Abstract
WNT7A (wingless-type MMTV integration site family, member 7A) is a known tumor suppressor gene of non-small cell lung carcinomas (NSCLC) and is frequently inactivated due to CpG-island hypermethylation in human cancers. The members of WNT family are involved in cell signaling and play crucial roles in cancer development. In the present work hypermethylation of the WNT7A gene was detected in 66% (29/44) of analyzed clear cell renal cell carcinomas (RCCs) using methyl-specific PCR (MSP). Moreover, bisulfite sequencing confirmed intensive hypermethylation of the 5′-CpG island of the WNT7A gene. Methylation analysis revealed positive correlations between tumor stage, Fuhrman nuclear grade and WNT7A hypermethylation. Additionally, restoration of WNT7A gene expression in the A498 cell line by 5-aza-2′-deoxycytidine treatment confirmed a direct contribution of hypermethylation in silencing of the WNT7A gene. High frequency of loss of heterozygosity (LOH) was demonstrated on chromosome 3p25 in regions surrounding the WNT7A gene. The frequent down-regulation of WNT7A gene expression was detected in 88% (15/17) of clear cell RCCs. We have also shown that the WNT7A gene possesses tumor suppression function by colony-formation and cell proliferation assays in RCC cell lines. In summary, the WNT7A gene is inactivated by genetic/epigenetic alterations in clear cell RCC and demonstrates tumor suppressor properties.
Introduction
Renal cell carcinoma (RCC) is the most common type of kidney cancer, responsible for 3% of human malignancies [1]. Clear cell RCC accounts for 70–75% of RCC and is distinguished by a set of genetic and epigenetic abnormalities [2]. It is known that inactivation of tumor suppressor genes is a frequent event for sporadic clear cell RCCs. DNA methylation and deletions are the most common mechanisms of inactivation of tumor suppressor genes in clear cell RCCs [3]–[6]. Moreover, it was shown that abnormalities of human chromosome 3 significantly contributed to clear cell RCCs development. Arai et al. identified chromosome 3 as one of the most affected by genetic/epigenetic alterations in clear cell RCCs [7], [8]. In particular, DNA methylation of promoter regions was shown for RASSF1, FHIT, LRRC3B, VHL and other well-characterized tumor suppressor genes in clear cell RCCs [9]–[12].
In previous work we have found that WNT7A associated locus is subjected to genetic/epigenetic alterations in set of RCC’s using NotI-microarray analysis [13]. The NotI-microarray technology allows to search for genetic (deletion, amplification) and epigenetic (DNA methylation) alterations of genes/loci simultaneously, due to the fact that NotI sites are frequently associated with promoter regions of genes [14]. This technology was used to search for such potential tumor suppressor genes like LRRC3B [15], [16], Fibulin3 [17], RBSP3 [18] and other genes [19].
WNT7A is a known tumor suppressor gene of non-small cell lung carcinomas (NSCLC) [20]–[22] and is frequently inactivated due to CpG-island hypermethylation in such human cancers as lung [19], [23], [24], pancreatic [25] and oral squamous cell carcinomas (OSCC) [26].
The members of the WNT family are involved in cell signaling through canonical [27] (β-catenin dependent) and non-canonical pathways such as Planar Cell Polarity [28] or Wnt/Calcium [29] (β-catenin independent). In the canonical pathway, interaction of WNT proteins with the Frizzle cell membrane receptor results in inhibition of glycogen synthase kinase 3 activity that acts as a negative regulator of β-catenin accumulation. Inhibition of glycogen synthase kinase 3 prevents proteasome-mediated degradation of β-catenin that results in cytoplasmic accumulation of β-catenin with subsequent translocation to the nucleus. The nuclear portion of β-catenin binds to the TCF/LEF family of transcription factors and induces transcription of target genes [30]. Noteworthy, the important role of WNT signaling in the mesenchymal-epithelial transition of metanephric progenitors and in the terminal epithelial differentiation during the kidney development was assumed [31], [32].
At present, the role of the WNT genes in carcinogenesis is rather controversial because several members such as WNT2 were shown to possess oncogenic features [33], while other members such as WNT5A were reported to act as tumor suppressors [34]. The behavior of the WNT7A gene in human cancer is tissue-specific. In lung cancer and leukemias WNT7A was characterized as a tumor suppressor gene [20]–[22], [35]. Additionally, it was shown that inactivation of WNT7A through DNA hypermethylation stabilizes the cancer phenotype of OSCC cell lines [26]. However, the WNT7A gene has oncogenic properties in ovarian cancer [36], [37].
In the present study we determined the genetic and epigenetic alterations of the WNT7A gene in clear cell RCCs. A correlation exists between genetic/epigenetic alterations and down-regulation of WNT7A gene expression. In addition, re-expression of the WNT7A gene in RCC cell lines inhibits colony formation and cell proliferation.
Materials and Methods
Ethics Statements
All patients gave written informed consent. The samples were collected in accordance with the Declaration of Helsinki and approved by the guidelines issued by the Ethic Committee of the Institute of Urology of the Academy of Medical Sciences, Kyiv, Ukraine.
Total RNA and Genomic DNA Isolation
Forty four tumor samples of clear cell RCCs with 32 non-malignant adjacent normal tissues were obtained from the Institute of Urology of the Academy of Medical Sciences, Kyiv, Ukraine (Table 1). The classification of the tumors based on the staging system of the American Joint Committee on Cancer (TNM) was used [38]. Genomic DNA was purified according to the protocol from Sambrook et al [39]. Total RNA was isolated with the RNeasy Mini Kit (Qiagen, Valencia, CA) following the manufacturer’s recommendations. The quality of the isolated RNA was assessed by electrophoresis.
