Abstract
Chikungunya virus (CHIKV) and Ross River virus (RRV) cause a debilitating, and often chronic, musculoskeletal inflammatory disease in humans. Macrophages constitute the major inflammatory infiltrates in musculoskeletal tissues during these infections. However, the precise macrophage effector functions that affect the pathogenesis of arthritogenic alphaviruses have not been defined. We hypothesized that the severe damage to musculoskeletal tissues observed in RRV or CHIKV-infected mice would promote a wound healing response characterized by M2-like macrophages. Indeed, we found that RRV and CHIKV-induced musculoskeletal inflammatory lesions, and macrophages present in these lesions, have a unique gene expression pattern characterized by high expression of arginase 1 and Ym1/Chi3l3 in the absence of FIZZ1/Relmα that is consistent with an M2-like activation phenotype. Strikingly, mice specifically deleted for Arg1 in neutrophils and macrophages had dramatically reduced viral loads and improved pathology in musculoskeletal tissues at late times post-RRV infection. These findings indicate that arthritogenic alphavirus infection drives a unique myeloid cell activation program in inflamed musculoskeletal tissues that inhibits virus clearance and impedes disease resolution in an Arg1-dependent manner.
INTRODUCTION
Chikungunya virus (CHIKV), Ross River virus (RRV), Mayaro virus, o'nyong-nyong virus and others are positive-sense, single-stranded RNA viruses in the Alphavirus genus of the family Togaviridae (1). These mosquito-transmitted alphaviruses cause a debilitating musculoskeletal inflammatory disease in humans and are emerging disease threats due to their ability to cause explosive epidemics. Past epidemics include a 1979-to-1980 epidemic of RRV disease in the South Pacific that involved more than 60,000 patients (2) and a 1959-to-1962 epidemic of o'nyong-nyong fever in Africa that involved at least 2 million infections (3). Since 2004, CHIKV has caused major epidemics in multiple countries in the Indian Ocean region with estimates on the order of 1–6 million cases (4) and, for the first time, has caused disease outbreaks in Europe and the Pacific Region (5–7). During these outbreaks, at least 106 CHIKV viremic travelers have been identified in the United States (8). The spread of the peri-urban mosquito Aedes (Ae.) albopictus into Europe and the Americas along with high viremia in infected travelers returning from areas with CHIKV activity increases the risk that this virus will continue to spread to new regions. In fact, more than 200 human cases of CHIKV disease in Italy were traced to a single infected traveler and CHIKV was subsequently detected in local Ae. albopictus mosquitoes (6, 9). In addition, the Pan American Health Organization and the Centers for Disease Control and Prevention recently released a preparedness guide anticipating the introduction of CHIKV in the Americas (10).
Clinical manifestations following infection with arthritis/myositis-associated alphaviruses are most commonly characterized by fever, intense pain in the peripheral joints, myalgias, and an impaired ability to ambulate (2, 11, 12). The joint pain associated with arthritogenic alphavirus infections is typically symmetrical with fingers, wrists, elbows, toes, ankles, and knees most commonly affected (13). A number of studies indicate that musculoskeletal pain lasts for months to years in a subset of persons infected with RRV or CHIKV; however, the cause of these long lasting symptoms is unclear (13–23). More severe disease including neurologic manifestations, myocarditis, and death have been reported. These atypical outcomes are associated with age and underlying medical conditions (24). There are currently no licensed antivirals or vaccines for any of the arthritis/myositis-associated alphaviruses; treatment is limited to supportive care with analgesics and anti-inflammatory drugs (2, 25).
Monocytes/macrophages constitute the major inflammatory infiltrates in musculoskeletal tissues of CHIKV or RRV-infected humans, non-human primates, and mice (26–33), and numerous studies have implicated macrophages in the pathogenesis of these infections. However, how macrophages are activated during alphavirus infection and the precise macrophage effector functions that affect the pathogenesis of arthritogenic alphaviruses have not been defined. Although macrophages at inflammatory sites often have a spectrum of overlapping phenotypes, subsets with distinct functions have been described (34–36). Classically-activated (M1) macrophages have a pro-inflammatory phenotype, high antigen presentation capacity, and contribute to host defense against infectious pathogens and tumors. In contrast, alternatively activated (M2) macrophages have high phagocytic capacity and promote tissue repair/remodeling during wound healing. In addition, M2 macrophages dampen inflammation and have immunoregulatory functions.
The activation of M2 or M2-like macrophages occurs in response to tissue damage in a variety of tissue types, including musculoskeletal tissues, and this response can be activated in the presence or absence of an infectious pathogen (37–42). These findings, and the findings that tissue repair mediated by Th2 cells is a primary defense against helminth infections (43), has led to the hypothesis that M2 macrophages are a component of a larger Th2-mediated response that evolved as a wound repair response (44). Consistent with this model, recent studies reported an important role for IL-4Rα-dependent M2 macrophages in the resolution of tissue damage during Respiratory Syncytial virus infection (45).
Interestingly, the expression of Arginase 1 (Arg1), a central metabolic enzyme of the liver that catalyzes the hydrolysis of L-arginine to urea and ornithine (46), is associated with M2 macrophages or M2-like cells (47, 48). In addition, the expression of Arg1 by human and murine monocytes/macrophages, neutrophils, and myeloid-derived suppressor cells (MDSCs) (49–51) has emerged as a major regulator of immune responses. Arg1 activity in myeloid cells impairs effective immunity against intracellular pathogens such as Mycobacterium tuberculosis and Toxoplasma gondii, exacerbates tumor growth by suppressing T cell function, and limits T cell-driven inflammatory tissue damage by suppressing effector T cell functions and promoting regulatory T cell activation (52–56).
Previous studies in mouse models have shown that arthritogenic alphavirus infection results in severe inflammation and tissue damage in skeletal muscle tissue, joint-associated tissues, and tendons (26, 27, 57, 58). We hypothesized that the severe damage to musculoskeletal tissues of RRV- or CHIKV-infected mice promotes a wound healing response characterized by M2-like macrophages. Given the immunosuppressive activity of these cells, we further reasoned that this wound healing response would impair the clearance of virus from inflamed tissues. Indeed, we found that Arg1 was dramatically induced in the musculoskeletal inflammatory lesions and infiltrating macrophages of RRV- and CHIKV-infected mice. Strikingly, mice specifically deleted for Arg1 in macrophages and neutrophils had dramatically reduced viral loads as well as improved tissue pathology at late, but not early, times post-RRV infection. Our findings suggest that Arg1 activity in myeloid cells has a critical role in regulating the clearance of arthritogenic alphaviruses from inflamed and injured musculoskeletal tissues.
MATERIALS AND METHODS
Viruses
The T48 stain of RRV was isolated from Aedes vigilax mosquitoes in Queensland, Australia (59). Prior to cDNA cloning, the virus was passaged 10 times in suckling mice followed by two passages on Vero cells (60). The SL15649 strain of CHIKV was isolated from a serum sample collected from a febrile patient in Sri Lanka in 2006. This virus was passaged 2 timed in Vero cells prior to cDNA cloning (27). Stocks of infectious RRV or CHIKV were generated from cDNAs as previously described (27, 28).
Cells
BHK-21 cells (ATCC CCL10) were grown as previously described (61). Bone marrow from the femur and tibia of C57BL/6 mice was harvested in RPMI 1640 (HyClone) with 10% fetal bovine serum (FBS). Following red blood cell lysis, bone marrow cells were plated in 10 cm dishes in DMEM (Sigma) containing 10% FBS, 5% horse serum, and 20% L-cell conditioned media for 6 days to differentiate into macrophages. Following differentiation, cells were scraped, counted, and plated in 48 well plates in DMEM containing 10% FBS. The following day, cells were treated with or without LPS (100 ng/ml; Sigma) and IFN-Y (15 ng/ml; BD Pharmingen) or recombinant mouse IL-4 (5 ng/ml; R &D Systems) for 24 hours. Cells were washed with PBS and then lysed in TRIzol Reagent (Life Technologies).
Mice
C57BL/6 WT (stock # 000664), LysMcre (stock # 004781) and Arg1-Flox/Flox (Arg1F/F) mice (stock # 008817) were obtained from The Jackson Laboratory and bred in house. LysMcre mice were generated by the insertion of the cre cDNA into the endogenous mouse M lysozyme gene (62). Arg1F/F mice were engineered to contain loxP sites on either side of exons 7 and 8 in the Arg1 gene (52). Animal husbandry and experiments were performed in accordance with all University of Colorado School of Medicine Institutional Animal Care and Use Committee guidelines. All mouse studies were performed in an animal biosafety level 3 laboratory. Twenty to twenty-four day old mice were used for all in vivo studies. Mice were inoculated in the left rear footpad with 103 pfu of virus in diluent (PBS/1% bovine calf serum) in a 10 μl volume. Mock-infected animals received diluent alone. Mice were monitored for disease signs and weighed at 24 hour intervals. Disease scores were determined by assessing grip strength, hind limb weakness, and altered gait as previously described (61). On the termination day of each experiment, mice were sacrificed by exsanguination, blood was collected, and mice were perfused by intracardial injection of 1× PBS or 4% paraformaldehyde. PBS-prefused tissues were removed by dissection and homogenized in TRIzol Reagent (Life Technologies) for RNA analysis or in RIPA buffer [50 mM Tris (pH 8), 150 mM NaCl, 1% NP-40, 0.5% sodium deoxycholate, 0.1% sodium dodecyl sulfate with protease inhibitor cocktail added (Roche)] for protein analysis with a MagNA Lyser (Roche). Alternatively, quadriceps muscles and spleens were dissected, minced, and incubated for 1.5 h with vigorous shaking at 37°C in digestion buffer (RPMI 1640, 10% FBS, 15 mM HEPES, 2.5 mg/ml collagenase Type 1 [Worthington Biochemical Corp.], 1.7 mg/ml DNase I [Roche], 1× gentamicin (Gibco), 1% pen/strep). Following digestion, cells were passed through a 100-μm cell strainer (BD Falcon) and banded on Lympholyte-M (Cedarlane) to isolate infiltrating leukocytes. Following red blood cell lysis (spleens only), cells were washed in wash buffer (1× PBS, 15 mM HEPES, 1× gentamicin, 1% pen/strep), and total viable cells were determined by trypan blue exclusion.
Flow cytometry and cell sorting
Leukocytes isolated from enzymatically-digested tissues were incubated with anti-mouse FcγRII/III (2.4G2; BD Pharmingen) for 20 min on ice to block nonspecific antibody binding and then stained in fluorescence-activated cell sorting (FACS) staining buffer (1× PBS, 2% FBS) with the following antibodies from eBioscience: anti-CD11b-allophycocyanin (APC) and anti-F4/80-fluorescein isothiocyanate (FITC). Cells were fixed overnight in 1% paraformaldehyde and analyzed on a LSR II using FACSDiva software (Becton Dickinson). For cell sorting, mice were sacrificed at 7 or 10 dpi and the quadriceps muscles were processed as described above. Cells were stained in FACS staining buffer with anti-CD11b-APC and anti-F4/80-FITC antibodies (eBioscience). CD11b+F4/80+ cells were sorted under BSL2 conditions on a FACSAria using FACSDiva software (Becton Dickinson). FlowJo software (Tree Star) was used for all analyses. For morphological analysis of FACS sorted CD11b+F4/80+ cells, a ThermoShandon Cytospin 4 was used and slides were stained with Wright-Geimsa stain (Sigma-Aldrich).
