Abstract
Relative to the atmosphere, much of the aerobic ocean is supersaturated with methane; however, the source of this important greenhouse gas remains enigmatic. Catabolism of methylphosphonic acid by phosphorus-starved marine microbes, with concomitant release of methane, has been suggested to explain this phenomenon, yet methylphosphonate is not a known natural product, nor has it been detected in natural systems. Further, its synthesis from known natural products would require unknown biochemistry. Here we show that the marine archaeon Nitrosopumilus maritimus encodes a pathway for methylphosphonate biosynthesis and that it produces cell-associated methylphosphonate esters. The abundance of a key gene in this pathway in metagenomic datasets suggests that methylphosphonate biosynthesis is relatively common in marine microbes, providing a plausible explanation for the methane paradox.
Methane plays a key role in the global carbon cycle and is a potent greenhouse gas. As such, knowledge of its sources and sinks is essential to climate change models and to understand the flow of carbon within the biosphere. An unsolved problem in this area is the observation that vast sections of the aerobic ocean are supersaturated with this gas, despite the fact that strictly anaerobic archaea are the only significant biological source of methane known (1). The amount of methane produced in these aerobic environments is substantial, comprising as much as 4% of the global methane budget (2). It has been suggested that anaerobic microenvironments within the aerobic ecosystem could allow production of methane by known methanogens; however, this is contested on a variety of grounds (for a discussion see (1, 3)). Recently, Karl et al. suggested a new model in which methane would be produced when aerobic marine microorganisms use methylphosphonic acid (MPn) as a source of phosphorus (2). The model is based on several observations: (i) a well-studied bacterial enzyme, carbon-phosphorus (C-P) lyase, produces methane from MPn (4), (ii), C-P lyase genes are abundant in marine microbes (5, 6), (iii) phosphonates comprise a significant fraction of the available phosphorus pool in marine systems (7, 8), and (iv) incubation of seawater microcosms with MPn leads to methane production (2). While this model is conceptually appealing, it has a significant missing link: MPn has never been detected in marine ecosystems, nor is it a known natural product. Moreover, based on known phosphonate biosynthetic pathways (9), it is difficult to see how MPn could be made without invoking unusual biochemistry.
With one exception, all known phosphonate biosynthetic pathways begin with formation of the C-P bond by the enzyme phosphoenolpyruvate mutase (Ppm) (9). We have used the ppm gene as a molecular marker to identify the genes responsible for synthesis of phosphonic acid antibiotics in numerous microorganisms (10-13). During the course of this work, we identified a putative phosphonate biosynthetic gene cluster in Nitrosopumilus maritimus, a member of the ubiquitous Group I marine Thaumarchaeota whose members are among the most abundant organisms in marine surface waters (14, 15). Based on the experimentally validated functions of homologous enzymes (10, 16, 17), it is very likely that N. maritimus has the capacity to synthesize 2-hydroxyethylphosphonate (HEP), which is a common intermediate in phosphonate biosynthetic pathways (Fig. S1A, Table S1). Immediately adjacent to the putative HEP biosynthetic genes is an operon encoding a putative oxidoreductase, two putative sulfatases and a protein of the cupin superfamily that we designated MpnS.
MpnS has weak homology to hydroxypropylphosphonate epoxidase (HppE) and hydroxyethylphosphonate dioxygenase (HepD), two enzymes that catalyze Fe(II)- and oxygen-dependent transformations of similar phosphonate substrates (Figs S1B & S2). Thus, we suspected that MpnS might be a similar phosphonate biosynthetic enzyme. To test this, we cloned and overexpressed the mpnS gene in Escherichia coli (18). Cell extracts containing MpnS stoichiometrically convert 13C-labelled HEP to a product whose retention time and molecular mass are identical to MPn in liquid chromatography-mass spectrometry (LC-MS) experiments (Figs. 1 & S3). Using purified MpnS protein and HEP labeled with 13C at either the 1- or 2- position, we conclusively showed that the products of the MpnS reaction are MPn and HCO3- (Fig. 1B & 1C). The MpnS-catalyzed reaction requires both Fe(II) and molecular oxygen, but does not require an exogenous electron donor. Thus, like HepD, MpnS is an Fe(II)-dependent oxygenase that cleaves the unactivated carbon-carbon bond of HEP. However, the two enzymes catalyze distinct reactions. In the HepD reaction, the reducing equivalents needed for incorporation of oxygen into the cleavage products are derived equally from the C-1 and C-2 carbons of HEP, whereas MpnS catalyzes the asymmetric oxidation of HEP, with all four electrons being derived from the C-2 carbon, affording the more reduced phosphonate product MPn.