Table 1. Clinical-pathological characteristics of clear cell RCC samples.
| Parameters | Means | ||
| Age (years) | 55 (22–78) | ||
| Sex (M/F) | 27/17 | ||
| Fuhrman nuclear grade | |||
| Grade 1 | 11 | ||
| Grade 2 | 18 | ||
| Grade 3 | 9 | ||
| Grade 4 | 6 | ||
| Tumor stage | |||
| Stage I | (T1N0M0) | 3 | |
| Stage II | (T2N0M0) | 29 | |
| Stage III | (T3N0M0) | 9 | |
| (T3N1M0) | 2 | ||
| Stage IV | (T3N0M2) | 1 | |
Cell Lines Culturing
Human RCC cell lines A498 and KRC/Y were obtained from Bank of cell lines of R. E. Kavetsky Institute of Experimental Pathology, Oncology and Radiobiology, National Academy of Science, (Kyiv, Ukraine) and Karolinska Institute (Stockholm, Sweden), respectively. A498 and KRC/Y cell lines were described earlier in our works [40], [41]. Cell lines A498 and KRC/Y were cultured in RPMI (Sigma-Aldrich, St. Louis, MO, USA) and IMDM media (Life Technology, Carlsbad, CA, USA), respectively. Media were supplemented with 10% fetal bovine serum and penicillin/streptomycin. Transfection of cell lines was performed using Lipofectamine 2000 (Life Technology), according to the manufacturer’s recommendations.
Methyl-specific PCR (MSP)
Bisulfite treatment of genomic DNA was performed with the EZ DNA Methylation kit (ZYMO Research, Orange, CA, USA) according to manufacturer’s protocol. Modified DNA (50 ng) was used for each PCR with primers described previously [25]: WNT7A M-F 5′-GTAGTTCGGCGTCGTTTTAC-3′, WNT7A M-R 5′-CGAAACCGTCTATCGATACG-3′, WNT7A, U-F 5′-TAGTTTGGTGTTGTTTTATGTTG-3′, WNT7A U-R 5′-CCCCAAAACCATCTATCAATAC-3′. PCRs were performed in the following conditions: 95°C - 4 min, then 35 cycles at 95°C - 15 sec, 59–62°C - 20 sec and 72°C - 30 sec, and the final extension at 72°C for 7 min. The PCR products were analyzed by electrophoresis in 10% polyacrylamide gels with subsequent ethidium bromide staining. M.SssI (NEB, Ipswich, MA, USA) methyltransferase-treated and untreated normal DNA was used as positive control in amplification with primers against methylated and unmethylated sequences, correspondingly. To verify the accuracy of the MSP, the PCR products were recovered from agarose gels, cloned in the pJET1.2-vector (Thermo Scientific, Fermentas) and sequenced. Four to five clones were sequenced for each sample.
Bisulfite Sequencing
Primers for bisulfite sequencing of the CpG-island of the WNT7A gene were designed around the MSP primers in the region +8 bp to +356 bp from the transcription start site (NC_000003.11, from 13921263 bp to 13921611 bp): WNT7A-BS For 5′-GGGGGTTGGAGGTAGTAG-3′ and WNT7A-BS Rev 5′-TTGTTTGGGTTATTTTTTTTTTAGTTTGGGT-3′. The PCR was carried out using 100 ng of bisulfite-treated DNA and the 1xSYBR Green Mix (Thermo Scientific, Fermentas) in the following conditions: 95°C - 10 min, then 40 cycles at 95°C - 15 sec, 58°C - 20 sec and 72°C - 60 sec, and the final extension at 72°C for 7 min. The PCR products were recovered from agarose gels, cloned in the pGEM-T easy vector (Promega, Madison, WI, USA) and sequenced. Around 8 clones were sequenced for each sample.
Quantitative Reverse Transcriptase PCR (qRT-PCR)
qRT-PCR was used to assess the change of WNT7A gene expression in tissue samples and RCC cell lines. Briefly, 2 µg of total RNA were treated with DNAse (Thermo Scientific, Fermentas) and transcribed into cDNA with an Oligo(dT)-primer using the First Strand cDNA Synthesis Kit (Thermo Scientific, Fermentas). qRT-PCR was performed at IQ5 real-time PCR detection system (BioRad, Hercules, CA, USA) at the following reaction conditions: 95°C - 10 min, than 40 cycles at 95°C - 15 sec, 62°C - 20 sec and 72°C - 30 sec. The TBP gene served as a reference gene. qRT-PCR was carried out with the previously reported primers [42]. Changes in WNT7A gene expression were calculated by the ΔΔCt method using the efficiency coefficient calculated according to Spiess et al [43].
Loss of Heterozygosity (LOH) Analysis
Detection of LOH of the microsatellite markers D3S2385, D3S1252, D3S2403 was carried out by amplification of the genomic DNA with Cy5-labeled primers and subsequent analysis by automated laser fluorescence system (Pharmacia Biotech, Uppsala, Sweden) [44]–[46]. The amplification reaction was performed using 1 U of DreamTaq (Thermo Scientific, Fermentas) in the following reaction conditions: 95°C - 4 min, 28 cycles at 95°C - 15 sec, 56–58°C - 20 sec, 72°C - 30 sec, and finally 72°C for 7 min. The fluorescence data were processed by Fragment Manager program (Pharmacia). Differences in peak intensity of alleles was calculated by two methods using the height and area of peaks [46], [47]. Decreased ratio of tumor allele intensity compared with normal and ratio less than 70% was accepted as a criterion for presence of LOH simultaneously for both methods of calculation of peak intensity [48]. Primer sequences were taken from the NCBI UniSTS database with the following accession numbers: D3S2385 - G08224, D3S1252 - L02085, D3S2403 - G08301.