Real-time RT-qPCR
RNA was isolated using a PureLink RNA Mini Kit (Life Technologies). For analysis of host gene expression, 0.2–1 microgram of total RNA was reverse-transcribed using Superscript III (Life Technologies), random oligo(dT) primers, and RNaseOUT. Real-time qPCR experiments were performed using Taqman© gene expression assays and a LightCycler 480 (Roche). 18S rRNA and hypoxanthine guanine phosporibosyl transferase (HPRT) were used as endogenous controls to normalize for input amounts of cDNA. The relative fold induction of amplified mRNA were determined by using the Ct method (63). For absolute quantification of viral RNA, a sequence-tagged (small caps) RRV-specific RT primer (4415-5'-ggcagtatcgtgaattcgatgcAACACTCCCGTCGACAACAGA-3') or CHIKV-specific primer (1036-5'-ggcagtatcgtgaattcgatgcCGTGTCGGTAGTCTTGCACAT-3') was used for reverse transcription. For RRV, a tag sequence-specific reverse primer (5'-GGCAGTATCGTGAATTCGATGC-3') was used with a RRV sequence-specific forward primer (RRV: 4346 5'-CCGTGGCGGGTATTATCAAT-3') and an internal Taqman probe (RRV: 4375 5'-ATTAAGAGTGTAGCCATCC-3') during qPCR to enhance specificity. For CHIKV, CHIKV sequence-specific forward (CHIKV: 874 5'-AAAGGGCAAACTCAGCTTCAC-3') and reverse (CHIKV: 961 5⍰-GCCTGGGCTCATCGTTATTC-3⍰) primers were used with an internal Taqman probe (CHIKV: 899 5'- CGCTGTGATACAGTGGTTTCGTGTG). To create standard curves, 10-fold dilutions from 108 to 100 copies of RRV or CHIKV genomic RNAs, synthesized in vitro, were spiked into RNA from BHK-21 cells and reverse transcription and qPCR were performed in an identical manner.
Mixed Leukocyte Reaction (MLR)
The capacity of defined macrophage populations to inhibit T cell reactivity in vitro was assessed by macrophage co-culture with naïve C57BL/6 T cells responding to allogeneic BALB/c splenic stimulator cells. The alloreactive MLR was performed largely as previously described (64). Briefly, 2 ×105 responding B6 lymph node cells were stimulated with 3 ×105 irradiated (2000R) allogeneic BALB/c spleen cells in quadruplicate cultures consisting of Eagle's MEM supplemented with 10% FCS in 96-well, flat-bottomed plates. To assess macrophage suppressive activity, graded numbers of the indicated macrophage populations were added to these MLR cultures. Ratios indicate the number of macrophage cells added relative to the number of responding lymph node cells. Total MLR culture volume was 200 μl after the addition of test macrophage cells or media. On day 4 of the MLR, cultures were assessed for proliferation by measuring the [3H]thymidine incorporation into DNA following a 6 hour pulse. Percent proliferation impacted by macrophage co-culture was assessed relative to untreated control responses of B6 responders alone to BALB/c stimulator cells.
Western blots
Protein lysates were separated by tris-HCl-buffered 10% SDS-polyacrylamide gel electrophoresis following by transfer to PVDF membranes. Membranes were blocked in 5% milk in PBS containing 0.1% Tween and incubated in the appropriate antibodies against the indicated proteins. GAPDH expression was used as a loading control. Anti-mouse Arg1 antibody (V-20) was obtained from Santa Cruz; anti-mouse GAPDH antibody (clone 71.1) was obtained from Sigma-Aldrich. Membranes were imaged on a Chemi-Dock XRS Plus imager (Bio-Rad) and bands for Arg1 and GAPDH were quantified using Bio-Rad ImageLab software.
Histological analysis
At the times indicated, mice were sacrificed, perfused by intracardial injection of 1× PBS, and the indicated tissues were dissected and fixed in 4% paraformaldehyde, pH 7.3. Tissues were embedded in paraffin and 5-μm sections were prepared. To assess histopathological changes such as tissue inflammation and damage, tissue sections were stained with hematoxylin and eosin (H&E) and evaluated by light microscopy. In all mice, presence, distribution, and severity of histologic lesions were blindly scored by two pathologists. Lesions consisted primarily of myocyte necrosis, myocte regeneration, inflammation, edema, and fibrosis. For all tissue changes, a scoring system was developed as follows: 0, absent; 1, minimal, less than 10% of tissue affected; 2, mild, 10–24% of tissue affected; 3, moderate, 25–39% of tissue affected; 4, marked, 40–59% of tissue affected; 5, severe, greater than 60% of tissue affected. Scores of histologic lesions for individual mice were plotted for analysis.
Immunohistochemistry
For immunohistochemical analysis of Arg1 expression, deparaffinized slides were blocked in normal serum (Vectastain ABC Kit, Vector Labs) for 20 min, then incubated in anti-Arg1 Ab (H-52, Santa Cruz) for one hour, followed by incubation in biotinylated secondary antibody for 30 min. For resolution of the staining, slides were incubated in Vectastain ABC reagent and developed in DAB Peroxidase Substrate Solution (Vector Labs). Slides were counter-stained with Mayer's hematoxylin (ThermoScientific).
Statistical analysis
All data were analyzed using GraphPad Prism 5 software. Data were evaluated for statistically significant differences using either a two-tailed, unpaired t test with or without Welch's correction or an analysis of variance (ANOVA) test followed by Tukeys's multiple comparison test. A P value < 0.05 was considered statistically significant. All differences not specifically indicated to be significant were not significant (P > 0.05).
RESULTS
Arg1 is highly expressed in inflammatory lesions of Ross River virus-infected mice
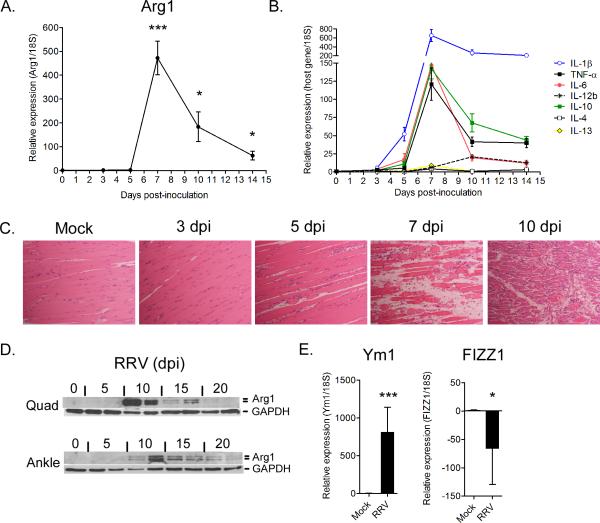
To study the pathogenesis of arthritogenic alphaviruses, we have developed mouse models of RRV- and CHIKV-induced disease where the major pathological outcomes, arthritis, myositis, and tenosynovitis, are consistent with the disease signs experienced by infected humans (26–28). To determine the extent to which the inflammation and damage to musculoskeletal tissues caused by infection with arthritogenic alphaviruses activates genes associated with wound repair or M2 macrophages, total RNA was isolated from quadriceps muscle tissue and ankle/foot tissues (data not shown) of PBS- or RRV-inoculated mice at various days post-inoculation (dpi) and analyzed by real-time RT-qPCR for the expression of Arg1, a signature gene of M2 macrophage activation in mice (Fig. 1A). At early times post-inoculation (3 and 5 dpi), Arg1 expression was undetectable or low in skeletal muscle tissue. At 7 dpi, a time point associated with the development of extensive skeletal muscle tissue inflammation and injury (28), the relative abundance of Arg1 transcripts increased dramatically relative to 18S rRNA expression (472-fold increase over mock-inoculated mice, P = 0.0005). Arg1 transcripts remained significantly elevated at 10 and 14 dpi (183-fold increase, P = 0.02 and 63-fold increase, P = 0.03 over mock-inoculated mice, respectively). Similar results were obtained when Arg1 expression data were normalized to HPRT expression (data not shown). To correlate the temporal induction of Arg1 with corresponding tissue inflammation and injury, we analyzed the same RNA samples for the expression of a variety of cytokines (Fig. 1B) and tissues sections derived from quadriceps muscles of uninfected (mock) or RRV-infected mice were H & E stained (Fig. 1C). The expression of IL-1β, TNF-α, IL-6, and IL-10 mRNAs exhibited an expression pattern similar to Arg1, with relatively low expression at 3 and 5 dpi (except for IL-1β mRNA which was high at 5 dpi), peak expression occurring at 7 dpi, and expression levels remaining elevated at both 10 and 14 dpi (Fig. 1B). In contrast, the relative expression of IL-4 and IL-13 transcripts was very low or undetectable at all time points examined. The expression of IL-12b mRNA in this tissue exhibited a distinct pattern, with very low expression until 10 dpi (Fig. 1B). The expression of Arg1, IL-1β, TNF-α, IL-6, and IL-10 correlated with cellular infiltration and damage of skeletal muscle tissue, which initiates at 5 dpi and is most prominent at 7, 10, and 14 dpi (Fig. 1C). Consistent with the RT-qPCR analyses, abundant Arg1 protein was detected by immunoblot analysis in both quadriceps muscle tissue and ankle joint tissue at 10 dpi and was still detectable at 20 dpi (Fig. 1D). These data demonstrate that Arg1 expression in tissues of RRV-infected mice correlates temporally with musculoskeletal tissue inflammation and injury. In addition, the expression of Arg1 correlated with the expression of IL-1β, TNF-α, IL-6, and IL-10, but not IL-4, IL-13, or IL-12b.
Figure 1. Arginase 1 is induced in inflamed musculoskeletal tissues of RRV-infected mice.
