Abstract
Measles remains a severe global health threat, and nearly 30 million new cases are reported annually. Although many studies have analyzed measles viruses (MV) at the epidemiologic and phylogenetic levels, no study has yet to integrate these two types of data. To this end, we isolated 16 wild-type MV strains China's Jilin province. The MV genotype H1 was the most prevalent strain. After sequencing the nucleoprotein (N) genes of these strains, a maximum clade credibility tree was constructed by the Bayesian Markov Chain Monte Carlo method using 450 MV strains from GenBank with epidemiological information. The MV N gene evolution rate was 1.127E-3. Analysis of the time of the most recent common ancestor (TMRCA) for genotypes A/B/C/G/H revealed that genotypes D and B had the largest and smallest TMRCA (45.86 and 26.63, respectively). The highest level of genetic diversity for the MV N gene occurred around the year 2000. Here in this study, we uncovered the MV genotypes circulating in China's Jilin Province and estimated the epidemiologic and phylogenetic relationship for the six different genotypes of MV.
Introduction
Measles virus (MV) is a negative-strand RNA paramyxovirus, and causes an acute infectious disease and a chronic neurological disorder, subacute sclerosing panencephalitis (SSPE) [1], [2]. Despite the development of vaccines against MV and widespread vaccination campaigns, measles remains one of the most contagious diseases and a leading cause of child death worldwide [3]. More than 30 million new cases are reported annually, the majority of which are in children. A staggering number of measles-related deaths (up to 95%) occur in developing countries [4], due to their limited healthcare resources China is no exception to this rule, despite its recent advances in economic standing. In Jilin Province, home to over 27 million individuals, measles remains the most deadly of all childhood rash/fever illnesses [5]. In 2009, the incidence of measles was still remarkably high, about 10 incidences/100,000 population [5].
The World Health Organization (WHO) has recognized eight clades of MV (designated A–H) that encompass 23 genotypes [6]. Some of the genotypes in these clades represent the prominent sporadic and outbreak-associated infections that have occurred across the globe over the past four decades: A, sporadic infections [7]; B, Africa-related genotypes [8]; D, genotypes implicated in outbreaks in the United States, Pakistan, India [9], and Japan [10]; and H1, the China-related genotype [11]. Thus far, the G genotype appears to be inactive [12]. At this point, a study to determine the MV genotype distribution of currently circulating MV strains will provide crucial insights into the epidemiologic and phylogenetic features of the disease in Jilin Province, thereby providing necessary information to create more effective vaccination strategies and transmission prevention.
The MV genome-encoded nucleoprotein (N) is critically involved in viral genome transcription and replication, frequently used for genotyping and phylogenetic analysis [13]. However, no study to date has reported on the relationship between the genetic characteristics of the various MV genotypes and their epidemiologic behavior. Such data would help to identify the source of virus for a particular region or population [14]. While next-generation sequencing technologies (such as Illumina's Solexa, 454's FLX, or Applied Biosystem's SOLiD) allow for rapid and in-depth genomic analysis, the newly developed bioinformatic methods (such as Bayesian [15]) allow for meaningful analysis of the evolutionary and epidemiological behavior of the sequencing data.The classic methods used to study system evolution include distance, maximum parsimony (MP), and maximum likelihood (ML) [16]. The newly proposed Bayesian method [15] not only retains the basic principle of the ML method but also introduces the Markov chain Monte Carlo method, which greatly reduces the calculation time. In addition, the Bayesian method uses the posterior probability to visually represent the phylogenetic relationships, thereby eliminating the need for bootstrapping.
In the current study, we isolated 16 measles viruses from 105 patient samples from five different cities in Jilin Province between 2005 and 2006. The N gene was sequenced from each MV sample and used to characterize the strain and perform epigenetic and phylogenetic analysis with the worldwide pool of MV strains published in GenBank (http://www.ncbi.nlm.nih.gov/nucleotide). Bayesian analyses were performed to construct a maximum clade credibility (MCC) tree and estimate the time of the most recent common ancestor (TMRCA) for all isolated and downloaded strains. Finally, Bayesian skyline plot analyses was used to reconstruct the past population history of MV by measuring the dynamics of N gene genetic diversity over time.