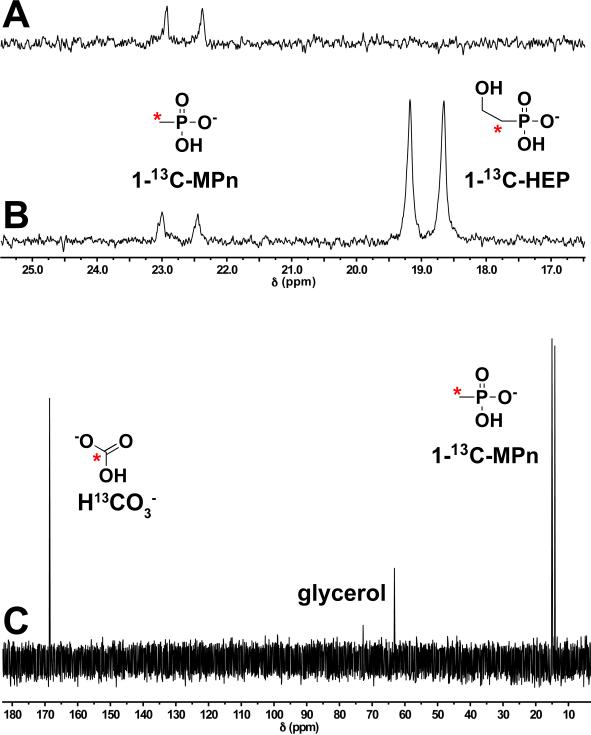
Fig. 1.
In vitro assay of MpnS activity. (A) Crude cell extract from an E. coli MpnS overexpression strain was incubated aerobically with 1-13C-HEP in the presence of Fe(II) and the phosphorus-containing products were examined using 31P NMR spectroscopy. After incubation for 1 hour a single product was observed as a doublet centered at 23.5 ppm. The mass and retention time of this product determined by LC-MS is consistent with this product being 1-13CMPn (Fig. S3). (B) Spiking of this reaction with the substrate, 1-13C-HEP produced a second doublet centered at 19 ppm, showing that the substrate was completely consumed in the initial reaction. (C) The identity of the reaction products was determined using 13C NMR after repeating the assay in a sealed vial using purified MpnS with a mixture of 1-13C-HEP and 2-13C-HEP as substrates. The C-2 labeled carbon of HEP is converted to 13C bicarbonate (H13CO -3), while the C-1-labelled carbon is converted to 1-13C-MPn. Bonding to phosphorus splits the 13C peak in the NMR spectrum. Thus, the C-1 peak is split and the C-2 peak is not. Glycerol, a component of the assay mixture, is also observed in the 13C spectrum. The 13C label is indicated by a red asterisk.
Having shown that MpnS catalyzes the synthesis of MPn in vitro, we asked whether N. maritimus synthesizes phosphonic acids using 31P NMR spectroscopy (Fig. 2A). The 1H-decoupled 31P spectrum of the soluble cell extract displayed two peaks in the 10-30 ppm range characteristic of phosphonic acids (19). The relative abundance of the two peaks varied with sample preparation and could be seen in both the soluble and cell debris fractions after sonication (Fig S4). Based on spiking of the sample with an authentic standard neither peak can be attributed to free methylphosphonate; however, because the phosphorus compounds are cell associated we expected them to be covalently linked to a larger, more complex molecule, thus changing the chemical shift in the 31P NMR spectrum. Accordingly, we conducted a series of 31P-1H Heteronuclear Multiple Bond Correlation (HMBC) experiments to identify the atoms linked to the P nuclei seen in the NMR spectra (Fig 2B). Because the behavior of phosphonate esters in such experiments is not well documented, we also synthesized and characterized a series of phosphonate esters to support our assignments (Figs S5-S7). Based on these experiments the 31P NMR peak at 28.7 ppm can be confidently assigned as an ester of methylphosphonate. Further support for this conclusion was provided by high-resolution mass spectrometry, which revealed the presence of free methylphosphonate after strong acid hydrolysis of N. maritimus cell debris (Fig 2C and S8). Based on these results and the gene context of the MpnS locus (Table S1), we suspect that N. maritimus synthesizes an exopolysaccharide decorated with methylphosphonate, similar to the HEP- and aminoethylphosphonate-modified polymers found in many bacteria and lower eukaryotes (20).