Three-four week-old C57BL/6J mice were mock-inoculated (0 dpi) or inoculated with 103 PFU of RRV in the left rear footpad. (A) RT-qPCR analysis of Arg1 mRNA expression in quadriceps muscles of mock-inoculated (n = 5) or RRV-inoculated mice at 3 (n= 5), 5 (n = 4), 7 (n = 7), 10 (n = 7), and 14 dpi (n = 5). Data are normalized to 18S rRNA levels and are expressed as the relative expression (n-fold increase) over Arg1 expression in quadriceps muscles of mock-inoculated mice. Each data point represents the arithmetic mean ± SEM. ***P < 0.001, *P < 0.05 as determined by unpaired t-tests with Welch's correction. (B) RT-qPCR analysis of IL-1β, TNF-α, Il-6, IL-12b, IL-10, IL-4, and IL-13 mRNA expression in quadriceps muscles of mock-inoculated (n = 5) or RRV-inoculated mice at 3 (n= 5), 5 (n = 4), 7 (n = 7), 10 (n = 7), and 14 dpi (n = 5). Data are normalized to 18S rRNA levels and are expressed as the relative expression (n-fold increase) over expression in quadriceps muscles of mock-inoculated mice. Each data point represents the arithmetic mean ± SEM. (C) 5-μm paraffin-embedded sections were generated from the quadriceps muscles of mock or RRV-infected mice at 3, 5, 7, and 10 dpi and H & E stained. Images are representative of 3–4 mice per group. (D) Immunoblot analysis of Arg1 expression in quadriceps muscles and ankle joint tissues of mock-inoculated (0 dpi) or RRV-inoculated mice at 5, 10, 15, and 20 dpi. GAPDH was used as a loading control. n = 2 mice per time point. (E) RT-qPCR analysis of Ym1 and FIZZ mRNA expression in quadriceps muscles of mock-inoculated (n = 5) or RRV-inoculated mice at 7 dpi (n = 7). Data are normalized to 18S rRNA levels and are expressed as the relative expression (n-fold increase) over expression in quadriceps muscles of mock-inoculated mice. Each data point represents the arithmetic mean ± SEM. ***P < 0.001, *P < 0.05 as determined by unpaired t-tests with Welch's correction.
In addition to Arg1, Ym1/Chi3l3 and FIZZ1/Relm-α are well-established markers of canonical M2 macrophage activation (65). Similar to Arg1 (Fig. 1A), we detected high levels of Ym1 expression in whole quad tissues of RRV-infected mice at 7 dpi (807-fold increase over mock-inoculated mice, P = 0.0007) (Fig. 1E). In contrast, FIZZ1 expression in the same tissues was actually reduced in RRV-infected mice relative to control mice (66-fold decrease compared to mock-inoculated mice, P = 0.03) (Fig. 1E). These findings suggest that at least a subset of macrophages infiltrating these tissues have a unique M2-like phenotype characterized by abundant expression of Arg1 and Ym1 in the absence of FIZZ1.
Arg1 is induced in CD11b+F4/80+ musculoskeletal tissue infiltrates
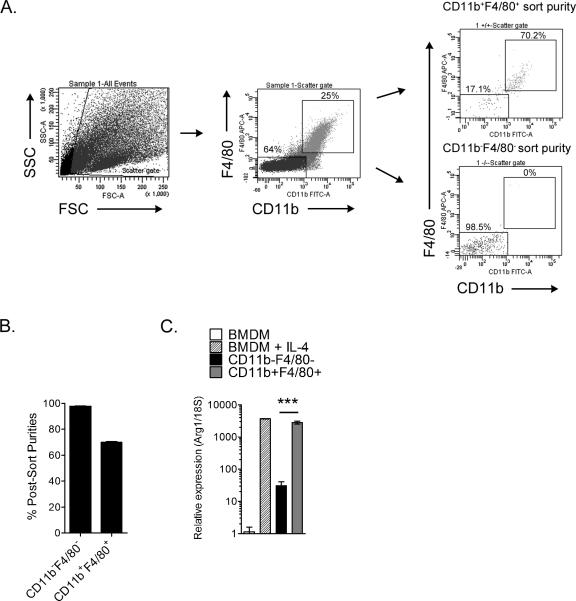
To identify Arg1-expressing cell type(s) within the musculoskeletal inflammatory lesions, we enzymatically-digested quadriceps muscles at 7 days post-RRV inoculation, isolated leukocytes, and FACS-sorted the CD11b+F4/80+ macrophage population and the CD11b−F4/80− cell population (Fig. 2A and 2B). RT-qPCR analysis of Arg1 expression in these different cell populations was compared to Arg1 expression in unstimulated and IL-4-stimulated bone marrow-derived macrophages (BMDMs) (Fig. 2C), as IL-4 is a cytokine known to strongly induce Arg1 expression in macrophages (66). As expected, IL-4 stimulation strongly induced Arg1 expression in BMDMs (Fig. 2C). The CD11b+F4/80+ macrophages sorted from inflamed quadriceps tissues of RRV-infected mice had high expression of Arg1 relative to the unstimulated BMDMs and significantly higher expression of Arg1 compared to the CD11b−F4/80−cells (92-fold increase over CD11b−F4/80− cells, P = 0.0028) (Fig. 2C). These findings suggest that at least a subset of the CD11b+F4/80+ macrophages are a major source of Arg1 expression in the inflamed musculoskeletal tissues of RRV-infected mice.
Figure 2. Tissue-infiltrating macrophages express Arginase 1.
Three-to-four week-old C57BL/6J mice were inoculated with 103 PFU of RRV in the left rear footpad. At 7 dpi, quadriceps muscles were enzymatically digested and CD11b−F4/80− and CD11b+F4/80+ cellular infiltrates were FACS-sorted. (A) Shown are representative dot plots of total cells, double-positive and double-negative populations, along with their post-sort purities. (B) Post-sort purities of CD11b−F4/80− (n = 4) and CD11b+F4/80+ (n = 4) populations. Each bar represents the arithmetic mean ± SEM. (C) RT-qPCR analysis of Arg1 mRNA expression in unstimulated or IL-4 stimulated (5 ng/ml) bone marrow-derived macrophages (BMDMs) (n = 3), FACS-sorted CD11b−F4/80− cells (n =4), and FACS-sorted CD11b+F4/80+ cells (n = 4). Data are normalized to 18S rRNA levels and are expressed as the relative expression (n-fold increase) over Arg1 expression in unstimulated BMDMs. Each data point represents the arithmetic mean ± SEM. ***P = 0.0028 as determined by an unpaired t-test with Welch's correction.
Interestingly, Arg1-expressing myeloid cells have been demonstrated to have immune suppressive activity (53, 54, 67). To test whether the Arg1-expressing CD11b+F4/80+ macrophages present in the inflamed musculoskeletal tissues of RRV-infected mice have functional properties consistent with Arg1-expressing myeloid cells, FACS-sorted CD11b+F4/80+ macrophages isolated from quadriceps muscles at 10 days post-RRV inoculation were evaluated for the ability to suppress T cell proliferation ex vivo in response to allogeneic (BALB/c) stimulator cells in a standard mixed leukocyte reaction (MLR). As shown in Fig. 3, CD11b+F4/80+ muscle tissue macrophages impaired T cell proliferation in a dose-dependent manner.
Figure 3. Macrophages isolated from musculoskeletal tissues suppress T cell proliferation ex vivo by a mechanism that is partially Arg1-dependent.
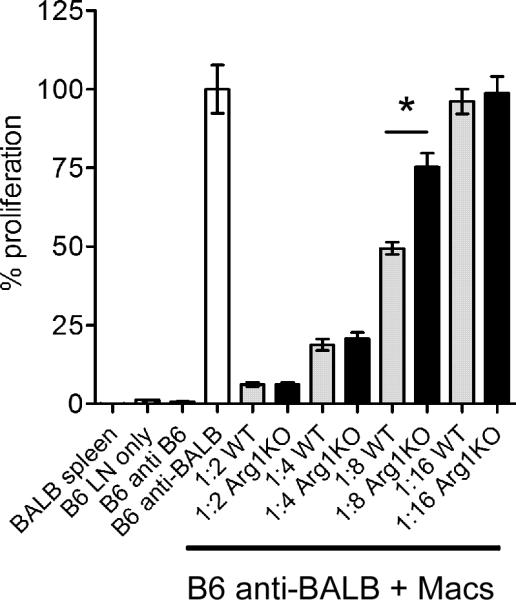
Three-to-four week-old C57BL/6J mice (n = 3) or LysMcre;Arg1F/F mice (n = 3) were inoculated with 103 PFU of RRV in the left rear footpad. At 10 dpi, quadriceps muscles were enzymatically digested and CD11b+F4/80+ cellular infiltrates were FACS-sorted and analyzed for immune suppressive activity in a mixed leukocyte reaction. C57BL/6J lymph node cells and irradiated BALB/c splenocytes were incubated in the absence or presence of FACS-sorted CD11b+F4/80+ cells at 1:2, 1:4, 1:8, and 1:16 ratios in the presence of tritiated thymidine. Control wells contained irradiated BALB/c splenocytes only, C57BL/6J lymph node cells only, or C57BL/6J lymph node cells incubated in the presence of C57BL/6J irradiated splenocytes. Thymidine incorporation was quantified after 4 days. Each data point represents the arithmetic mean ± SEM and is representative of two independent experiments. *P < 0.05 as determined by ANOVA followed by Tukey's multiple comparison test.
Arg1 is highly expressed in inflammatory lesions of chikungunya virus-infected mice
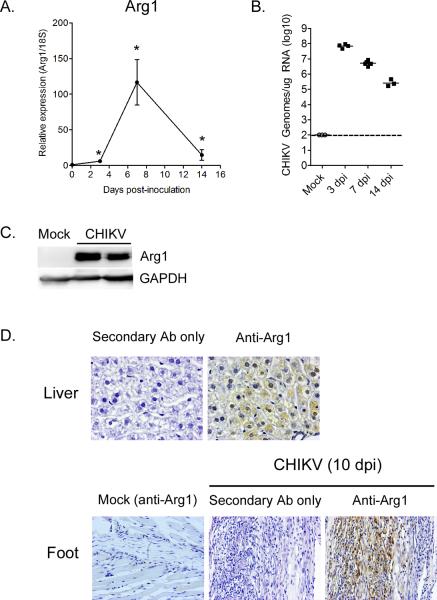
We recently developed a mouse model of CHIKV-induced musculoskeletal inflammatory disease based on subcutaneous inoculation of mice with a low passage clinical isolate of CHIKV (27). We utilized this model to determine the extent to which Arg1 induction in musculoskeletal inflammatory lesions is unique to RRV infection or if it is common to arthritogenic alphavirus infection. Relative to uninfected mice, Arg1 mRNA expression in the left ankle/foot of CHIKV-infected mice was relatively low at 3 dpi (5.9-fold increase over mock-inoculated mice, P = 0.024), dramatically induced at 7 dpi (116.5-fold increase over mock-inoculated mice, P = 0.02), and remained significantly elevated at 14 dpi (14.6-fold increase over mock-inoculated mice, P = 0.012) (Fig. 4A). CHIKV RNA loads in the left ankle/foot at these time points were highest at 3 dpi in comparison to 7 and 14 dpi (Fig. 4B). Thus, similar to RRV-infected mice, the expression of Arg1 correlated with injury and infiltration of CHIKV-infected left ankle/foot tissues with CD11b+F4/80+ macrophages, neutrophils, Natural Killer (NK) cells, and T cells, which is low at 3 dpi and high at 7 dpi as previously reported (27). Consistent with the RT-qPCR analyses, Arg1 protein was highly expressed at 10 dpi in ankle/foot tissue of CHIKV-infected mice (Fig. 4C). In addition, Arg1 protein was readily detected by immunohistochemistry in tissue sections generated at 10 dpi from the ankle/foot of CHIKV-infected mice but not mock-inoculated control mice (Fig. 4D). These findings indicate that high Arg1 expression in inflamed musculoskeletal tissues is a common characteristic of arthritogenic alphavirus infection.
