-4. [DOI] [PubMed] [Google Scholar]
- Le Bras S, Loyer N, Le Borgne R. The multiple facets of ubiquitination in the regulation of notch signaling pathway. Traffic. 2011;12:49–61. doi: 10.1111/j.1600-0854.2010.01126.x. [DOI] [PubMed] [Google Scholar]
- Leslie JD, Ariza-McNaughton L, Bermange AL, McAdow R, Johnson SL, Lewis J. Endothelial signalling by the Notch ligand Delta-like 4 restricts angiogenesis. Development. 2007;134:839–844. doi: 10.1242/dev.003244. [DOI] [PubMed] [Google Scholar]
- Li HS, Wang D, Shen Q, Schonemann MD, Gorski JA, Jones KR, Temple S, Jan LY, Jan YN. Inactivation of Numb and Numblike in embryonic dorsal forebrain impairs neurogenesis and disrupts cortical morphogenesis. Neuron. 2003;40:1105–1118. doi: 10.1016/s0896-6273(03)00755-4. [DOI] [PubMed] [Google Scholar]
- Louvi A, Artavanis-Tsakonas S. Notch signalling in vertebrate neural development. Nat Rev Neurosci. 2006;7:93–102. doi: 10.1038/nrn1847. [DOI] [PubMed] [Google Scholar]
- Lu B, Jan L, Jan YN. Control of cell divisions in the nervous system: symmetry and asymmetry. Annu Rev Neurosci. 2000;23:531–556. doi: 10.1146/annurev.neuro.23.1.531. [DOI] [PubMed] [Google Scholar]
- Macara IG. Parsing the polarity code. Nat Rev Mol Cell Biol. 2004;5:220–231. doi: 10.1038/nrm1332. [DOI] [PubMed] [Google Scholar]
- Malatesta P, Hartfuss E, Götz M. Isolation of radial glial cells by fluorescent-activated cell sorting reveals a neuronal lineage. Development. 2000;127:5253–5263. doi: 10.1242/dev.127.24.5253. [DOI] [PubMed] [Google Scholar]
- McCaffrey LM, Macara IG. The Par3/aPKC interaction is essential for end bud remodeling and progenitor differentiation during mammary gland morphogenesis. Genes Dev. 2009;23:1450–1460. doi: 10.1101/gad.1795909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Megason SG, Fraser SE. Digitizing life at the level of the cell: high-performance laser-scanning microscopy and image analysis for in toto imaging of development. Mech Dev. 2003;120:407–420. doi: 10.1016/j.mod.2003.07.005. [DOI] [PubMed] [Google Scholar]
- Miyata T. Development of three-dimensional architecture of the neuroepithelium: role of pseudostratification and cellular ‘community’. Dev Growth Differ. 2008;50(Suppl 1):S105–112. doi: 10.1111/j.1440-169X.2007.00980.x. [DOI] [PubMed] [Google Scholar]
- Miyata T, Kawaguchi A, Okano H, Ogawa M. Asymmetric inheritance of radial glial fibers by cortical neurons. Neuron. 2001;31:727–741. doi: 10.1016/s0896-6273(01)00420-2. [DOI] [PubMed] [Google Scholar]
- Miyata T, Kawaguchi A, Saito K, Kawano M, Muto T, Ogawa M. Asymmetric production of surface-dividing and non-surface-dividing cortical progenitor cells. Development. 2004;131:3133–3145. doi: 10.1242/dev.01173. [DOI] [PubMed] [Google Scholar]
- Mizutani K, Yoon K, Dang L, Tokunaga A, Gaiano N. Differential Notch signalling distinguishes neural stem cells from intermediate progenitors. Nature. 2007;449:351–355. doi: 10.1038/nature06090. [DOI] [PubMed] [Google Scholar]
- Mueller T, Wullimann MF. BrdU-, neuroD (nrd)- and Hu-studies reveal unusual non-ventricular neurogenesis in the postembryonic zebrafish forebrain. Mech Dev. 2002;117:123–135. doi: 10.1016/s0925-4773(02)00194-6. [DOI] [PubMed] [Google Scholar]
- Murciano A, Zamora J, López-Sánchez J, Frade JM. Interkinetic nuclear movement may provide spatial clues to the regulation of neurogenesis. Mol Cell Neurosci. 2002;21:285–300. doi: 10.1006/mcne.2002.1174. [DOI] [PubMed] [Google Scholar]
- Noctor SC, Flint AC, Weissman TA, Dammerman RS, Kriegstein AR. Neurons derived from radial glial cells establish radial units in neocortex. Nature. 2001;409:714–720. doi: 10.1038/35055553. [DOI] [PubMed] [Google Scholar]
- Noctor SC, Martinez-Cerdeno V, Ivic L, Kriegstein AR. Cortical neurons arise in symmetric and asymmetric division zones and migrate through specific phases. Nat Neurosci. 2004;7:136–144. doi: 10.1038/nn1172. [DOI] [PubMed] [Google Scholar]
- Ossipova O, Ezan J, Sokol SY. PAR-1 phosphorylates Mind bomb to promote vertebrate neurogenesis. Dev Cell. 2009;17:222–233. doi: 10.1016/j.devcel.2009.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park HC, Kim C-H, Bae Y, Yeo S, et al. Analysis of upstream elements in the HuC promoter leads to the establishment of transgenic zebrafish with fluorescent neurons. Dev Biol. 2000;227:279–293. doi: 10.1006/dbio.2000.9898. [DOI] [PubMed] [Google Scholar]