Restoration of WNT7A Gene Expression by 5-aza-2′-deoxycytidine Treatment in the A498 Renal Cell Carcinoma Cell Line
For this purpose the A498 cells were treated with 5 µM 5-aza-2′-deoxycytidine (Sigma-Aldrich) for 5 days. A498 cells treated by solvent for 5-aza-2′-deoxycytidine was used as mock control. The medium was replaced daily. After the treatment, total RNA and genomic DNA were isolated. To assess the effect of drug treatment of the A498 cells on the expression and methylation status of the WNT7A gene, qRT-PCR and MSP were used as mentioned above. MSP was carried out with the equal amount of bisulfite treated DNA obtained from 5-aza-2′-deoxycytidine and mock treated A498 cells. To detect expression of WNT7A and TBP genes, qRT-PCR was carried out for 30 and 24 cycles respectively. Level of the TBP expression was used as an internal control.
Colony Formation and Cell Proliferation Tests
For colony formation tests, A498 and KRC/Y cells were transfected with pcDNA3.1-WNT7A and pcDNA3.1-empty vectors. The level of WNT7A expression in cell lines after transfection by pcDNA3.1-WNT7A and pcDNA3.1-empty vectors was assessed by qRT-PCR as mentioned above. Cells (40,000-50,000 cells per well) were seeded in 6-well plates the day following transfection in triplicates. Selection on the 400µg/mL of G418 (Sigma-Aldrich) was started 48 h after transfection. Cells were stained by crystal violet after 2 weeks of G418 selection and number of colonies was counted. The experiment was performed in triplicate.
To perform cell proliferation tests, 1000–1500 cells per well were seeded in 96-well plates 24 h after transfection. The number of cells was counted using the Cell Quantification kit (CCK-8) (Sigma-Aldrich) at 0 h, 24 h, 48 h, 72 h and 96 h after plating according to the manufacturer’s recommendations. During cell proliferation tests cells were grown in medium without G418.
Statistical Analysis
Statistical analysis was performed using STATISTICA 7.0 program (StatSoft Inc, Tulsa, OK, USA). The expression level for different stages of tumors was compared with the t-test for independent groups. The value p<0.05 was considered as a statistically significant difference. The nonparametric Mann-Whitney U Test was used to calculate difference between samples with the methylation or LOH status and the clinical-pathological characteristics. The difference was considered as significant if p<0.05. The Spearman’s rank correlation coefficient was used to calculate correlation between decrease of the gene expression and the hypermethylation/LOH status. The value rs which corresponds to the value p<0.05 was considered as a statistically significant correlation.
Results
Determination of WNT7A Methylation Status in Clear Cell RCC and Restoration of WNT7A Gene Expression after 5-aza-2′-deoxycytidine Treatment
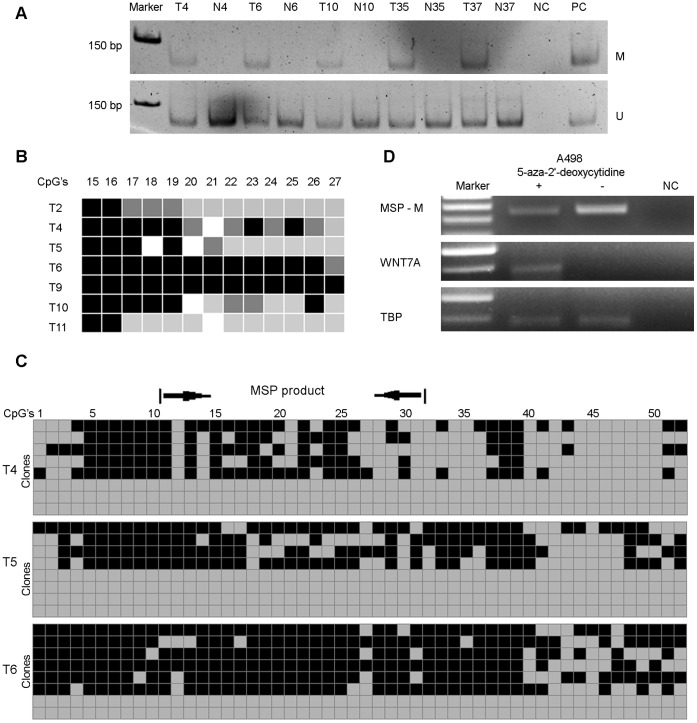
To examine the presence of epigenetic alterations of the WNT7A gene in clear cell RCCs, the methylation status of the 5′-CpG island of the WNT7A gene was first assessed by MSP. The methylation status of the WNT7A gene promoter was examined in 44 clear cell RCCs and 28 adjacent non-malignant renal tissues (See Table S1 in the supplemental material). The PCR products with specific primers for methylated WNT7A were detected in 66% (29/44) of the clear cell RCCs analyzed. No DNA methylation was detected in the non-malignant adjacent renal tissues. The PCR products with specific primers for unmethylated WNT7A were detected in all samples analyzed. To check the specificity of the MSP, the PCR products of 7 tumor samples with an identified hypermethylated WNT7A gene were sequenced. Data of sequencing confirmed the results of MSPs. Representative MSPs and sequencing of MSP-products are presented in Figure 1A and 1B.
Figure 1. Study of WNT7A gene methylation status in clear cell RCC.
(A). Representative MSP analysis of the WNT7A gene by using methylated (M) and unmethylated (U) specific primers, PC – positive control, M.Sssi treated or untreated normal DNA, NC – negative control (H20), T4, T6, T10, T35, T37: tumor samples, N4, N6, N10, N35, N37: normal samples. (B). Sequencing of MSP products; white squares - 0–19% methylation at the CpG dinucleotide, grey squares - 20–59% methylation at the CpG dinucleotide, dark grey squares - 60–79% methylation at the CpG dinucleotide, black squares - 80–100% methylation at the CpG dinucleotide, T2, T4, T5, T6, T9, T10, T11: tumor samples. (C). Methylation status of the fifty two CpG dinucleotides of the WNT7A 5′-CpG island in tumor samples with a methylated WNT7A gene, where each CpG dinucleotide is shown by either a black square when methylated or a grey square when unmethylated; arrows indicate position of MSP primers, T4, T5, T6 are tumor samples. (D). Restoration of WNT7A expression by 5-aza-2′-deoxycytidine treatment of the A498 cell line, MSP-M - methylation analysis of the WNT7A gene by using methylated specific primers, NC – negative control (H20).