Results and Discussion
Phylogenetic analysis
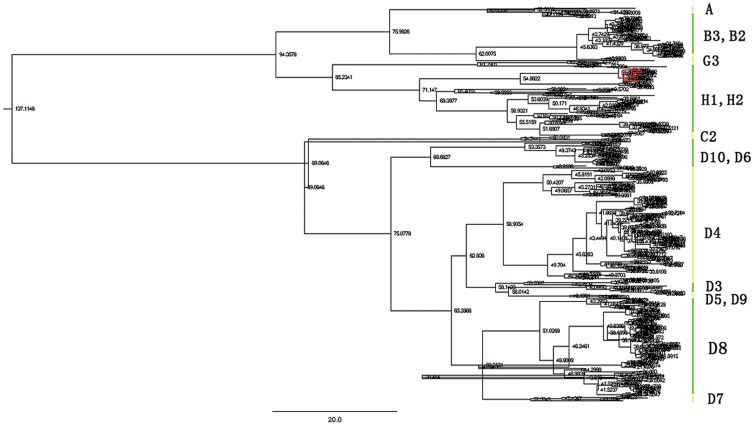
Of the 82 throat swabs and 23 urine samples from suspected MV patients, 16 were successfully isolated with Vero/SLAM cells and tested positive for the MV N gene by RT-PCR. Sequencing revealed that the N gene amplicons from each sample were 451 bp. RPD rigorous recombination analysis showed a lack of recombination in the N gene region from any of the samples. The N coding region sequences from our 16 patient samples and the 450 downloaded from GenBank were used to create a maximum clade credibility tree (Figure 1). The phylogenetic analysis revealed two clusters represented by the 466 MV N sequences. The first cluster contained genotypes A, B, G, H, and C. The second cluster contained only genotype D, implying that genotype D has a distinctive evolutionary profile. In addition, the 16 newly isolated MV wild-type strains showed the closest relation to genotype H1. This is not surprising since the H1 strain is related to previous outbreaks in China. Based on these results, we defined the epidemic strains of Jilin Province as H1 (indicated in red in Figure 1).
Figure 1. The maximum clade credibility tree was estimated by Bayesian analysis of nucleoprotein gene sequences with ∼450 bp of 466 measles virus strains.
The posterior probabilities of the key nodes are depicted above the respective nodes. Samples isolated by our laboratory (n = 16, red lines) were analyzed with the other worldwide strains that were downloaded form GenBank. The green and yellow vertical lines indicate different genotypes. The original file containing more details is available as File S2, which can be opened with FigTree v1.3.1 software to be amplified for more details.
The MV genome-encoded nucleoprotein (N) is critically involved in viral genome transcription and replication, and mediates formation of the MV helical nucleocapsid [13]. The N gene is frequently used for genotyping and phylogenetic analysis of MV. However, no study to date has reported on the relationship between the genetic characteristics of the various MV genotypes and their epidemiologic behavior. Such data would help to identify the source of virus for a particular region or population [14].
Evolutionary rate and TMRCA of each MV genotype
To date, 23 genotypes of the measles virus have been reported, many of which were identified after and near the year 2000 [17], [18]. To understand the evolutionary behavior of MV both worldwide and in northeastern China, we estimated the dates of origin of each genotype using the Bayesian relaxed molecular clock method. The N gene evolutionary rate was estimated to be 1.127E-3 substitutions/site/year (Table 1). The oldest and youngest genotypes of MV were determined to be D (TMRCA: 45.86) and A (TMRCA: 20.28), respectively. According to our analysis, D and A emerged in 1964.1 and 1989.7, respectively. Table 1 summarizes the times of first report for each genotype included in our analysis. The first report times for genotypes B/D/G/H were several years after the emergence time determined by our analysis. However, the report times for genotypes A/C were far before the time we estimated. We theorize that this inconsistency may be due to the fact that very few sequences of genotypes A/C have been reported, especially during the earliest years of their appearance in the literature.
Table 1. Evolutionary characteristics of measles virus genotypes based on the nucleoprotein gene, using the uncorrelated log-normal relaxed clock model, implemented in BEAST.
| MV genotype | Location, year reported | HBV TMRCA (years; 95% HPD) | Emergence time |
| Substitution rate (CR)* 1.127 (0.925–1.329) | |||
| TMRCA (A) | Northern Ireland, 1956 [19] | 20.281 (16.169–26.164) | 1989.7 |
| TMRCA (B) | Cameroon, 1983 [20] | 26.631 (13.764–41.201) | 1983.4 |
| TMRCA (C) | Northern Ireland, 1955 [19] | 23.072 (18.161–33.029) | 1986.9 |
| TMRCA (D) | Northern Ireland, 1960s | 45.862 (31.214–64.844) | 1941.1 |
| TMRCA (G) | USA, 1983 [20] | 25.371 (16.131–38.058) | 1984.6 |
| TMRCA (H) | China, 1993 [21] | 34.949 (25.673–46.931) | 1975.1 |
| TMRCA (JiLin) | - | 6.872 (5.845–7.932) | 2003.1 |
Substitution rates are expressed as 10−3 substitutions per site per year.