- Petersen PH, Zou K, Hwang JK, Jan YN, Zhong W. Progenitor cell maintenance requires numb and numblike during mouse neurogenesis. Nature. 2002;419:929–934. doi: 10.1038/nature01124. [DOI] [PubMed] [Google Scholar]
- Pierfelice T, Alberi L, Gaiano N. Notch in the vertebrate nervous system: an old dog with new tricks. Neuron. 2011;69:840–855. doi: 10.1016/j.neuron.2011.02.031. [DOI] [PubMed] [Google Scholar]
- Rasin MR, Gazula VR, Breunig JJ, Kwan KY, Johnson MB, Liu-Chen S, Li HS, Jan LY, Jan YN, Rakic P, Sestan N. Numb and Numbl are required for maintenance of cadherin-based adhesion and polarity of neural progenitors. Nat Neurosci. 2007;10:819–827. doi: 10.1038/nn1924. [DOI] [PubMed] [Google Scholar]
- Reugels AM, Boggetti B, Scheer N, Campos-Ortega JA. Asymmetric localization of Numb:EGFP in dividing neuroepithelial cells during neurulation in Danio rerio. Dev Dyn. 2006;235:934–948. doi: 10.1002/dvdy.20699. [DOI] [PubMed] [Google Scholar]
- Rhyu MS, Jan LY, Jan YN. Asymmetric distribution of numb protein during division of the sensory organ precursor cell confers distinct fates to daughter cells. Cell. 1994;76:477–491. doi: 10.1016/0092-8674(94)90112-0. [DOI] [PubMed] [Google Scholar]
- Sauer FC. Mitosis in the neural tube. J Comp Neurol. 1935;62:377–405. [Google Scholar]
- Shitamukai A, Konno D, Matsuzaki F. Oblique radial glial divisions in the developing mouse neocortex induce self-renewing progenitors outside the germinal zone that resemble primate outer subventricular zone progenitors. J Neurosci. 2011;31:3683–3695. doi: 10.1523/JNEUROSCI.4773-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spana EP, Doe CQ. The prospero transcription factor is asymmetrically localized to the cell cortex during neuroblast mitosis in Drosophila. Development. 1995;121:3187–3195. doi: 10.1242/dev.121.10.3187. [DOI] [PubMed] [Google Scholar]
- Spana EP, Doe CQ. Numb antagonizes Notch signaling to specify sibling neuron cell fates. Neuron. 1996;17:21–26. doi: 10.1016/s0896-6273(00)80277-9. [DOI] [PubMed] [Google Scholar]
- Tallafuss A, Trepman A, Eisen JS. DeltaA mRNA and protein distribution in the zebrafish nervous system. Dev Dyn. 2009;238:3226–3236. doi: 10.1002/dvdy.22136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tawk M, Araya C, Lyons DA, Reugels AM, Girdler GC, Bayley PR, Hyde DR, Tada M, Clarke JD. A mirror-symmetric cell division that orchestrates neuroepithelial morphogenesis. Nature. 2007;446:797–800. doi: 10.1038/nature05722. [DOI] [PubMed] [Google Scholar]
- Temple S. Division and differentiation of isolated CNS blast cells in microculture. Nature. 1989;340:471–473. doi: 10.1038/340471a0. [DOI] [PubMed] [Google Scholar]
- Temple S. The development of neural stem cells. Nature. 2001;414:112–117. doi: 10.1038/35102174. [DOI] [PubMed] [Google Scholar]
- Thisse C, Thisse B. High Throughput Expression Analysis of ZF-Models Consortium Clones. ZFIN Direct Data Submission. 2005 ( http://zfin.org)
- von Trotha JW, Campos-Ortega JA, Reugels AM. Apical localization of ASIP/PAR-3:EGFP in zebrafish neuroepithelial cells involves the oligomerization domain CR1, the PDZ domains, and the C-terminal portion of the protein. Dev Dyn. 2006;235:967–977. doi: 10.1002/dvdy.20715. [DOI] [PubMed] [Google Scholar]
- Williams SE, Beronja S, Pasolli HA, Fuchs E. Asymmetric cell divisions promote Notch-dependent epidermal differentiation. Nature. 2011;470:353–358. doi: 10.1038/nature09793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeo SY, Kim M, Kim HS, Huh TL, Chitnis AB. Fluorescent protein expression driven by her4 regulatory elements reveals the spatiotemporal pattern of Notch signaling in the nervous system of zebrafish embryos. Dev Biol. 2007;301:555–567. doi: 10.1016/j.ydbio.2006.10.020. [DOI] [PubMed] [Google Scholar]
- Yong KJ, Yan B. The relevance of symmetric and asymmetric cell divisions to human central nervous system diseases. J Clin Neurosci. 2011;18:458–463. doi: 10.1016/j.jocn.2010.08.023. [DOI] [PubMed] [Google Scholar]
- Yoon K, Gaiano N. Notch signaling in the mammalian central nervous system: insights from mouse mutants. Nat Neurosci. 2005;8:709–715. doi: 10.1038/nn1475. [DOI] [PubMed] [Google Scholar]
- Yoon K, Koo B, Im S, Jeong H, Ghim J, Kwon M, Moon J, Miyata T, Kong Y. Mind bomb 1-expressing intermediate progenitors generate notch signaling to maintain radial glial cells. Neuron. 2008;58:519–531. doi: 10.1016/j.neuron.2008.03.018. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.