Secondly to verify that MSP determines methylation status of WNT7A 5′-CpG-island correctly, bisulfite sequencing was performed for 3 tumor samples that had revealed methylated WNT7A 5′-CpG island according to the MSP data. Bisulfite sequencing showed that MSP accurately reflects the methylation status of the WNT7A 5′-CpG island in the samples selected (Figure 1C).
To further assess whether hypermethylation of the WNT7A 5′-CpG island might be directly responsible for WNT7A silencing, the A498 cell line was treated with the DNA methyltransferase inhibitor 5-aza-2′-deoxycytidine. As expected this led to decreased WNT7A methylation and restored WNT7A expression (Figure 1D).
Hypermethylation of the WNT7A gene is significantly higher in tumors at advanced stages (III–IV) than in tumors at early stages (I–II) (p = 0.003). The methylation status of the WNT7A gene showed a correlation with the Fuhrman nuclear grade of clear cell RCC: grades (1–2) vs grades (3–4) (p = 0.037). Moreover, WNT7A methylation was observed more frequently in patients, older than 50 years (p = 0.012) than in younger patients (Table 2). No correlation was found between the status of WNT7A methylation and gender.
Table 2. Association of clinical-pathological characteristics and hypermethylation/LOH status of the WNT7A gene in clear cell RCCs.
| Parameters | Methylated | p*-value | LOH | |
| Age | <50 | 38% (5/13) | 0.012 | 86% (6/7) |
| >50 | 77% (24/31) | 85% (17/20) | ||
| Fuhrman nuclear grades 1–2 | 55%(16/29) | 0.037 | 94% (16/17) | |
| Fuhrman nuclear grades 3–4 | 87%(13/15) | 70% (7/10) | ||
| Stages I–II | 53%(17/32) | 0.003 | 89% (16/18) | |
| Stages III–IV | 100%(12/12) | 78% (7/9) | ||
p-value is referred to correlation between clinical-pathological characteristics and hypermethylation status.
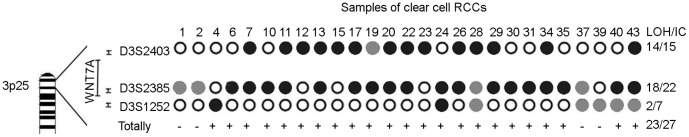
LOH Analysis of Polymorphic Markers D3S2385, D3S2403 and D3S1252 in Clear Cell RCC
A copy number study of the chromosome 3p25 region surrounding the WNT7A gene was performed using the microsatellite marker analysis. Detection of LOH was performed in 28 samples of clear cell RCCs and the adjacent non-malignant tissues for the polymorphic repeats D3S2403, D3S2385 and D3S1252. The frequency of loss for the above-mentioned markers was 93% (14/15), 82% (18/22) and 29% (2/7) of informative cases, respectively. Among the analyzed samples, 27 cases were informative for at least one LOH marker. Overall, we detected 23 (85%) samples that contained at least one LOH and 4 samples without LOH. In 12 samples one LOH was detected, and in 11 samples - two LOH simultaneously. Data from LOH assays are presented in Figure 2 and Table S1.
Figure 2. The LOH assays: status of the informative cases of clear cell RCC for the 3q25 region surrounding the WNT7A gene.
D3S2403, D3S2385, D3S1252– microsatellite markers, white circles - homozygotes (non informative cases), grey circles – absence of the LOH, black circles – presence of the LOH, IC - informative cases, “+” - LOH positive sample, “−” - LOH negative sample.
Expression of the WNT7A Gene in Clear Cell RCC
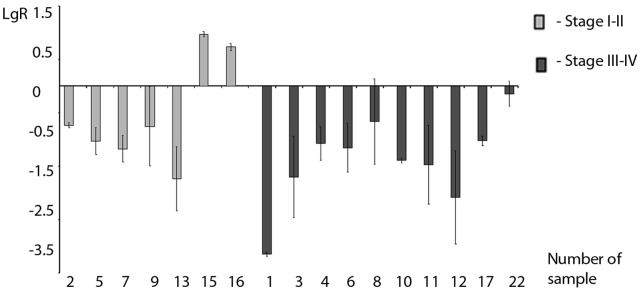
We examined whether the expression of the WNT7A gene was correlated with DNA methylation/presence of LOH in clear cell RCCs. For this purpose, the expression of the WNT7A gene in 17 clear cell renal cell carcinomas was determined by qRT-PCR. A decrease of gene expression was detected in 88% of clear cell RCCs (15/17 samples) (Figure 3 and Table S1). A correlation was detected between the decrease of WNT7A gene expression and hypermethylation of the WNT7A gene or the presence of LOH (rs = 0.917, p<0.05). The mean values of expression in the form of the logarithmic ratio of tumor/normal tissue for samples with stages I–II (−0.53+/−1.0) and stages III–IV (−1.54+/−0.8) tend to be different (p = 0.09).
Figure 3. Panel of WNT7A gene expression in clear cell RCC samples.
R – values of expression in the form of the logarithmic ratio of tumor/normal tissue of the WNT7A gene relatively to the TBP gene.