The TMRCA for the 16 MV strains from JiLin province was 6.872 (5.845–7.932, 95% CR). Their most recent common ancestor was estimated to have appeared in 2003. This result corresponded to the fact that measles cases in Jilin Province showed a distinct increase in 2005, and reached 18 incidences/100,000 population in 2006 [5]. Considering that viruses usually require a 2–4 year interval to cause epidemic outbreaks [22], the 2005 outbreak of measles in Jilin Province may have in fact been caused by this strain.
Genetic diversity (g) analysis with the Bayesian skyline plot
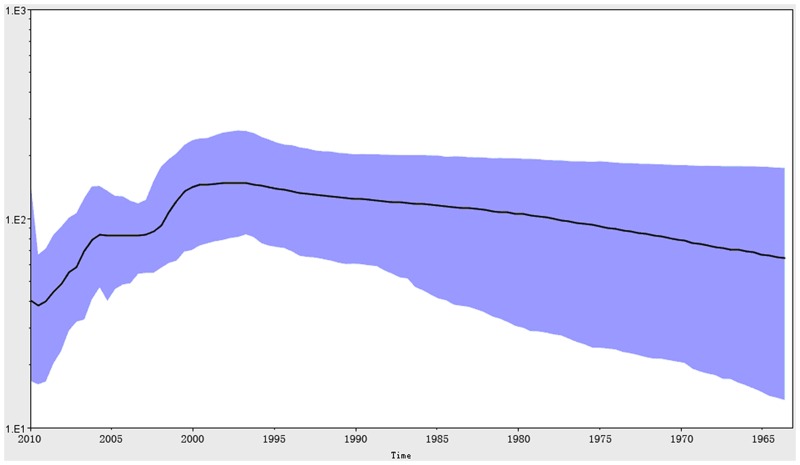
We used Bayesian skyline plot analysis [15] to reconstruct the past population history of MV by measuring the dynamics of N gene genetic diversity over time (Figure 2). The highest level of genetic diversity (g) was observed for the N gene in 2009, suggesting that a remarkable amount of MV genetic diversity was occurring at that time. The WHO's statistics data on measles reported cases have indicated that the overall number of MV infection cases in a region is reduced in parallel to improved coverage of measles vaccine immunization. However, in Jilin Province, the number of MV infection cases rose sharply between 1999 and 2001, despite the fact that the region's immunization rate remained high [23]. Furthermore, the incidence of measles in Jilin Province substantially increased in 2009, to about 10 incidences/100,000 population. Again, within those years, the government's immunization program, consisting of routine immunization and supplementary immunization, had continued [5].
Figure 2. The genetic diversity dynamics of the measles virus N gene estimated by a Bayesian skyline plot through time.
The horizontal axis is in units of years, and the vertical axis is Neτ (the product of the effective population size and the generation length in radiocarbon years). The thick solid line is the median estimate, and the dashed lines show the 95% HPD limits. The plot for N gene shows a rise in relative genetic diversity occurring about 10 years ago, in 2000. Very low genetic diversity existed in the early 1970s and in 2010.
According to our Bayesian skyline plot analyses, the MV N gene genetic diversity showed a slight increase from the 1970s to 2000, reaching the highest genetic diversity near 2000. The outbreaks of MV in Jilin Province may be related to the increase in genetic diversity at those times. This phenomenon may also underlie other MV outbreaks across the globe. As the MV genetic diversity in Jilin Province has reduced since 2009, this may be a good time to make a strong increase in China's immunization campaign. Similar permissive periods may exist for other countries, and will be identifiable by the integrated epidemiologic and phylogenetic approach used in our current study. In this way, we may be able to achieve the global target of a 95% reduction in measles mortality from the level seen in 2000 by 2015, as set forth by the World Health Assembly in May 2010.