Figure 4. Arginase 1 is induced in inflamed musculoskeletal tissues of CHIKV-infected mice.
Three-to-four week old C57BL/6J mice were mock-inoculated (0 dpi) or inoculated with 103 PFU of CHIKV in the left rear footpad. (A) RT-qPCR analysis of Arg1 mRNA expression in the left ankle/foot tissue of mock-inoculated (n = 5) or CHIKV-inoculated mice at 3 (n= 6), 7 (n = 6), and 14 dpi (n = 8). Data are normalized to 18S rRNA levels and are expressed as the relative expression (n-fold increase) over Arg1 expression in left ankle/foot tissue of mock-inoculated mice. Each data point represents the arithmetic mean ± SEM. *P < 0.05 as determined by unpaired t-tests with Welch's correction. (B) CHIKV genomes in the left foot/ankle of mock or CHIKV-infected mice at 3, 7, and 14 dpi were quantified by absolute RT-qPCR as described in the materials and methods. Horizontal bars indicate the mean and the dashed line indicates the limit of detection. (C) Immunoblot analysis of Arg1 expression in ankle/foot tissues of mock-inoculated or CHIKV-inoculated mice at 10 dpi. GAPDH was used as a loading control. Data are representative of two independent experiments. (D) 5-μm sections generated from the liver (upper panel) or ankle/foot (lower panel) at 10 dpi were stained for Arg1 expression by immunohistochemistry. Images are representative of three mice per group.
LysM-Cre mediated deletion of Arg1 ablates RRV and CHIKV-induced Arg1 expression
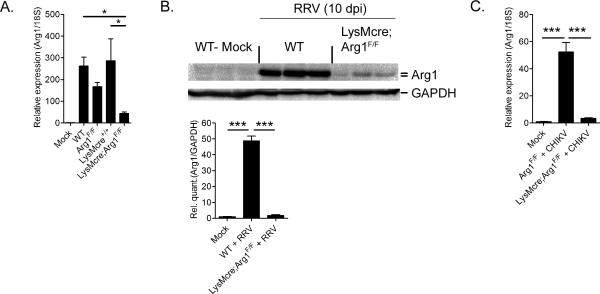
Due to the established role of myeloid cell Arg1 activity in wound repair and regulation of immune and inflammatory responses (50), we sought to specifically investigate the role of myeloid cell Arg1 during infection with arthritogenic alphaviruses. We bred Lysozyme M Cre-expressing mice (LysMcre) (62) with mice possessing loxP sites on either side of exons 7 and 8 in the Arg1 gene (Arg1F/F) (52) to generate mice in which Arg1 expression is specifically ablated from macrophages and neutrophils (LysMcre;Arg1F/F), as previously reported (52). In contrast to BMDMs generated from Arg1F/F mice, Arg1 protein expression was dramatically reduced in BMDMs from LysMcre;Arg1F/F mice following treatment with IL-4 (Fig. S1). Importantly, LysMcre;Arg1F/F mice expressed Arg1 to similar levels in the liver as control mice (Fig. S1), indicating that this conditional KO deletes Arg1 specifically from myeloid cells. We next tested the extent to which genetic ablation of Arg1 in macrophages and neutrophils prevented the induction of Arg1 at the sites of inflammation following RRV infection. Compared to RRV-infected Arg1-sufficient mice, RRV-infected LysMcre;Arg1F/F mice had significantly reduced Arg1 mRNA (Fig. 5A) and protein (Fig. 5B) levels in quadriceps muscles at 7 and 10 dpi, respectively. Similarly, Arg1 expression at 7 dpi was dramatically reduced in the inflamed ankle/foot tissue of CHIKV-infected LysMcre;Arg1F/F mice compared to Arg1-sufficient mice (Fig. 5C). These data suggest that infiltrating macrophages and/or neutrophils are the predominant source of Arg1 expression in the musculoskeletal tissues of RRV and CHIKV-infected mice.
Figure 5. Expression of Arginase 1 in inflamed tissues of RRV- or CHIKV-infected mice is ablated in LysMcre;Arg1F/F mice.
(A) RT-qPCR analysis of Arg1 mRNA expression in quadriceps muscles of mock (n = 3) or RRV-infected WT (n = 7), Arg1F/F (n = 6), LysMCre+/+ (n = 4), and LysMcre;Arg1F/F (n = 5) mice at 7 dpi. Data are normalized to 18S rRNA levels and are expressed as the relative expression (n-fold increase) over Arg1 expression in quadriceps tissue of mock-inoculated mice. Each data point represents the arithmetic mean ± SEM. *P < 0.05 as determined by ANOVA followed by Tukey's multiple comparison test. (B) Immunoblot analysis of Arg1 protein expression in quadriceps muscles of uninfected (mock) or RRV-infected WT and LysMcre;Arg1F/F mice at 10 dpi (n = 3 mice/group). GAPDH was used as a loading control. Arg1 and GAPDH band intensities were quantified and Arg1 expression was normalized to GAPDH and expressed as the fold increase over Arg1 expression in mock-infected mice. ***P = < 0.001 as determined by ANOVA followed by Tukey's multiple comparison test. (C) RT-qPCR analysis of Arg1 mRNA expression in the left ankle/foot tissue of mock-inoculated Arg1-sufficient mice (n = 5), CHIKV-inoculated Arg1F/F mice (n= 6), or CHIKV-inoculated LysMcre;Arg1F/F (n = 6) at 7 dpi. Data are normalized to 18S rRNA levels and are expressed as the relative expression (n-fold increase) over Arg1 expression in left ankle/foot tissue of mock-inoculated mice. Each data point represents the arithmetic mean ± SEM and are cumulative from two independent experiments. ***P < 0.001 as determined by ANOVA followed by Tukey's multiple comparison test.
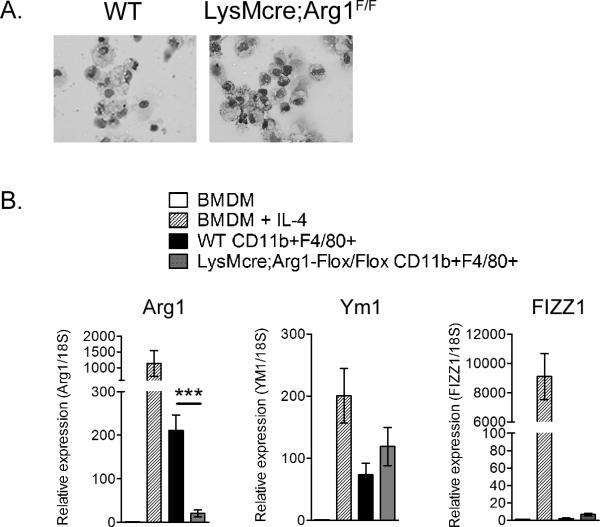
Previous studies in humans and mice, including differential cell counts on Giemsa-stained cytospins of collagenase-digested murine quadriceps muscles at various times post-RRV infection [a procedure that has been successfully used to isolate viable neutrophils (68)], have demonstrated that musculoskeletal tissue infiltrates associated with RRV infection are primarily mononuclear (26, 28, 33, 58, 69–72). To confirm that LysMcre-mediated ablation of Arg1 prevented Arg1 expression in macrophages at the sites of RRV-induced musculoskeletal tissue inflammation and injury, we isolated inflammatory infiltrates from enzymatically-digested quadriceps muscles at 10 days post-RRV inoculation and FACS-sorted CD11b+F4/80+ macrophages. As shown in Fig. 6A, the CD11b+F4/80+ cells FACS-sorted from WT [avg. 82% purity (range 75%–95%)] and LysMcre;Arg1F/F mice [avg. 82% purity (range 77%–85%)] were morphologically indistinguishable. RT-qPCR analysis of Arg1 expression in these cells was compared to Arg1 expression in unstimulated and IL-4-stimulated bone marrow-derived macrophages (BMDMs) (Fig. 6B). Again, CD11b+F4/80+ macrophages sorted from inflamed quadriceps tissues of RRV-infected WT mice had high expression of Arg1 relative to the unstimulated BMDMs (Fig. 6B). In contrast, CD11b+F4/80+ macrophages FACS-sorted from LysMcre;Arg1F/F mice had significantly reduced Arg1 expression compared to CD11b+F4/80+ macrophages FACS-sorted from control mice (Fig. 6B), indicating that Arg1 is efficiently deleted from the tissue infiltrating inflammatory macrophages in LysMcre;Arg1F/F mice. These findings further confirm that one major source of Arg1 expression in the musculoskeletal inflammatory lesions of RRV-infected mice is tissue-infiltrating macrophages. To test whether deletion of Arg1 from CD11b+F4/80+ infiltrates had an impact on functionality, we tested the ability of FACS-sorted CD11b+F4/80+ macrophages from control mice and LysMcre;Arg1F/F mice to suppress T cell proliferation in an MLR and found that suppressive activity was, in part, Arg1-dependent (Fig. 3).
Figure 6. Gene expression of CD11b+F4/80+ tissue-infiltrating macrophages.
(A) Wright-Giemsa staining of FACS-sorted CD11b+F4/80+ cells isolated from quadriceps muscles of RRV-infected WT or LysMcre;Arg1F/F mice at 10 dpi. (B) RT-qPCR analysis of Arg1, Ym1, and FIZZ1 expression in unstimulated or IL-4 stimulated (5 ng/ml) bone marrow-derived macrophages (BMDMs) (n = 3) and FACS-sorted CD11b+F4/80+ cells isolated from quadriceps muscles of RRV-infected WT (n =4) or LysMcre;Arg1F/F mice (n = 4) at 10 dpi. Data are normalized to 18S rRNA levels and are expressed as the relative expression (n-fold increase) over expression in unstimulated BMDMs. Each data point represents the arithmetic mean ± SEM. ***P = 0.007 as determined by unpaired, two-tailed t-tests with Welch's correction.
Our analysis of gene expression in whole tissue had revealed a gene expression pattern characterized by high Arg1 and Ym1 in the absence of FIZZ1 (Fig.1). To explore the extent to which this gene expression pattern reflected the gene expression pattern of tissue-infiltrating macrophages, and the extent to which deletion of Arg1 impacted the expression of these genes, we quantified the expression of Ym1 and FIZZ1 in FACS-sorted CD11b+F4/80+ cells. CD11b+F4/80+ macrophages sorted from RRV-infected WT mice and LysMcre;Arg1F/F mice had similar expression levels of Ym1 and little to no expression of FIZZ1 relative to the unstimulated BMDMs (Fig. 6B). These results indicate that deletion of Arg1 did not alter expression of other genes typically associated with the polarization of murine macrophages towards an M2 phenotype and suggest that at least some of the macrophages infiltrating musculoskeletal tissues of RRV-infected mice have a unique M2-like gene expression pattern.