Colony Formation and Proliferation Tests
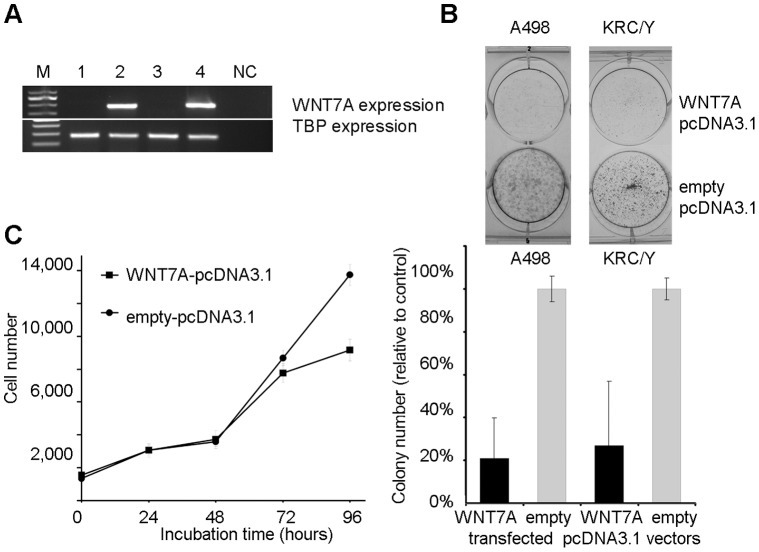
The effect of WNT7A re-expression on colony formation of the A498 and KRC/Y cell lines was investigated. A high level of the WNT7A mRNA was detected in the A498 and KRC/Y cells after transfection with the WNT7A-pcDNA3.1 vector in comparison with the empty vector (Figure 4A). The ectopic expression of the WNT7A gene in the A498 and KRC/Y cell lines led to a significant reduction in colony number (p<0.05). The number of A498 WNT7A-pcDNA3.1 and KRC/Y WNT7A-pcDNA3.1 colonies was 20.8% and 26.8% from the number of A498 Empty-pcDNA3.1 and KRC/Y Empty-pcDNA3.1 colonies respectively (Figure 4B).
Figure 4. Suppressive effect of WNT7A gene re-expression in RCC cell lines.
Effect of WNT7A gene re-expression (A) on colony formation (B) for the A498, KRC/Y cell lines, and (C) cell proliferation assays for the A498 cell line; M – marker, 1 and 2– A498 cells were transfected by empty-pcDNA3.1 and WNT7A-pcDNA3.1 vectors, 3 and 4– KRC/Y cells were transfected by empty-pcDNA3.1 and WNT7A-pcDNA3.1 vectors, NC – negative control (H20). All experiments were performed in triplicate. Representative results are shown.
To further investigate the effect of WNT7A on cell proliferation and survival we performed the cell proliferation test for the A498 cell line. There was a significant negative effect of WNT7A expression on cell growth in comparison to cells transfected with an empty vector (p<0.05) (Figure 4C).
Discussion
A large number of tumor suppressor genes are inactivated through DNA hypermethylation of the promoter regions in a wide range of cancers [49]–[51]. Moreover studying of genetic and epigenetic alterations is a powerful tool in searching for novel tumor suppressor genes [52], [53].
To perform a detailed analysis of rearrangements of the WNT7A gene in clear cell RCC, the methylation status of the 5′-CpG island of the WNT7A gene and the presence of deletions in the locus that corresponds to the WNT7A gene were studied. MSP indeed revealed the hypermethylation (66%) of the WNT7A gene promoter in clear cell RCC. In comparison, the WNT7A gene was hypermethylated in pancreatic carcinomas (71%) [25] and OSCC (78%) [26]. Additionally, it was shown that WNT7A is higher methylated in NSCLC tissue compared to matched normal lung tissues [19], [23], [24]. Thus promoter hypermethylation acts as the main mechanism of the WNT7A silencing in a wide range of cancer types.
To investigate the genetic alterations of the WNT7A gene locus, the microsatellite markers analysis of this region on chromosome 3p25 was also performed. The WNT7A gene is located between markers D3S2385, D3S2403 and D3S1252. A loss of heterozygosity at least with one marker was found in 85% (23/27) of informative cases. Moreover, for the first time we have shown a loss of the microsatellite markers D3S2385 and D3S2403 in cancer. It should be recalled that the D3S1252 marker was lost in 14% of informative cases of head and neck carcinomas [54].
To verify the correlation between hypermethylation/LOH presence and gene expression, the level of the WNT7A gene expression was investigated. Hypermethylation of 5′-CpG island of the WNT7A gene/LOH presence coincided with decreased expression of the gene in 88% (15/17) of selected clear cell RCC samples. The methylation/LOH status and expression of WNT7A gene have been studied on the same set of samples. Also, we observed that WNT7A expression was restored after 5-aza-2′-deoxycytidine treatment of the RCC cell line. In addition, it was reported that expression of the WNT7A gene is frequently reduced in lung cancer [55], and that restoration of WNT7A gene expression led to growth inhibition of NSCLC cell lines [22]. Importantly, we have found that decreased WNT7A expression positively correlates with tumor progression.
A statistically significant correlation exists between the WNT7A hypermethylation status and some of the clinical-pathological characteristics. The WNT7A gene is more frequently methylated in tumors at advanced stages (III–IV) and high nuclear grades (3–4) than in tumors at early stages (I–II) and low nuclear grades (1–2) of clear cell RCC (Table 2). Similar data were demonstrated in OSCC where methylation of the WNT7A gene is characteristic of tumors at advanced stages [26]. At the same time, we did not detect any statistically significant difference of frequency of microsatellite marker loss and any clinical-pathological characteristics.