Materials and Methods
Study population
Ethics statement: This study was approved by the independent ethics committee (IEC) of Jilin University. A written informed consent was obtained from the parents after we described the study to them. To evaluate the distribution profile of MV genotypes in Jilin province, 16 MV-positive samples (Table 2) were obtained from 105 patients admitted to our hospital with typical measles symptoms between 2005 and 2006. All 16 samples were stored at −20°C until further use. The geographic distribution of these samples was: Changchun (n = 6), Jilin City (n = 3), SiPing (n = 3), YanBian (n = 1), TongHua (n = 1), and SongYuan (n = 2).
Table 2. Summary of natural measles virus strains isolated in Jilin Province, China in 2005–2006.
| No. | Age | City | Specimen type |
| 1 | 20 m | Changchun | Throat swab |
| 2 | 23 m | Jilin City | Throat swab |
| 3 | 21 y | Jilin City | Throat swab |
| 4 | 6 m | Yanbian | Throat swab |
| 5 | 21 y | Tonghua | Urine |
| 6 | 27 m | Siping | Throat swab |
| 7 | 31 m | Siping | Urine |
| 8 | 21 m | Siping | Urine |
| 9 | 23 y | Changchun | Throat swab |
| 10 | 6 m | Songyuan | Throat swab |
| 11 | 26 m | Changchun | Throat swab |
| 12 | 29 m | Changchun | Throat swab |
| 13 | 23 y | Changchun | Throat swab |
| 14 | 15 y | Songyuan | Throat swab |
| 15 | 14 y | Changchun | Throat swab |
| 16 | 20 m | Jilin City | Throat swab |
m, months; y, years.
RNA extraction, reverse transcription-polymerase chain reaction (RT-PCR), and sequencing
The MV samples were amplified in Vero/SLAM (signaling lymphocyte-activation molecule) cells (internal laboratory stock cells) as previously described [24]. Total MV RNA was extracted from the infected cell suspension by using the MiniBEST Viral RNA/DNA Extraction Kit (Ver.4.0; TaKaRa, Shiga, Japan) and following the manufacturer's instructions. The RNA was used as template for RT-PCR amplification of the nucleoprotein (N) gene with the TaKaRa One Step RNA PCR Kit (AMV) and gene-specific primers (forward: 5′-GCT ATG CCA TGG GAG TAG GAG TGG-3′, reverse: 5′-GGC CTC TCG CAC CTA GTC TAG-3′ [25]). RT-PCR products were purified and sent for sequencing at the Beijing Genomics Institute (Shenzhen, China) using a PRISM™ 3730 DNA Sequencer (Applied Biosystems, Inc., Carlsbad, CA, USA).
Sequence collection and phylogenetic analyses
Sequence collection
A total of 476 N gene sequences were downloaded from GenBank (http://www.ncbi.nlm.nih.gov/nucleotide). Of those, 450 had known collection dates (range: 1970–2010), genotypes, and isolate country, and were retrieved for analysis. The accession numbers of these sequences can be found in File S1. These nucleotide sequences were isolated mainly from large measles outbreaks and sporadic cases that occurred globally over the last four decades.
Alignment processing and recombination detection
The N sequences were aligned using MEGA software (Ver.5.0 [26]), and edited with the SEAL sequence simulation and alignment evaluation software (http://tree.bio.ed.ac.uk/software/seal/). To perform the phylogenetic analysis, the missing nucleotides were coded as “missing characters” in the nexus block. To test for the presence of recombination in the N gene, sequences were screened using the Recombination Detection Program (RDP3) software package [27]. The highest multiple-comparison-corrected p-value cutoff was set at 0.01. In all cases, the best-fit model of nucleotide substitution was determined using the MODELTEST program [28].
Bayesian Markov Chain Monte Carlo (MCMC) evolutionary analysis
Bayesian analysis, using the MCMC approach, was performed to construct a maximum clade credibility tree. Convergence was inspected by Tracer (v1.5, http://beast.bio.ed.ac.uk/Tracer), with uncertainties addressed as 95% highest probability density (HPD) intervals. The results were summarized using the TreeAnnotator program with the MCC tree model. Finally, FigTree was used to graphically display the molecular phylogenies. The TMRCA was estimated for six MV genotypes (A–D, G, H) to deduce the oldest and most recently emerged genotypes. The Bayesian skyline plot (BSL) was used to measure the dynamics of N gene genetic diversity over time [15].