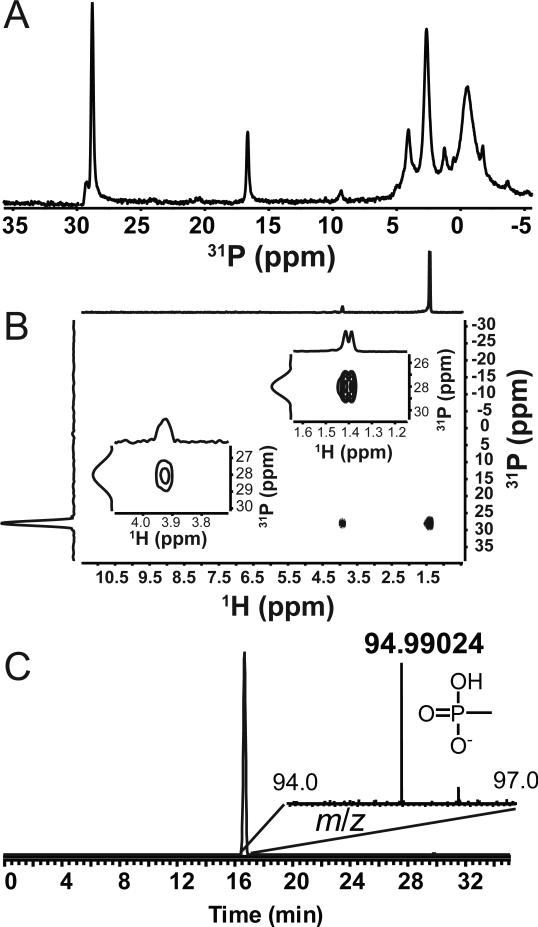
Fig. 2.
In vivo production of methylphosphonate esters by N. maritimus. (A) A cell extract of N. maritimus was prepared by sonication of whole cells as described. After removal of the cell debris by centrifugation, the supernatant was examined by 31P NMR spectroscopy revealing at least two compounds with chemical shifts in the range typical of phosphonic acids. (B) The two-dimensional HMBC NMR spectrum of N. maritimus cell extract. Comparison of the proton splitting patterns (shown in the insets) to those of model compounds (Figs S6 & S7) clearly shows that the P compound at 28 ppm in the 31P dimension is a methylphosphonate ester. The doublet of the proton at 1.4 ppm coupled to the phosphorus is diagnostic for a methyl group bonded directly to phosphorus, i.e. a methylphosphonate moiety. (C) High resolution LC-MS analysis showing the presence of free methylphosphonate after strong acid hydrolysis of N. maritimus cell debris. The extracted ion chromatogram centered around m/z 94.99035 (exact monoisotopic mass of methylphosphonate [M-H]-) with Fourier-transform mass spectrum and ion structure is shown in the inset. The chromatographic and MS fragmentation pattern is identical to an authentic MPn standard (Fig S8).