Based on our data we assume that the WNT7A gene could be a potential tumor suppressor gene of clear cell RCC. To support this possibility the tumor suppressor properties of the WNT7A gene in RCC cell lines were investigated. For this purpose, the WNT7A gene was re-expressed in RCC cell lines A498 and KRC/Y. This led to a significant reduction in colony number in both cell lines. These findings are similar to data obtained previously concerning re-expression of WNT7A in NSCLC [21], [22]. In addition, re-expression of WNT7A significantly reduced the proliferation rate of the A498 cell line. Thus, the WNT7A gene does indeed possess tumor suppressor properties in RCCs.
In summary, genetic and epigenetic alterations play a key role in silencing of the WNT7A gene in clear cell RCC. Moreover, restoration of WNT7A expression inhibits the growth of RCC cell lines. Therefore, we propose that inactivation of the WNT7A gene may play an important role in the development of clear cell RCC.
Supporting Information
Clinical-pathological characteristics and methylation, LOH, expression status of the WNT7A gene in clear cell RCC samples.
(DOC)
Acknowledgments
We thank Dr. S.A. Kravchenko for technical support with automated laser fluorescence system. We thank Dr. Yu Kudryavets (R. E. Kavetsky Institute of Experimental Pathology, Oncology and Radiobiology, National Academy of Science, Kyiv, Ukraine) for kindly providing us the A498 cell line. We thank Dr. Anne-Lise Haenni for critical reading of this manuscript.
Funding Statement
This work was supported by the State Fond of Fundamental Research (grant No F46/457-2011). E.R. Zabarovsky was supported by research grants from the Swedish Cancer Society, the Swedish Institute and the Swedish Research Council. A. Kondratov was partially supported by a travel fellowship from EACR for research in the MTC department of Karolinska Institute (Sweden, Stockholm). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Jemal A, Siegel R, Xu J,Ward E (2010) Cancer statistics 2010. Cancer J Clin : 277–300. [DOI] [PubMed]
- 2. Arai E, Kanai Y (2011) Genetic and epigenetic alterations during renal carcinogenesis. Int J Clin Exp Pathol 4: 58–73. [PMC free article] [PubMed] [Google Scholar]
- 3.McRonald FE, Morris MR, Gentle D, Winchester L, Baban D, et al. (2009) CpG methylation profiling in VHL related and VHL unrelated renal cell carcinoma. Mol Cancer. 8:31 Available: http://www.molecular-cancer.com/content/8/1/31. Accessed June 3 2009. [DOI] [PMC free article] [PubMed]
- 4. Morris MR, Ricketts C, Gentle D, Abdulrahman M, Clarke N, et al. (2010) Identification of candidate tumour suppressor genes frequently methylated in renal cell carcinoma. Oncogene 29(14): 2104–2117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Glavac D, Ravnik-Glavac M, Ovcak Z, Masera A (1996) Genetic changes in the origin and development of renal cell carcinoma (RCC). Pflugers Arch. 431: 193–4. [DOI] [PubMed] [Google Scholar]
- 6. Sanz-Casla MT, Maestro ML, del Barco V, Zanna I, Moreno J, et al. (2003 ) Loss of heterozygosity and methylation of p16 in renal cell carcinoma. Urol Res 31: 159–162. [DOI] [PubMed] [Google Scholar]
- 7. Arai E, Ushijima S, Tsuda H, Fujimoto H, Hosoda F, et al. (2008) Genetic Clustering of Clear Cell Renal Cell Carcinoma Based on -Comparative Genomic Hybridization: Its Association with DNAMethylationAlteration and Patient Outcome. Clin Cancer Res 14: 5531–5539. [DOI] [PubMed] [Google Scholar]
- 8.Arai E, Chiku S, Mori T, Gotoh M, Nakagawa T, et al. (2012) Single-CpG-resolution methylome analysis identifies clinicopathologically aggressive CpG islandmethylator phenotype clear cell renal cell carcinomas. Carcinogenesis. 2012 May 18. doi: 10.1093/carcin/bgs177. [DOI] [PMC free article] [PubMed]
- 9. Morrissey C, Martinez A, Zatyka M, Agathanggelou A, Honorio S, et al. (2001) Epigenetic inactivation of the RASSF1A 3p21.3 tumor suppressor gene in both clear cell and papillary renal cell carcinoma. Cancer Res 61: 7277–7281. [PubMed] [Google Scholar]
- 10. Kvasha S, Gordiyuk V, Kondratov A, Ugryn D, Zgonnyk Y, et al. (2008) Hypermethylation of the 5′CpG island of the FHIT gene in clear cell renal carcinomas. Cancer Lett 265: 250–257. [DOI] [PubMed] [Google Scholar]
- 11. Kondratov AG, Stoliar LA, Kvasha SM, Gordiyuk VV, Zgonnyk YM, et al. (2012) Methylation pattern of the putative tumor-suppressor gene LRRC3B promoter in clear cell renal cell carcinomas. Mol Med Report 5(2): 509–512. [DOI] [PubMed] [Google Scholar]
- 12. Clifford SC, Prowse AH, Affara NA, Buys CH, Maher ER (1998) Inactivation of the von Hippel-Lindau (VHL) tumour suppressor gene and allelic losses at chromosome arm 3p in primary renal cell carcinoma: evidence for a VHL-independent pathway in clear cell renal tumourigenesis. Genes Chromosomes Cancer 22(3): 200–209. [DOI] [PubMed] [Google Scholar]
- 13.Skrypkina IYa, Kashuba VI, Gordiyuk VV, Saraev V, Zubko Yu, et al. (2006) Identification of changes in gene loci potentially associated with renal cancer by novel technique of NotI microarrays. Reports of NAS Ukraine: 188–192. [PubMed]
- 14. Li J, Protopopov A, Wang F, Sentchenko V, Petushkov V, et al. (2002) NotI subtraction and NotI-specific microarrays to detect copy number and methylation changes in whole genomes. Proc. Natl. Acad. Sci. USA 99: 10724–10729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Dunwell TL, Hesson LB, Pavlova T, Zabarovska V, Kashuba V, et al. (2009) Epigenetic analysis of childhood acute lymphoblastic leukemia. Epigenetics. 4(3): 185–193. [DOI] [PubMed] [Google Scholar]