The (GTR+G) substitution model was chosen in accordance with the results from the MODELTEST analysis, due to the fact that the generalized time reversible (GTR) approach is considered the most general, neutral, independent approach that accounts for finite-sites and is time-reversible [29]. The “G” value represents the gamma distribution, which is a two-parameter family of continuous probability distributions. Furthermore, the molecular clock model of the Relaxed Clock: Uncorrelated Log-normal was selected for the Bayesian analysis based on the fact that this method assumes no a priori correlation between a lineage's rate of evolution and that of its ancestor. Ten million MCMC runs were sufficient to achieve the convergence of all parameters (effective sampling size >200). Each Bayesian MCMC analysis was run for 20 million states and sampled at every 10,000 states. Posterior probabilities were calculated with a burn-in of 2 million states and checked for convergence using Tracer (v1.5 [30]).
Supporting Information
Access Number and sequences. A total of 450 N gene sequences were downloaded from GenBank with known collection dates (range: 1970–2010), genotypes, and isolate country, and were retrieved for analysis.
(TXT)
Original file of MCC tree build with MV N gene, which can be opened with FigTree v1.3.1 software to be amplified for more details.
(RAR)
Acknowledgments
We would like to extend our special thanks to the two anonymous reviewers for their helpful comments on our manuscript. We also would like to express our sincere thanks to Professor Luquan Ren, Key Laboratory of Bionic Engineering (Ministry of Education, China), Jilin University, for his special help in finishing this paper.
Funding Statement
This work was supported by National Basic Research Program of China (973 program, 2011CB512003) and Graduate Innovation Fund of Jilin University(Project No. 20121125). It was also supported in part by National Natural Science Foundation of China (81071424) and The Ministry of Education Major Science and Technology Innovation Program Seed Foundation (707020). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Hilleman MR (2001) Current overview of the pathogenesis and prophylaxis of measles with focus on practical implications. Vaccine 20: 651–665. [DOI] [PubMed] [Google Scholar]
- 2. Schneider-Schaulies S, ter Meulen V (2002) Measles virus and immunomodulation: molecular bases and perspectives. Expert Rev Mol Med 4: 1–18. [DOI] [PubMed] [Google Scholar]
- 3. Comert S, Vitrinel A, Gursu HA, Deniz NC, Akin Y (2006) Subacute sclerosing panencephalitis presenting as acute disseminated encephalomyelitis. Indian J Pediatr 73: 1119–1121. [DOI] [PubMed] [Google Scholar]
- 4. WHO (2005) Marburg haemorrhagic fever, Angola–update. Wkly Epidemiol Rec 80: 125–126 Accessed 2012 Feb 1. [PubMed] [Google Scholar]
- 5.ChinaCDC. Available: http://wwwchinacdccn/n272442/n272530/index.html. Accessed 2012 February 1.
- 6. WHO (2005) New genotype of measles virus and update on global distribution of measles genotypes. Wkly Epidemiol Rec 80: 347–351. [PubMed] [Google Scholar]
- 7. Niedermeyer HP, Gantumur T, Neubert WJ, Arnold W (2007) Measles virus and otosclerosis. Adv Otorhinolaryngol 65: 86–92. [DOI] [PubMed] [Google Scholar]
- 8. El Mubarak HS, van de Bildt MW, Mustafa OA, Vos HW, Mukhtar MM, et al. (2002) Genetic characterization of wild-type measles viruses circulating in suburban Khartoum, 1997–2000. J Gen Virol 83: 1437–1443. [DOI] [PubMed] [Google Scholar]
- 9. Rota PA, Liffick SL, Rota JS, Katz RS, Redd S, et al. (2002) Molecular epidemiology of measles viruses in the United States, 1997–2001. Emerg Infect Dis 8: 902–908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kubo H, Iritani N, Seto Y (2003) Co-circulation of two genotypes of measles virus and mutual change of the prevailing genotypes every few years in Osaka, Japan. J Med Virol 69: 273–278. [DOI] [PubMed] [Google Scholar]