The data presented above suggest that N. maritimus produces a cell-associated methylphosphonate ester via an MpnS-dependent biosynthetic pathway. To link this finding to the larger marine environment we screened the Global Oceanic Survey (GOS) metagenomic dataset (21) for the presence of MpnS homologs. We also searched for homologs of the related HepD and HppE proteins. Initially we screened the assembled GOS scaffolds, finding forty-six MpnS and twenty HepD homologs using a BLASTP cutoff value of 10-10 (Table S2). No HppE homologs were observed. Importantly, none of the HepD homologs were identified using N. maritimus MpnS as the query sequence; likewise none of the MpnS homologs were identified using HepD as a query. Thus, BLASTP clearly differentiates between the two homologous groups, supporting the assignment of the recovered sequences as MpnS and HepD proteins, respectively. To independently support these functional assignments we constructed maximum likelihood phylogenetic trees including biochemically validated MpnS, HepD and HppE proteins (Fig 3A & S9). We also used a hierarchical clustering method to examine all putative and validated MpnS, HepD and HppE proteins (Fig S10). In both cases robust support for the functional assignments was obtained. Thus, we conclude that the recovered GOS MpnS homologs are likely to be methylphosphonate synthases.
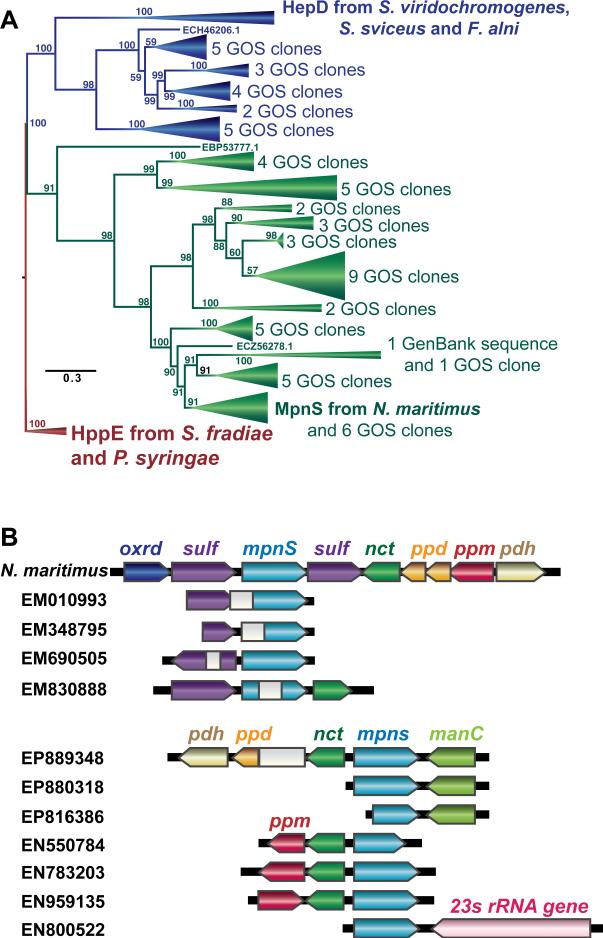
Fig. 3.
(A) The evolutionary relationships of biochemically characterized MpnS, HepD and HppE proteins (shown in bold) and homologs recovered from Genbank and the GOS metagenomic dataset was inferred using maximum likelihood analysis as described. Bootstrap values from 100 replicates are shown at the nodes. Robust bootstrap support for the tree shows that the method clearly differentiates MpnS (green), HepD (blue) and HppE (red) proteins. The full tree with all individual homologs shown is presented in Fig S9. (B) The gene content of large scaffolds containing the GOS MpnS homologs is compared to the mpnS locus of N. maritimus. The grey boxes represent sequencing gaps between paired-end reads.
Additional support for the function of the MpnS homologs was revealed by analysis of neighboring genes found in GOS DNA scaffolds (Fig 3B, Table S3). Many of the nearby ORFs are homologous to those found in the N. maritimus gene cluster, including the phosphonate biosynthetic genes ppm, ppd, and pdh, as well as homologs of the sulfatases and nucleotidyl transferase genes, suggesting that the GOS scaffolds encode genes for the synthesis of similar MPn esters. Several other genes found on the scaffolds provide evidence for the identity of the organisms in which they are found. One of the scaffolds includes a 23S rRNA gene that can be confidently placed within the SAR11 clade between Pelagibacter species (Fig S11), while two of the manC genes are nearly identical to ones found in Pelagibacter sp. HTCC7211. Although the mpnS gene is absent in sequenced Pelagibacter genomes, these data strongly support the conclusion that some members of this genus have the capacity to synthesize MPn.