- 16. Haraldson K, Kashuba VI, Dmitriev AA, Senchenko VN, Kudryavtseva AV, et al. (2012) LRRC3B gene is frequently epigenetically inactivated in several epithelial malignancies and inhibit cell growth and replication. Biochimie 94(5): 1151–1157. [DOI] [PubMed] [Google Scholar]
- 17. Law EW, Cheung AK, Kashuba VI, Pavlova TV, Zabarovsky ER, et al. (2012) Anti-angiogenic and tumor-suppressive roles of candidate tumor-suppressor gene, Fibulin-2, in nasopharyngeal carcinoma. Oncogene 31(6): 728–738. [DOI] [PubMed] [Google Scholar]
- 18.Senchenko VN, Anedchenko EA, Kondratieva TT, Krasnov GS, Dmitriev AA, et al. (2010) Simultaneous down-regulation of tumor suppressor genes RBSP3/CTDSPL, NPRL2/G21 and RASSF1A in primary non-small cell lung cancer. BMC Cancer 10:75. Available: http://www.biomedcentral.com/1471-2407/10/75. Accessed March 1 2010. [DOI] [PMC free article] [PubMed]
- 19. Dmitriev AA, Kashuba VI, Haraldson K, Senchenko VN, Pavlova TV, et al. (2012) Genetic and epigenetic analysis of non-small cell lung cancer with NotI-microarrays. Epigenetics 7(5): 502–13. [DOI] [PubMed] [Google Scholar]
- 20. Ohira T, Gemmill RM, Ferguson K, Kusy S, Roche J, et al. (2003) WNT7a induces E-cadherin in lung cancer cells. Proc Natl Acad Sci USA 100(18): 10429–10434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Winn RA, Marek L, Han SY, Rodriguez K, Rodriguez N, et al. (2005) Restoration of Wnt-7a expression reverses non-small cell lung cancer cellular transformation through frizzled-9-mediated growth inhibition and promotion of cell differentiation. J Biol Chem. 2005 280(20): 19625–19634. [DOI] [PubMed] [Google Scholar]
- 22. Winn RA, Van Scoyk M, Hammond M, Rodriguez K, Crossno JT Jr, et al. (2006) Antitumorigenic effect of Wnt 7a and Fzd 9 in non-small cell lung cancer cells is mediated through ERK-5-dependent activation of peroxisome proliferator-activated receptor gamma. J Biol Chem. 281(37): 26943–26950. [DOI] [PubMed] [Google Scholar]
- 23.Tennis MA, Vanscouyk MM, Wilson LA, Kelley N, Winn RA (2012) Methylation of WNT7a is modulated by DNMT1 and cigarette smoke condensate in non-small cell lung cancer. Plos One 7(3); e32921. doi:10.1371/journal.pone.0032921. [DOI] [PMC free article] [PubMed]
- 24. Tennis MA, Vanscoyk M, Freeman S, Winn RA (2012) Promoter hypermethylation leads to loss of wnt7a in non-small cell lung cancer. Proc Am Thorac Soc 9 (2): 83–4. [Google Scholar]
- 25. Sato N, Fukushima N, Maitra A, Matsubayashi H, Yeo CJ, et al. (2003) Discovery of novel targets for aberrant methylation in pancreatic carcinoma using high-throughput microarrays. Cancer Res 63(13): 3735–3742. [PubMed] [Google Scholar]
- 26. Kurasawa Y, Kozaki K, Pimkhaokham A, Muramatsu T, Ono H, et al. (2011) Stabilization of phenotypic plasticity through mesenchymal-specific DNA hypermethylation in cancer cells. Oncogene 31(15): 1963–1974. [DOI] [PubMed] [Google Scholar]
- 27. Buechling T, Boutros M (2011) WNT signaling at and above the receptor level. Curr Top Dev Biol 97: 21–53. [DOI] [PubMed] [Google Scholar]
- 28. Katoh M (2005) WNT/PCP signaling pathway and human cancer. Oncol Rep 14(6): 1583–1588. [PubMed] [Google Scholar]
- 29. De A (2011) WNT/Ca2+ signaling pathway: a brief overview. Acta Biochim Biophys Sin 43(10): 745–756. [DOI] [PubMed] [Google Scholar]
- 30. Nelson WJ, Nusse R (2004) Convergence of Wnt, beta-catenin, and cadherin pathways. Science 303(5663): 1483–1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Schmidt-Ott, Kai M. Jonathan Barasch (2008) WNT/β-catenin signaling in nephron progenitors and their epithelial progeny. Kidney Int 74(8): 1004–1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kispert A, Vainio S, McMahon AP (1998) Wnt-4 is a mesenchymal signal for epithelial transformation of metanephric mesenchyme in the developing kidney. Development 125: 4225–4234. [DOI] [PubMed] [Google Scholar]
- 33. Katoh M (2003) WNT2 and human gastrointestinal cancer. Int J Mol Med. 12(5): 811–816. [PubMed] [Google Scholar]
- 34.Liang H, Chen Q, Coles AH, Anderson SJ, Pihan G, et al. (2003) Wnt5a inhibits B cell and functions as a tumor suppressor in hematopoietic tissue. Cell : 349–360. [DOI] [PubMed]
- 35.Ochoa-Hernandez AB, Ramos-Solano M, Meza-Canales ID, Garcia-Castro B, Rosales-Reynoso MA, et al. (2012) Peripheral T-lymphocytes express WNT7A and its restoration in leukemia-derived lymphoblasts inhibits cell proliferation. BMC Cancer 12: 60. Available: http://www.biomedcentral.com/1471-2407/12/60. Accessed 2012 February 7. [DOI] [PMC free article] [PubMed]
- 36. Yoshioka S, King ML, Ran S, Okuda H, Maclean JA 2nd, et al (2012) WNT7A Regulates Tumor Growth and Progression in Ovarian Cancer through the WNT/β-Catenin Pathway. Mol Cancer Res 10(3): 469–482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Merritt MA, Parsons PG, Newton TR, Martyn AC, Webb PM, et al. (2009) Expression profiling identifies genes involved in neoplastic transformation of serous ovarian cancer. BMC Cancer 9:378. Available: http://www.biomedcentral.com/1471-2407/9/378. Accessed 2009 October 23 [DOI] [PMC free article] [PubMed]
- 38.Psutka SP, Eisner BH (2011) Nonradiological treatment for renal tumors. In: Mueller P, Adam A, ed(s). Interventional oncology; A practical guide for the interventional radiologist. Springer. 122 p.