Impact of LysM-Cre mediated deletion of Arg1 on acute RRV disease
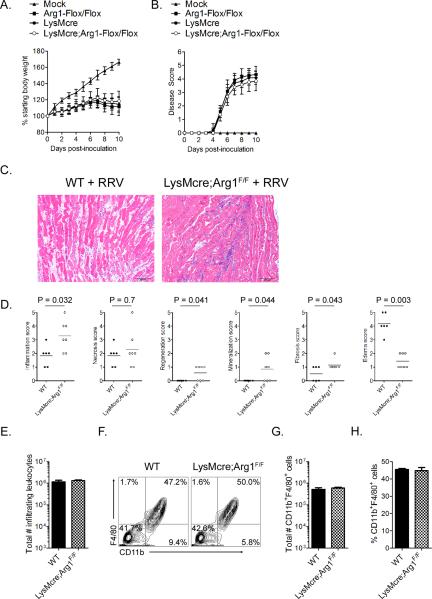
To assess the effects of LysMcre-specific ablation of Arg1 on acute RRV-induced disease (days 1–10 post-infection), LysMcre;Arg1F/F mice and control Arg1-sufficient mice were inoculated with virus diluent only (mock) or RRV and evaluated for multiple parameters of acute disease. Control mice and LysMcre;Arg1F/F mice had no significant differences in RRV-induced effects on weight gain (Fig. 7A) or the development of musculoskeletal disease as evidenced by defects in hind limb gripping ability and an altered gait (Fig. 7B). Histologic changes in skeletal muscle tissue of infected mice during this stage, evaluated in a blinded manner as described in the material and methods, varied in severity but was generally characterized by acute myofiber necrosis accompanied by edema and inflammation (Fig. 7C and 7D). At 7 dpi, necrotic myofibers were often homogeneously bright eosinophilic with loss of cross-striations, fragmented sarcoplasm and pyknotic or karyorrhectic nuclei (Fig. 7C). Fine basophilic granular sarcoplasmic mineralization was frequently observed. Accompanying acutely injured myofibers was tissue edema, which was characterized by separation of tissue elements by clear spaces. Inflammatory infiltrates ranged in severity and cell type but were predominated by macrophages with many fewer admixed neutrophils. The right gastrocnemius muscles of mock-infected mice were all scored as 0 for all of the histologic changes studied (data not shown). Inflammation scores were elevated in RRV-infected LysMcre;Arg1F/F mice compared to RRV-infected WT mice (Fig. 7D, panel 1, P = 0.03), suggesting that the absence of Arg1 in macrophages and neutrophils may have increased the distribution of inflammation throughout the tissue. In contrast, edema was significantly more prevalent in tissues of RRV-infected WT mice compared to RRV-infected LysMcre;Arg1F/F mice (Fig. 7D, panel 6, P = 0.003). As expected, histopathology scores for regeneration, mineralization, and fibrosis, which are typically associated with the subacute/chronic stages of tissue inflammation, were low in all mice at 7 dpi (Fig. 7D).
Figure 7. Acute RRV-induced disease is unaltered in LysMcre;Arg1F/F mice.
Three-to-four week-old Arg1F/F (n = 24), LysMCre+/+ (n = 25), or LysMcre;Arg1F/F (n = 29) mice were inoculated with 103 PFU of RRV by injection in the left rear footpad and assessed for (A) weight gain and (B) musculoskeletal disease signs including loss of gripping ability and altered gait at 24-hour intervals. Each data point represents the arithmetic mean ± standard deviation (SD). Data are combined from 6–8 independent experiments. No significant differences were detected as determined by repeated measures ANOVA. (C) At 7 dpi, 5-μm paraffin-embedded sections were generated from the gastrocnemius muscle and stained with H&E. Images are representative of 5–7 mice per group. (D) H&E stained sections from 7 dpi were scored in a blinded manner for the degree of inflammation, necrosis, regeneration, mineralization, fibrosis, and edema based on the following scale for percent of tissue affected: 0: absent (0%), 1: minimal (<10%), 2: mild (11–25%), 3: moderate (26–40%), 4: marked (41–60%), and 5: severe (>60%). P values were determined by Mann-Whitney tests. (E) At 7 dpi, infiltrating leukocytes were isolated from enzymatically digested quadriceps muscles of RRV-infected WT (n = 6) or LysMcre;Arg1F/F (n = 8) mice as described in material and methods and the total cell number was determined. Each bar represents the arithmetic mean ± SEM from two independent experiments. No statistically significant differences were detected by an unpaired t-test. (F) Total leukocytes isolated from enzymatically digested quadriceps muscles of RRV-infected WT (n = 6) or LysMcre;Arg1F/F (n = 8) mice at 7 dpi were analyzed by flow cytometry for the expression of CD11b and F4/80. Shown are representative contour plots. (G) Total number and (H) percentages of CD11b+F4/80+ cells isolated from enzymatically digested quadriceps muscles of RRV-infected WT (n = 6) or LysMcre;Arg1F/F (n = 8) mice at 7 dpi. Each bar represents the arithmetic mean ± SEM from two independent experiments. No statistically significant differences were detected by unpaired t-tests.
The assessment of histologic changes in skeletal muscle tissue suggested that LysMcre-mediated deletion of Arg1 may have altered cellular infiltration of tissues during the acute stage of disease. Furthermore, macrophages have been implicated in functioning as pathogenic effectors during the early acute stage of RRV and CHIKV-induced disease (57, 58, 70). To determine whether the number of macrophages or other cellular infiltrates in muscle tissue was altered in LysMcre;Arg1F/F mice, the total numbers of infiltrating leukocytes and CD11b+F4/80+ macrophages in musculoskeletal tissue at 7 dpi were quantified by flow cytometry. Similar total numbers of inflammatory infiltrates (Fig. 7E), as well as the total number and percentages of CD11b+F4/80+ macrophages (Fig. 7F–7H) were isolated from inflamed quadriceps muscles of RRV-infected WT mice and LysMcre;Arg1F/F mice. These findings indicate that genetic ablation of Arg1 in macrophages and neutrophils does not alter the initial recruitment of macrophages to the sites of inflammation that is associated with the development of acute RRV-induced musculoskeletal disease.
LysM-Cre mediated deletion of Arg1 enhances clearance of RRV from musculoskeletal tissues and reduces tissue pathology at late times post-infection
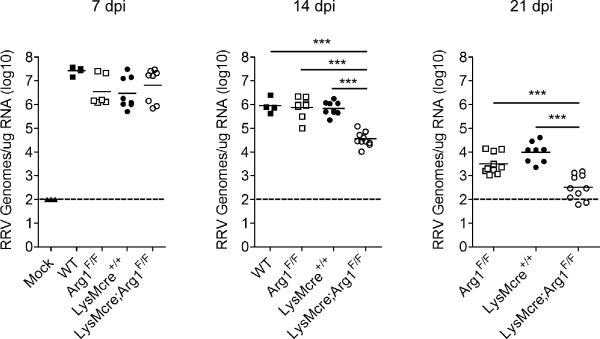
Arg1 activity in tumor-associated macrophages, MDSCs, and macrophages responding to parasitic infections function in immune suppression (50). We found that CD11b+F4/80+ cells FACS-sorted from inflamed muscle tissue suppressed T cell proliferation ex vivo by a mechanism that was, in part, Arg1-dependent (Fig. 3). Thus, we hypothesized that the high levels of Arg1 expression detected at the sites of musculoskeletal tissue inflammation and damage would inhibit the clearance of RRV from these sites. To investigate this hypothesis, control or LysMcre;Arg1F/F mice were inoculated with RRV and viral burdens in quadriceps muscles at 7, 14, and 21 dpi were quantified by RT-qPCR (Fig. 8). At 7 dpi, RRV RNA levels were similar in infected control and LysMcre;Arg1F/F mice, further confirming that specific ablation of Arg1 in macrophages and neutrophils does not substantially affect acute RRV replication or disease. In contrast, LysMcre;Arg1F/F mice had dramatically reduced RRV RNA levels at 14 dpi (21-fold decrease, P < 0.001) and 21 dpi (17-fold decrease, P < 0.001) compared to control mice. These findings indicate that specific ablation of Arg1 in macrophages and neutrophils enhances virus clearance from damaged and inflamed tissues in the context of infection with RRV.
Figure 8. Specific deletion of Arg1 in macrophages enhances clearance of RRV from musculoskeletal tissues.
Three-to-four week-old C57BL/6J WT (7 dpi n = 4; 14 dpi n = 4), Arg1F/F (7 dpi n = 6; 14 dpi n = 8; 21 dpi n = 10), LysMCre+/+ (7 dpi n = 8; 14 dpi n = 9; 21 dpi n = 8), and LysMcre;Arg1F/F (7 dpi n = 8; 14 dpi n = 11; 21 dpi n = 10) mice were inoculated with 103 PFU of RRV by injection in the left rear footpad. At 7, 14, and 21 dpi, mice were sacrificed, perfused by intracardial injection with 1× PBS, and total RNA was isolated from the right quadriceps muscles. RRV genomes were quantified by absolute RT-qPCR as described in the materials and methods. Horizontal bars indicate the mean and dashed lines indicate the limit of detection. *** P ≤ 0.001 as determined by ANOVA followed by Tukey's multiple comparison test.