Relatives of Nitrosopumilus and Pelagibacter are among the most abundant organisms in the sea, with global populations estimated at 1028 for both ammonia-oxidizing Thaumarchaeota (14) and members of the SAR11 clade (22). Thus, the observation of mpnS in some members of these genera is consistent with the idea that MPn synthesis is prevalent in marine systems. To provide direct support for this notion, we measured the abundance of the mpnS gene relative to the abundance of typical single-copy genes as previously described (23). We also quantified the occurrence of the ppm gene to provide an estimate the relative occurrence of phosphonate synthesis in general (Table S4). Based on these data, we estimate that ca. 16% of marine microbes are capable of phosphonate biosynthesis, while 0.6% have the capacity to synthesize MPn. Because the GOS samples are confined to the upper few meters of the ocean, extrapolation of this analysis to the deeper ocean should be viewed with some skepticism. Nevertheless, the upper 200 m of the world's oceans are thought to contain ca. 3.6 ×1028 microbial cells with an average generation time of ca. two weeks (24). Thus, even with the relatively modest abundance of MPn biosynthesis suggested by our data, it seems quite possible that these cells could provide sufficient MPn precursor to account for the observed methane production in the aerobic ocean via the C-P lyase dependent scenario suggested by Karl et al (2).
Supplementary Material
Acknowledgments
This work was supported by the National Institutes of Health (GM PO1 GM077596 and F32 GM095024), the Howard Hughes Medical Institute and the National Science Foundation NSF (MCB-0604448, OCE-1046017 and MCB-0920741). Its contents are solely the responsibility of the authors and do not necessarily represent the official views of the NIGMS, NIH, NSF or HHMI.
References and Notes
- 1.Rogers JE, Whitman WB, editors. Microbial production and consumption of greenhouse gases : methane, nitrogen oxides, and halomethanes. American Society for Microbiology; Washington, D.C.: 1991. [Google Scholar]
- 2.Karl DM, et al. Nat. Geosci. 2008;1:473. [Google Scholar]
- 3.Reeburgh WS. Chem. Rev. 2007;107:486. doi: 10.1021/cr050362v. [DOI] [PubMed] [Google Scholar]
- 4.Daughton CG, Cook AM, Alexander M. FEMS Microbiol. Lett. 1979;5:91. [Google Scholar]
- 5.Martinez A, Tyson GW, DeLong EF. Environ. Microbiol. 2010;12:222. doi: 10.1111/j.1462-2920.2009.02062.x. [DOI] [PubMed] [Google Scholar]
- 6.Ilikchyan IN, McKay RML, Zehr JP, Dyhrman ST, Bullerjahn GS. Environ. Microbiol. 2009;11:1314. doi: 10.1111/j.1462-2920.2009.01869.x. [DOI] [PubMed] [Google Scholar]
- 7.Clark LL, Ingall ED, Benner R. Am. J. Sci. 1999;299:724. [Google Scholar]
- 8.Clark LL, Ingall ED, Benner R. Nature. 1998;393:426. [Google Scholar]
- 9.Metcalf WW, van der Donk WA. Annu. Rev. Biochem. 2009;78:65. doi: 10.1146/annurev.biochem.78.091707.100215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Borisova SA, Circello BT, Zhang JK, van der Donk WA, Metcalf WW. Chem. Biol. 2010 Jan 29;17:28. doi: 10.1016/j.chembiol.2009.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Blodgett JA, Zhang JK, Metcalf WW. Antimicrob. Agents and Ch. 2005;49:230. doi: 10.1128/AAC.49.1.230-240.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Eliot AC, et al. Chem Biol. 2008;15:765. doi: 10.1016/j.chembiol.2008.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Circello BT, Eliot AC, Lee JH, van der Donk WA, Metcalf WW. Chem Biol. 2010;17:402. doi: 10.1016/j.chembiol.2010.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Karner MB, DeLong EF, Karl DM. Nature. 2001;409:507. doi: 10.1038/35054051. [DOI] [PubMed] [Google Scholar]
- 15.Konneke M, et al. Nature. 2005;437:543. doi: 10.1038/nature03911. [DOI] [PubMed] [Google Scholar]
- 16.Shao Z, et al. J. Biol. Chem. 2008;283:23161. doi: 10.1074/jbc.M801788200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Seidel HM, Freeman S, Seto H, Knowles JR. Nature. 1988;335:457. doi: 10.1038/335457a0. [DOI] [PubMed] [Google Scholar]
- 18.Materials and methods are available as supplementary material on Science Online.