- 39.Sambrook J, Fritsch EF, Maniatis T (1989) Molecular cloning: a laboratory manual. New York: Cold Spring Harbor. 181 p.
- 40. Alimov A, Kost-Alimova M, Liu J, Li C, Bergerheim U, et al. (2000) Combined LOH/CGH analysis proves the existence of interstitial 3p deletions in renal cell carcinoma. Oncogene 19(11): 1392–1389. [DOI] [PubMed] [Google Scholar]
- 41. Wang F, Grigorieva EV, Li J, Senchenko VN, Pavlova TV, et al. (2008) HYAL1 and HYAL2 Inhibit Tumour Growth In Vivo but Not In Vitro. PLoS One. Aug 22 3(8): e3031 doi:10.1371/journal.pone.0003031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tsutomu N, Misao NI, Toshie T (2007) Effects of sulfated hyaluronan on keratinocyte differentiation and Wnt Notch gene expression. Biomaterials :844–850. [DOI] [PubMed]
- 43.Spiess AN, Feig C,Ritz C (2008) accurate sigmoidal fitting of real-time PCR data by introducing a parameter for asymmetry.BMC Bioinformatics : 221–233. [DOI] [PMC free article] [PubMed]
- 44. Chenz X, Bonnefoi H, Diebold-Berger S, Lyautey J, Lederrey C, et al. (1999) Detecting Tumor-related Alterations in Plasma or Serum DNA of Patients Diagnosed with Breast Cancer. Clinical Cancer Research 5: 2297–2303. [PubMed] [Google Scholar]
- 45. Elo JP, Harkonen P, Kyllonen AP, Lukkarinen O, Poutanen M, et al. (1997) Loss of Heterozygosity at 16q24.1-q24.2. Significantly Associated with Metastatic and Aggressive Behavior of Prostate Cancer. Cancer Research 57: 3356–3359. [PubMed] [Google Scholar]
- 46. Ramburan A, Chetty R, Hadley GP, NaidooR, Govender D (2004) Microsatellite analysis of the DCC gene in nephroblastomas: pathologic correlations and prognostic implications. Modern Pathology 17: 89–95. [DOI] [PubMed] [Google Scholar]
- 47. Goessl C, Heicappell R, Munker R, Anker P, Stroun M, et al. (1998) Microsatellite analysis of plasma DNA from patients with clear cell renal carcinoma. Cancer Res 58(20): 4728–4732. [PubMed] [Google Scholar]
- 48. Corcoran MM, Rasool O, Liu Y, Iyengar A, Grander D, et al. (1998) Detailed molecular delineation of 13q14.3 loss in B-cell chronic lymphocytic leukemia. Blood 91(4): 1382–1390. [PubMed] [Google Scholar]
- 49. Sebova K, Zmetakova I, Bella V, Kajo K, Stankovicova I, et al. (2011) RASSF1A and CDH1 hypermethylation as potential epimarkers in breast cancer. Cancer Biomark. 10(1): 13–26. [DOI] [PubMed] [Google Scholar]
- 50. Onay H, Pehlivan S, Koyuncuoglu M, Kirkali Z, Ozkinay F (2009) Multigene methylation analysis of conventional renal cell carcinoma. Urol Int 83(1): 107–12. [DOI] [PubMed] [Google Scholar]
- 51. Hesson LB, Cooper WN, Latif F (2007) Evaluation of the 3p21.3 tumour-suppressor gene cluster. Oncogene 26(52): 7283–301. [DOI] [PubMed] [Google Scholar]
- 52. Morris MR, Ricketts CJ, Gentle D, McRonald F, Carli N, et al. (2011) Genome-wide methylation analysis identifies epigenetically inactivated tumour suppressor genes in renal cell carcinoma. Oncogene 30(12): 1390–401. [DOI] [PubMed] [Google Scholar]
- 53. Zabarovsky ER, Lerman MI, Minna JD (2002) Tumor suppressor genes on chromosome 3p involved in the pathogenesis of lung and other cancers. 21(45): 6915–6935. [DOI] [PubMed] [Google Scholar]
- 54. Rowley H, Jones A, Spandidos D, Field J (1996) Definition of a tumor suppressor gene locus on the short arm of chromosome 3 in squamous cell carcinoma of the head and neck by means of microsatellite markers. Arch Otolaryngol Head Neck Surg. 122(5): 497–501. [DOI] [PubMed] [Google Scholar]
- 55. Calvo R, West J, Franklin W, Erickson P, Bemis L, et al. (2000) Altered HOX and WNT7A expression in human lung cancer. Proc Natl Acad Sci USA. 97(23): 12776–12781. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Clinical-pathological characteristics and methylation, LOH, expression status of the WNT7A gene in clear cell RCC samples.
(DOC)