- 11. Zhang Y, Zhu Z, Rota PA, Jiang X, Hu J, et al. (2007) Molecular epidemiology of measles viruses in China, 1995–2003. Virol J 4: 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. de Swart RL, Wertheim-van Dillen PM, van Binnendijk RS, Muller CP, Frenkel J, et al. (2000) Measles in a Dutch hospital introduced by an immuno-compromised infant from Indonesia infected with a new virus genotype. Lancet 355: 201–202. [DOI] [PubMed] [Google Scholar]
- 13. Curran J, Kolakofsky D (1999) Replication of paramyxoviruses. Adv Virus Res 54: 403–422. [DOI] [PubMed] [Google Scholar]
- 14. Rota PA, Featherstone DA, Bellini WJ (2009) Molecular epidemiology of measles virus. Curr Top Microbiol Immunol 330: 129–150. [DOI] [PubMed] [Google Scholar]
- 15. Drummond AJ, Rambaut A, Shapiro B, Pybus OG (2005) Bayesian coalescent inference of past population dynamics from molecular sequences. Mol Biol Evol 22: 1185–1192. [DOI] [PubMed] [Google Scholar]
- 16. Blair C, Murphy RW Recent trends in molecular phylogenetic analysis: where to next? J Hered 102: 130–138. [DOI] [PubMed] [Google Scholar]
- 17. Jin L, Sun YJ, Ge L, Brown DW (1998) Characterization of a new genotype of measles virus detected in China and England. Epidemiol Infect 121: 691–697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Chibo D, Riddell M, Catton M, Birch C (2002) Novel measles virus genotype, East Timor and Australia. Emerg Infect Dis 8: 735–737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Jin L, Beard S, Hunjan R, Brown DW, Miller E (2002) Characterization of measles virus strains causing SSPE: a study of 11 cases. J Neurovirol 8: 335–344. [DOI] [PubMed] [Google Scholar]
- 20. Rota PA, Bloom AE, Vanchiere JA, Bellini WJ (1994) Evolution of the nucleoprotein and matrix genes of wild-type strains of measles virus isolated from recent epidemics. Virology 198: 724–730. [DOI] [PubMed] [Google Scholar]
- 21. Xu W, Tamin A, Rota JS, Zhang L, Bellini WJ, et al. (1998) New genetic group of measles virus isolated in the People's Republic of China. Virus Res 54: 147–156. [DOI] [PubMed] [Google Scholar]
- 22. Tee KK, Lam TT, Chan YF, Bible JM, Kamarulzaman A, et al. (2010) Evolutionary genetics of human enterovirus 71: origin, population dynamics, natural selection, and seasonal periodicity of the VP1 gene. J Virol 84: 3339–3350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.WHO. Available: http://www.who.int/immunization_monitoring/diseases/measles/en/index.html. Accessed 2012 February 1.
- 24. Tatsuo H, Ono N, Tanaka K, Yanagi Y (2000) SLAM (CDw150) is a cellular receptor for measles virus. Nature 406: 893–897. [DOI] [PubMed] [Google Scholar]
- 25. Zhu Z, Zhang Y, Xu S, Yu P, Tian X, et al. (2009) Outbreak of acute respiratory disease in China caused by B2 species of adenovirus type 11. J Clin Microbiol 47: 697–703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Tamura K, Peterson D, Peterson N, Stecher G, Nei M, et al. MEGA5: Molecular Evolutionary Genetics Analysis using Maximum Likelihood, Evolutionary Distance, and Maximum Parsimony Methods. Mol Biol Evol [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Martin DP (2009) Recombination detection and analysis using RDP3. Methods Mol Biol 537: 185–205. [DOI] [PubMed] [Google Scholar]
- 28. Posada D, Crandall KA (1998) MODELTEST: testing the model of DNA substitution. Bioinformatics 14: 817–818. [DOI] [PubMed] [Google Scholar]
- 29. Tavaré S (1986) Some probabilistic and statistical problems in the analysis of DNA sequences. In Some mathematical questions in biology- DNA sequence analysis. Providence, RI: Amer Math 17: 57–86. [Google Scholar]
- 30. Alvarado Mora MV, Romano CM, Gomes-Gouvea MS, Gutierrez MF, Botelho L, et al. (2011) Molecular characterization of the Hepatitis B virus genotypes in Colombia: a Bayesian inference on the genotype F. Infect Genet Evol 11: 103–108. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Access Number and sequences. A total of 450 N gene sequences were downloaded from GenBank with known collection dates (range: 1970–2010), genotypes, and isolate country, and were retrieved for analysis.
(TXT)
Original file of MCC tree build with MV N gene, which can be opened with FigTree v1.3.1 software to be amplified for more details.
(RAR)