- 19.Tebby JC, editor. CRC handbook of phosphorus-31 nuclear magnetic resonance data. CRC Press; Boca Raton: 1991. [Google Scholar]
- 20.Hilderbrand RL, editor. The Role of Phosphonates in Living Systems. CRC Press; Boca Raton: 1983. [Google Scholar]
- 21.Yooseph S, et al. PLoS. Biol. 2007;5:432. [Google Scholar]
- 22.Morris RM, et al. Nature. 2002;420:806. doi: 10.1038/nature01240. [DOI] [PubMed] [Google Scholar]
- 23.Howard EC, Sun S, Biers EJ, Moran MA. Environ. Microbiol. 2008;10:2397. doi: 10.1111/j.1462-2920.2008.01665.x. [DOI] [PubMed] [Google Scholar]
- 24.Whitman WB, Coleman DC, Wiebe WJ. Proc. Natl. Acad. Sci. USA. 1998;95:6578. doi: 10.1073/pnas.95.12.6578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dunwell JM, Culham A, Carter CE, Sosa-Aguirre CR, Goodenough PW. Trends Biochem. Sci. 2001;26:740. doi: 10.1016/s0968-0004(01)01981-8. [DOI] [PubMed] [Google Scholar]
- 26.Liu PH, et al. J. Am. Chem. Soc. 2001;123:4619. doi: 10.1021/ja004153y. [DOI] [PubMed] [Google Scholar]
- 27.Cicchillo RM, et al. Nature. 2009;459:871. doi: 10.1038/nature07972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Higgins LJ, Yan F, Liu PH, Liu HW, Drennan CL. Nature. 2005;437:838. doi: 10.1038/nature03924. [DOI] [PubMed] [Google Scholar]
- 29.Nair SK, van der Donk WA. Arch. Biochem. Biophys. 2011;505:13. doi: 10.1016/j.abb.2010.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kuemin M, van der Donk WA. Chem. Commun. 2010;46:7694. doi: 10.1039/c0cc02958k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Whitteck JT, et al. Angew. Chem., Int. Ed. Engl. 2007;46:9089. doi: 10.1002/anie.200703810. [DOI] [PubMed] [Google Scholar]
- 32.Martens-Habbena W, Berube PM, Urakawa H, de la Torre JR, Stahl DA. Nature. 2009;461:976. doi: 10.1038/nature08465. [DOI] [PubMed] [Google Scholar]
- 33.Papadopoulos JS, Agarwala R. Bioinformatics. 2007;23:1073. doi: 10.1093/bioinformatics/btm076. [DOI] [PubMed] [Google Scholar]
- 34.Stamatakis A. Bioinformatics. 2006;22:2688. doi: 10.1093/bioinformatics/btl446. [DOI] [PubMed] [Google Scholar]
- 35.Olson SA. Brief Bioinform. 2002;3:87. doi: 10.1093/bib/3.1.87. [DOI] [PubMed] [Google Scholar]
- 36.de Hoon MJ, Imoto S, Nolan J, Miyano S. Bioinformatics. 2004;20:1453. doi: 10.1093/bioinformatics/bth078. [DOI] [PubMed] [Google Scholar]
- 37.Saldanha AJ. Bioinformatics. 2004;20:3246. doi: 10.1093/bioinformatics/bth349. [DOI] [PubMed] [Google Scholar]
- 38.Desper R, Gascuel O. Mol. Biol. Evol. 2004;21:587. doi: 10.1093/molbev/msh049. [DOI] [PubMed] [Google Scholar]
- 39.Markowitz VM, et al. Nuc Acids Res. 2010;38:D382. doi: 10.1093/nar/gkp887. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.