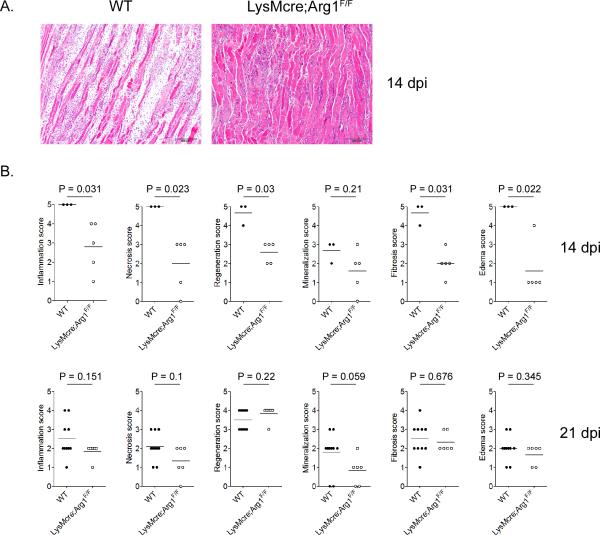
We next investigated the extent to which the genetic deletion of Arg1 and the enhanced clearance of RRV in LysMcre;Arg1F/F mice were associated with alterations in tissue pathology. Tissue sections generated from the gastrocnemius muscles of uninfected mice and RRV-infected mice at 14 and 21 dpi were H & E stained and evaluated in a blinded manner as described in the materials and methods. The right gastrocnemius muscles of mock-infected mice were all scored as 0 by blinded pathologists for all of the histologic changes studied (data not shown). Histologic changes in infected mice at these time points consisted of chronic changes of myofiber regeneration, inflammation and fibrosis. In contrast to 7 dpi, inflammatory infiltrates consisted of increased numbers of lymphocytes and plasma cells, in addition to numerous macrophages, visualized with areas of fibrosis and myofiber repair. Fibrosis varied in severity and was typically observed as expansion of the interstitium by proliferating or hypertrophied fibroblasts producing scant mature collagen. Regenerating myofibers were observed as indicated by hypertrophied myocytes with centralized nuclei which were arranged in rows. At 14 dpi, inflammation, necrosis, regeneration, fibrosis, and edema were significantly more severe in RRV-infected WT mice compared to LysMcre;Arg1F/F mice (Fig. 9A and 9B), which correlated with the higher viral loads detected in Arg1-sufficient mice (Fig. 8). By 21 dpi, no statistically significant differences in muscle tissue pathology were observed in RRV-infected WT mice compared to RRV-infected LysMcre;Arg1F/F mice (Fig. 9B). Taken together, these findings suggest that the Arg1-dependent inhibition of RRV clearance by tissue-infiltrating myeloid cells results in an extended duration of high viral loads and associated tissue pathology.
Figure 9. Inflammatory tissue pathology at late times following RRV infection.
Three week-old C57BL/6J WT, Arg1F/F, LysMCre+/+, and LysMcre;Arg1F/F mice were mock-inoculated or inoculated with 103 PFU of RRV by injection in the left rear footpad. At 14 and 21 dpi, 5-μm paraffin-embedded sections were generated from the gastrocnemius muscle and stained with H&E. (A) At 14 dpi, 5-μm paraffin-embedded sections were generated from the gastrocnemius muscle and stained with H&E. Images are representative of 5–7 mice per group. (B) H&E stained sections from 14 and 21 dpi were scored in a blinded manner for the degree of inflammation, necrosis, regeneration, mineralization, fibrosis, and edema based on the following scale for percent of tissue affected: 0: absent (0%), 1: minimal (<10%), 2: mild (11–25%), 3: moderate (26–40%), 4: marked (41–60%), and 5: severe (>60%). P values were determined by Mann-Whitney tests.
DISCUSSION
In this study, we discovered that the musculoskeletal inflammatory lesions that develop during infection with RRV and CHIKV are characterized by abundant expression of Arg1 in tissue-infiltrating macrophages. Arg1 expression and activity in myeloid cells has emerged as a major regulator of anti-tumor immune responses, wound repair, fibrosis, and host responses to parasitic infections (49–51, 73). However, few studies have evaluated the function of Arg1 during virus infections. Here, we show that specific ablation of Arg1 in macrophages and neutrophils dramatically enhances the clearance of RRV from musculoskeletal tissues and diminishes muscle tissue pathology at late times post-RRV infection, establishing a critical role for myeloid cell Arg1 in the pathogenesis of these infections.
Previous studies in humans and mice demonstrated that musculoskeletal tissue infiltrates associated with RRV infection are primarily mononuclear (26, 28, 33, 58, 69–72), and studies of RRV and CHIKV infection in mice have implicated macrophages in contributing to disease severity. Treatment of mice with macrophage toxic agents prior to virus inoculation (57, 58, 70), or inhibition of macrophage recruitment (74, 75), reduced acute disease signs associated with RRV or CHIKV infection. These studies suggested that macrophages function as pathogenic effectors during the acute stage of these virus-induced inflammatory diseases; however, specific macrophage activation programs and effector mechanisms that mediate these effects have not been defined.
Macrophages have heterogenous phenotypes and may exert various, even opposite, functions depending on their activation state and anatomical location (34, 35). We hypothesized that RRV/CHIKV infection and the associated inflammation and tissue damage activates a wound healing response characterized by activation of M2 or M2-like macrophages. Indeed, our gene expression studies revealed a dramatic increase in Arg1 and Ym1 expression coordinate in time with peak inflammatory tissue pathology, suggesting that tissue damage may drive the polarization of at least some of the macrophages infiltrating these tissues toward an M2-like phenotype. Consistent with this notion, gene expression analyses of FACS-sorted muscle tissue infiltrates from RRV-infected WT mice indicated that CD11b+F4/80+ macrophages expressed abundant Arg1 and Ym1. In addition, LysMcre-mediated deletion of Arg1 dramatically reduced Arg1 expression in inflamed musculoskeletal tissues of RRV and CHIKV-infected mice. Taken together, these data suggest that macrophages and/or neutrophils are the predominant Arg1-expressing cells at the sites of inflammation in mice infected with arthritogenic alphaviruses.
Interestingly, the Arg1-expressing macrophages present in RRV-induced musculoskeletal inflammatory lesions of WT C57BL/6 mice arise in an inflammatory environment with little to no expression of IL-4 or IL-13 mRNA. Furthermore, based on their gene expression pattern, these cells do not appear to fit into the canonical definition of M2/alternatively activated macrophages which are defined as macrophages that have been stimulated with IL-4/IL-13 via the IL-4Rα chain (35). In contrast to our analyses, which detected expression of Arg1 and Ym1 in the absence of FIZZ1 in both whole tissue and FACS-sorted CD11b+F4/80+ macrophages, IL-4/IL-13-stimulated macrophages typically co-express the prototypic murine M2 genes Arg1, Ym1, and FIZZ1. Similarly, macrophages in mice infected with various parasites, macrophages from RSV-infected mice, endotoxin-tolerant macrophages, and tumor-associated macrophages skewed toward M2 polarization also co-express Arg1, Ym1, and FIZZ1 (37, 45, 76–78). More recently, studies have identified an IL-4/IL-13/STAT6-independent, STAT3-dependent pathway by which macrophages are activated to express Arg1 in the absence of both Ym1 and FIZZ1 (79), demonstrating that unique signaling pathways can uncouple Arg1 expression in macrophages from other canonical M2 genes. We found that expression of Arg1 in inflamed muscle tissue of RRV-infected mice correlated with the expression of mRNAs encoding STAT3-activating cytokines including IL-6 and IL-10, suggesting that Arg1 may be induced in RRV-infected mice by a STAT3-dependent pathway. An additional STAT6-independent pathway leading to induction of Arg1 was identified following treatment of macrophages with macrophage stimulating protein (MSP) (80). Future studies in our laboratory are aimed at defining the specific signaling pathways that regulate Arg1 expression in the context of RRV and CHIKV infection.
As mentioned above, we found that macrophages isolated from musculoskeletal inflammatory lesions of RRV-infected mice expressed high levels of Arg1 and Ym1 with little to no expression of FIZZ1. This expression pattern is similar to the gene expression pattern of MDSCs that infiltrate the heart in response to Trypanosoma cruzi infection (81). Thus, the CD11b+F4/80+Arg1+ cells present in the inflamed musculoskeletal tissues of RRV/CHIKV-infected mice may share overlapping features with mononuclear MDSCs. Arginase activity, nitric oxide, and reactive oxygen species production are the main effectors of the immunosuppressive activity of MDSCs (82). We found that Arg1-expressing macrophages sorted from the inflamed quadriceps of RRV-infected mice suppressed T cell proliferation ex vivo by a mechanism that was, in part, Arg1-dependent. These findings suggest that the macrophages present in RRV-induced musculoskeletal inflammatory lesions have immunosuppressive activity similar to MDSCs.
Conditional deletion of Arg1 from macrophages and neutrophils had minimal impact on the development of RRV-induced acute disease, including effects on body weight, outward disease signs, cellular infiltration of tissues, and viral loads at 7 dpi. These findings are consistent with our gene expression analyses that indicated Arg1 expression in musculoskeletal tissues was low until 7 dpi. Strikingly, LysMcre-mediated deletion of Arg1 had a dramatic impact on viral loads at late times post-inoculation. These findings suggest that Arg1 activity in tissue-infiltrating myeloid cells inhibits viral clearance. In addition, the reduced viral loads in LysMcre;Arg1F/F mice was associated with improved skeletal muscle tissue pathology, suggesting that the enhanced clearance of RRV in these mice reduced the duration of tissue damage. Additionally, deletion of Arg1 did not alter the expression of other genes typically associated with murine macrophages polarized towards an M2 phenotype, suggesting that the enhanced viral clearance in LysMcre;Arg1F/F mice is due to the loss of Arg1.
Although we do not yet know the exact mechanisms by which Arg1 in macrophages and/or neutrophils inhibits RRV clearance, previous investigations on alphavirus clearance from the central nervous system demonstrated that both anti-viral antibodies and T cells are critical for the clearance of infectious virus and viral RNA (83–86). Arg1 activity in myeloid cell populations has been shown to directly suppress T cell effector functions in various contexts, primarily by depletion of the amino acid L-arginine, the substrate of Arg1 (53, 54, 67). Depletion of L-arginine from the microenvironment inhibits T-cell effector functions and proliferation via several mechanisms, including decreasing the expression of the CD3ζ-chain and preventing the expression of the cell cycle regulators cyclin D3 and cyclin-dependent kinase 4 (87). Ongoing studies in our lab are investigating the hypothesis that Arg1-expressing myeloid cells inhibit anti-viral effector T cells at the sites of inflammation.
Human RRV and CHIKV disease is characterized by a prolonged disease course, with most cases requiring weeks to months for complete resolution of musculoskeletal pain and inflammation. Taken together, our findings suggest that Arg1 activity in tissue-infiltrating macrophages and neutrophils may contribute to the duration and/or severity of the disease and could promote the development of chronic disease that is reported by a subset of RRV/CHIKV-infected persons. Thus, therapeutically targeting Arg1 activity or induction could inhibit the duration of disease. Arg1 is a central metabolic enzyme in the liver that catalyzes the hydrolysis of L-arginine to urea and ornithine (46). Thus, systemic inhibition of arginase would likely be associated with toxicity. However, treatment of dystrophin-deficient mdx mice, a model of Duchenne muscular dystrophy (DMD), with the Arg1 substrate, L-arginine, improved skeletal muscle function (88, 89). Similarly, L-arginine supplementation restored T cell function and reduced parasite load and associated pathology in a mouse model of Leishmania major infection (67). Thus, treatment of RRV- or CHIKV-infected mice with L-arginine could enhance the anti-viral T cell response as well as further improve skeletal muscle repair. Alternatively, inhibition of Arg1 induction in tissue-infiltrating myeloid cells could be beneficial. Thus, future studies to elucidate the pathways that regulate Arg1 expression in myeloid cells in the context of alphavirus infections are critical to inform therapeutic strategies.
In sum, the role of Arg1 in infectious diseases is not well known. To date, parasitic infections are the most common models used to study the role of myeloid cell Arg1 in infectious disease pathogenesis (90). In contrast, very few studies have evaluated the function of myeloid cell Arg1 during viral infections. Our studies have identified that Arg1-expressing myeloid cells dramatically inhibit RRV clearance and promote tissue pathology at late times post-infection. These findings suggest that Arg1-expressing myeloid cells function as pathogenic effectors at late stages of disease and that therapeutics that target Arg1 or the induction of Arg1 could limit the severity or duration of these debilitating virus-induced diseases.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Matthew Maulis for valuable assistance with tissue processing and embedding. We thank Philip Pratt and Sonia Soto for assistance with flow cytometry and flow sorting, respectively.
This research was supported by NIH-NIAID research grant K22 AI079163 and grant funding from the Arthritis National Research Foundation awarded to T.E.M. K.S. was supported by NIH-NIAID training grant T32 AI052066 and NIH-NCRR Colorado CTSI grant TL1 RR025778.
REFERENCES
- 1.Powers AM, Brault AC, Shirako Y, Strauss EG, Kang W, Strauss JH, Weaver SC. Evolutionary relationships and systematics of the alphaviruses. J Virol. 2001;75:10118–10131. doi: 10.1128/JVI.75.21.10118-10131.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Harley D, Sleigh A, Ritchie S. Ross River virus transmission, infection, and disease: a cross-disciplinary review. Clin Microbiol Rev. 2001;14:909–932. doi: 10.1128/CMR.14.4.909-932.2001. table of contents. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Williams MC, Woodall JP, Gillett JD. O'nyong-Nyong Fever: An Epidemic Virus Diesease in East Africa. Vii. Virus Isolations from Man and Serological Studies up to July 1961. Trans R Soc Trop Med Hyg. 1965;59:186–197. doi: 10.1016/0035-9203(65)90080-5. [DOI] [PubMed] [Google Scholar]
- 4.Weaver SC, Reisen WK. Present and future arboviral threats. Antiviral Res. 2010;85:328–345. doi: 10.1016/j.antiviral.2009.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Grandadam M, Caro V, Plumet S, Thiberge JM, Souares Y, Failloux AB, Tolou HJ, Budelot M, Cosserat D, Leparc-Goffart I, Despres P. Chikungunya virus, southeastern France. Emerg Infect Dis. 2011;17:910–913. doi: 10.3201/eid1705.101873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rezza G, Nicoletti L, Angelini R, Romi R, Finarelli AC, Panning M, Cordioli P, Fortuna C, Boros S, Magurano F, Silvi G, Angelini P, Dottori M, Ciufolini MG, Majori GC, Cassone A. Infection with chikungunya virus in Italy: an outbreak in a temperate region. Lancet. 2007;370:1840–1846. doi: 10.1016/S0140-6736(07)61779-6. [DOI] [PubMed] [Google Scholar]
- 7.ProMed-mail. Chikungunya: New Caledonia. [Accessed 3 August 2011];ProMED-mail. 2011 May;25 20110525. 1595: [Google Scholar]
- 8.Gibney KB, Fischer M, Prince HE, Kramer LD, St George K, Kosoy OL, Laven JJ, Staples JE. Chikungunya fever in the United States: a fifteen year review of cases. Clin Infect Dis. 2011;52:e121–126. doi: 10.1093/cid/ciq214. [DOI] [PubMed] [Google Scholar]
- 9.Watson R. Chikungunya fever is transmitted locally in Europe for first time. Bmj. 2007;335:532–533. doi: 10.1136/bmj.39332.708738.DB. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Oliva O, San Martin JL, Nasci RS. Preparedness and Response for Chikungunya Virus: Introduction in the Americas. PAHO; Washington, D.C.: 2011. pp. 1–161. [Google Scholar]
- 11.Pinheiro FP, Freitas RB, Travassos da Rosa JF, Gabbay YB, Mello WA, LeDuc JW. An outbreak of Mayaro virus disease in Belterra, Brazil. I. Clinical and virological findings. Am J Trop Med Hyg. 1981;30:674–681. doi: 10.4269/ajtmh.1981.30.674. [DOI] [PubMed] [Google Scholar]
- 12.Staples JE, Breiman RF, Powers AM. Chikungunya fever: an epidemiological review of a re-emerging infectious disease. Clin Infect Dis. 2009;49:942–948. doi: 10.1086/605496. [DOI] [PubMed] [Google Scholar]
- 13.Sissoko D, Malvy D, Ezzedine K, Renault P, Moscetti F, Ledrans M, Pierre V. Post-Epidemic Chikungunya Disease on Reunion Island: Course of Rheumatic Manifestations and Associated Factors over a 15-Month Period. PLoS Negl Trop Dis. 2009;3:e389. doi: 10.1371/journal.pntd.0000389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Borgherini G, Poubeau P, Jossaume A, Gouix A, Cotte L, Michault A, Arvin-Berod C, Paganin F. Persistent arthralgia associated with chikungunya virus: a study of 88 adult patients on reunion island. Clin Infect Dis. 2008;47:469–475. doi: 10.1086/590003. [DOI] [PubMed] [Google Scholar]
- 15.Brighton SW, Prozesky OW, de la Harpe AL. Chikungunya virus infection. A retrospective study of 107 cases. S Afr Med J. 1983;63:313–315. [PubMed] [Google Scholar]
- 16.Larrieu S, Pouderoux N, Pistone T, Filleul L, Receveur MC, Sissoko D, Ezzedine K, Malvy D. Factors associated with persistence of arthralgia among Chikungunya virus-infected travellers: report of 42 French cases. J Clin Virol. 2010;47:85–88. doi: 10.1016/j.jcv.2009.11.014. [DOI] [PubMed] [Google Scholar]
- 17.Simon F, Parola P, Grandadam M, Fourcade S, Oliver M, Brouqui P, Hance P, Kraemer P, Ali Mohamed A, de Lamballerie X, Charrel R, Tolou H. Chikungunya infection: an emerging rheumatism among travelers returned from Indian Ocean islands. Report of 47 cases. Medicine (Baltimore) 2007;86:123–137. doi: 10.1097/MD/0b013e31806010a5. [DOI] [PubMed] [Google Scholar]
- 18.Kularatne SA, Weerasinghe SC, Gihan C, Wickramasinghe S, Dharmarathne S, Abeyrathna A, Jayalath T. Epidemiology, clinical manifestations, and long-term outcomes of a major outbreak of chikungunya in a hamlet in sri lanka, in 2007: a longitudinal cohort study. J Trop Med. 2012;2012:639178. doi: 10.1155/2012/639178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Harley D, Bossingham D, Purdie DM, Pandeya N, Sleigh AC. Ross River virus disease in tropical Queensland: evolution of rheumatic manifestations in an inception cohort followed for six months. Med J Aust. 2002;177:352–355. doi: 10.5694/j.1326-5377.2002.tb04836.x. [DOI] [PubMed] [Google Scholar]
- 20.Condon RJ, Rouse IL. Acute symptoms and sequelae of Ross River virus infection in South-Western Australia: a follow-up study. Clin Diagn Virol. 1995;3:273–284. doi: 10.1016/s0928-0197(94)00043-3. [DOI] [PubMed] [Google Scholar]
- 21.Mylonas AD, Brown AM, Carthew TL, McGrath B, Purdie DM, Pandeya N, Vecchio PC, Collins LG, Gardner ID, de Looze FJ, Reymond EJ, Suhrbier A. Natural history of Ross River virus-induced epidemic polyarthritis. Med J Aust. 2002;177:356–360. doi: 10.5694/j.1326-5377.2002.tb04837.x. [DOI] [PubMed] [Google Scholar]
- 22.Selden SM, Cameron AS. Changing epidemiology of Ross River virus disease in South Australia. Med J Aust. 1996;165:313–317. doi: 10.5694/j.1326-5377.1996.tb124989.x. [DOI] [PubMed] [Google Scholar]
- 23.Westley-Wise VJ, Beard JR, Sladden TJ, Dunn TM, Simpson J. Ross River virus infection on the North Coast of New South Wales. Aust N Z J Public Health. 1996;20:87–92. doi: 10.1111/j.1467-842x.1996.tb01343.x. [DOI] [PubMed] [Google Scholar]
- 24.Economopoulou A, Dominguez M, Helynck B, Sissoko D, Wichmann O, Quenel P, Germonneau P, Quatresous I. Atypical Chikungunya virus infections: clinical manifestations, mortality and risk factors for severe disease during the 2005–2006 outbreak on Reunion. Epidemiol Infect. 2009;137:534–541. doi: 10.1017/S0950268808001167. [DOI] [PubMed] [Google Scholar]
- 25.Pialoux G, Gauzere BA, Jaureguiberry S, Strobel M. Chikungunya, an epidemic arbovirosis. Lancet Infect Dis. 2007;7:319–327. doi: 10.1016/S1473-3099(07)70107-X. [DOI] [PubMed] [Google Scholar]
- 26.Morrison TE, Fraser RJ, Smith PN, Mahalingam S, Heise MT. Complement contributes to inflammatory tissue destruction in a mouse model of Ross River virus-induced disease. J Virol. 2007;81:5132–5143. doi: 10.1128/JVI.02799-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Morrison TE, Oko L, Montgomery SA, Whitmore AC, Lotstein AR, Gunn BM, Elmore SA, Heise MT. A mouse model of chikungunya virus-induced musculoskeletal inflammatory disease: evidence of arthritis, tenosynovitis, myositis, and persistence. Am J Pathol. 2011;178:32–40. doi: 10.1016/j.ajpath.2010.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Morrison TE, Whitmore AC, Shabman RS, Lidbury BA, Mahalingam S, Heise MT. Characterization of Ross River virus tropism and virus-induced inflammation in a mouse model of viral arthritis and myositis. J Virol. 2006;80:737–749. doi: 10.1128/JVI.80.2.737-749.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hoarau JJ, Jaffar Bandjee MC, Krejbich Trotot P, Das T, Li-Pat-Yuen G, Dassa B, Denizot M, Guichard E, Ribera A, Henni T, Tallet F, Moiton MP, Gauzere BA, Bruniquet S, Jaffar Bandjee Z, Morbidelli P, Martigny G, Jolivet M, Gay F, Grandadam M, Tolou H, Vieillard V, Debre P, Autran B, Gasque P. Persistent chronic inflammation and infection by Chikungunya arthritogenic alphavirus in spite of a robust host immune response. J Immunol. 2010;184:5914–5927. doi: 10.4049/jimmunol.0900255. [DOI] [PubMed] [Google Scholar]
- 30.Ozden S, Huerre M, Riviere JP, Coffey LL, Afonso PV, Mouly V, de Monredon J, Roger JC, El Amrani M, Yvin JL, Jaffar MC, Frenkiel MP, Sourisseau M, Schwartz O, Butler-Browne G, Despres P, Gessain A, Ceccaldi PE. Human muscle satellite cells as targets of Chikungunya virus infection. PLoS One. 2007;2:e527. doi: 10.1371/journal.pone.0000527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Labadie K, Larcher T, Joubert C, Mannioui A, Delache B, Brochard P, Guigand L, Dubreil L, Lebon P, Verrier B, de Lamballerie X, Suhrbier A, Cherel Y, Le Grand R, Roques P. Chikungunya disease in nonhuman primates involves long-term viral persistence in macrophages. J Clin Invest. 2010;120:894–906. doi: 10.1172/JCI40104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Soden M, Vasudevan H, Roberts B, Coelen R, Hamlin G, Vasudevan S, La Brooy J. Detection of viral ribonucleic acid and histologic analysis of inflamed synovium in Ross River virus infection. Arthritis Rheum. 2000;43:365–369. doi: 10.1002/1529-0131(200002)43:2<365::AID-ANR16>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- 33.Fraser JR, Cunningham AL, Clarris BJ, Aaskov JG, Leach R. Cytology of synovial effusions in epidemic polyarthritis. Aust N Z J Med. 1981;11:168–173. doi: 10.1111/j.1445-5994.1981.tb04226.x. [DOI] [PubMed] [Google Scholar]
- 34.Gordon S, Taylor PR. Monocyte and macrophage heterogeneity. Nat Rev Immunol. 2005;5:953–964. doi: 10.1038/nri1733. [DOI] [PubMed] [Google Scholar]
- 35.Murray PJ, Wynn TA. Protective and pathogenic functions of macrophage subsets. Nat Rev Immunol. 2011;11:723–737. doi: 10.1038/nri3073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Biswas SK, Mantovani A. Macrophage plasticity and interaction with lymphocyte subsets: cancer as a paradigm. Nat Immunol. 2010;11:889–896. doi: 10.1038/ni.1937. [DOI] [PubMed] [Google Scholar]
- 37.Loke P, Gallagher I, Nair MG, Zang X, Brombacher F, Mohrs M, Allison JP, Allen JE. Alternative activation is an innate response to injury that requires CD4+ T cells to be sustained during chronic infection. J Immunol. 2007;179:3926–3936. doi: 10.4049/jimmunol.179.6.3926. [DOI] [PubMed] [Google Scholar]
- 38.Duffield JS, Forbes SJ, Constandinou CM, Clay S, Partolina M, Vuthoori S, Wu S, Lang R, Iredale JP. Selective depletion of macrophages reveals distinct, opposing roles during liver injury and repair. J Clin Invest. 2005;115:56–65. doi: 10.1172/JCI22675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shechter R, London A, Varol C, Raposo C, Cusimano M, Yovel G, Rolls A, Mack M, Pluchino S, Martino G, Jung S, Schwartz M. Infiltrating blood-derived macrophages are vital cells playing an anti-inflammatory role in recovery from spinal cord injury in mice. PLoS Med. 2009;6:e1000113. doi: 10.1371/journal.pmed.1000113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nahrendorf M, Swirski FK, Aikawa E, Stangenberg L, Wurdinger T, Figueiredo JL, Libby P, Weissleder R, Pittet MJ. The healing myocardium sequentially mobilizes two monocyte subsets with divergent and complementary functions. J Exp Med. 2007;204:3037–3047. doi: 10.1084/jem.20070885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ruffell D, Mourkioti F, Gambardella A, Kirstetter P, Lopez RG, Rosenthal N, Nerlov C. A CREB-C/EBPbeta cascade induces M2 macrophage-specific gene expression and promotes muscle injury repair. Proc Natl Acad Sci U S A. 2009;106:17475–17480. doi: 10.1073/pnas.0908641106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Villalta SA, Deng B, Rinaldi C, Wehling-Henricks M, Tidball JG. IFN-gamma promotes muscle damage in the mdx mouse model of Duchenne muscular dystrophy by suppressing M2 macrophage activation and inhibiting muscle cell proliferation. J Immunol. 2011;187:5419–5428. doi: 10.4049/jimmunol.1101267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chen F, Liu Z, Wu W, Rozo C, Bowdridge S, Millman A, Van Rooijen N, Urban JF, Jr., Wynn TA, Gause WC. An essential role for T(H)2-type responses in limiting acute tissue damage during experimental helminth infection. Nat Med. 2012;18:260–266. doi: 10.1038/nm.2628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Allen JE, Wynn TA. Evolution of Th2 immunity: a rapid repair response to tissue destructive pathogens. PLoS Pathog. 2011;7:e1002003. doi: 10.1371/journal.ppat.1002003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shirey KA, Pletneva LM, Puche AC, Keegan AD, Prince GA, Blanco JC, Vogel SN. Control of RSV-induced lung injury by alternatively activated macrophages is IL-4R alpha-, TLR4-, and IFN-beta-dependent. Mucosal Immunol. 2010;3:291–300. doi: 10.1038/mi.2010.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Morris SM., Jr. Recent advances in arginine metabolism: roles and regulation of the arginases. Br J Pharmacol. 2009;157:922–930. doi: 10.1111/j.1476-5381.2009.00278.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mantovani A, Sozzani S, Locati M, Allavena P, Sica A. Macrophage polarization: tumor-associated macrophages as a paradigm for polarized M2 mononuclear phagocytes. Trends Immunol. 2002;23:549–555. doi: 10.1016/s1471-4906(02)02302-5. [DOI] [PubMed] [Google Scholar]
- 48.Martinez FO, Helming L, Gordon S. Alternative activation of macrophages: an immunologic functional perspective. Annu Rev Immunol. 2009;27:451–483. doi: 10.1146/annurev.immunol.021908.132532. [DOI] [PubMed] [Google Scholar]
- 49.Bronte V, Zanovello P. Regulation of immune responses by L-arginine metabolism. Nat Rev Immunol. 2005;5:641–654. doi: 10.1038/nri1668. [DOI] [PubMed] [Google Scholar]
- 50.Munder M. Arginase: an emerging key player in the mammalian immune system. Br J Pharmacol. 2009;158:638–651. doi: 10.1111/j.1476-5381.2009.00291.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rodriguez PC, Ochoa AC. Arginine regulation by myeloid derived suppressor cells and tolerance in cancer: mechanisms and therapeutic perspectives. Immunol Rev. 2008;222:180–191. doi: 10.1111/j.1600-065X.2008.00608.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.El Kasmi KC, Qualls JE, Pesce JT, Smith AM, Thompson RW, Henao-Tamayo M, Basaraba RJ, Konig T, Schleicher U, Koo MS, Kaplan G, Fitzgerald KA, Tuomanen EI, Orme IM, Kanneganti TD, Bogdan C, Wynn TA, Murray PJ. Toll-like receptor-induced arginase 1 in macrophages thwarts effective immunity against intracellular pathogens. Nat Immunol. 2008;9:1399–1406. doi: 10.1038/ni.1671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pesce JT, Ramalingam TR, Mentink-Kane MM, Wilson MS, El Kasmi KC, Smith AM, Thompson RW, Cheever AW, Murray PJ, Wynn TA. Arginase-1-expressing macrophages suppress Th2 cytokine-driven inflammation and fibrosis. PLoS Pathog. 2009;5:e1000371. doi: 10.1371/journal.ppat.1000371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rodriguez PC, Quiceno DG, Zabaleta J, Ortiz B, Zea AH, Piazuelo MB, Delgado A, Correa P, Brayer J, Sotomayor EM, Antonia S, Ochoa JB, Ochoa AC. Arginase I production in the tumor microenvironment by mature myeloid cells inhibits T-cell receptor expression and antigen-specific T-cell responses. Cancer Res. 2004;64:5839–5849. doi: 10.1158/0008-5472.CAN-04-0465. [DOI] [PubMed] [Google Scholar]
- 55.Zea AH, Rodriguez PC, Atkins MB, Hernandez C, Signoretti S, Zabaleta J, McDermott D, Quiceno D, Youmans A, O'Neill A, Mier J, Ochoa AC. Arginase-producing myeloid suppressor cells in renal cell carcinoma patients: a mechanism of tumor evasion. Cancer Res. 2005;65:3044–3048. doi: 10.1158/0008-5472.CAN-04-4505. [DOI] [PubMed] [Google Scholar]
- 56.Bronte V, Serafini P, De Santo C, Marigo I, Tosello V, Mazzoni A, Segal DM, Staib C, Lowel M, Sutter G, Colombo MP, Zanovello P. IL-4-induced arginase 1 suppresses alloreactive T cells in tumor-bearing mice. J Immunol. 2003;170:270–278. doi: 10.4049/jimmunol.170.1.270. [DOI] [PubMed] [Google Scholar]
- 57.Gardner J, Anraku I, Le TT, Larcher T, Major L, Roques P, Schroder WA, Higgs S, Suhrbier A. Chikungunya virus arthritis in adult wild-type mice. J Virol. 2010;84:8021–8032. doi: 10.1128/JVI.02603-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lidbury BA, Rulli NE, Suhrbier A, Smith PN, McColl SR, Cunningham AL, Tarkowski A, van Rooijen N, Fraser RJ, Mahalingam S. Macrophage-derived proinflammatory factors contribute to the development of arthritis and myositis after infection with an arthrogenic alphavirus. J Infect Dis. 2008;197:1585–1593. doi: 10.1086/587841. [DOI] [PubMed] [Google Scholar]
- 59.Doherty R. l., Whitehead RH, Gorman BM, O'Gower AK. The isolation of a third group A arbovirus in Australia, with preliminary observations on its realtionships to epidemic polyarthritis. Aust. J. Sci. 1963;26:183–184. [Google Scholar]
- 60.Kuhn RJ, Niesters HG, Hong Z, Strauss JH. Infectious RNA transcripts from Ross River virus cDNA clones and the construction and characterization of defined chimeras with Sindbis virus. Virology. 1991;182:430–441. doi: 10.1016/0042-6822(91)90584-x. [DOI] [PubMed] [Google Scholar]
- 61.Jupille HJ, Oko L, Stoermer KA, Heise MT, Mahalingam S, Gunn BM, Morrison TE. Mutations in nsP1 and PE2 are critical determinants of Ross River virus-induced musculoskeletal inflammatory disease in a mouse model. Virology. 2011;410:216–227. doi: 10.1016/j.virol.2010.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Clausen BE, Burkhardt C, Reith W, Renkawitz R, Forster I. Conditional gene targeting in macrophages and granulocytes using LysMcre mice. Transgenic Res. 1999;8:265–277. doi: 10.1023/a:1008942828960. [DOI] [PubMed] [Google Scholar]
- 63.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 64.Nicolls MR, Coulombe M, Beilke J, Gelhaus HC, Gill RG. CD4-dependent generation of dominant transplantation tolerance induced by simultaneous perturbation of CD154 and LFA-1 pathways. J Immunol. 2002;169:4831–4839. doi: 10.4049/jimmunol.169.9.4831. [DOI] [PubMed] [Google Scholar]
- 65.Gordon S, Martinez FO. Alternative activation of macrophages: mechanism and functions. Immunity. 2010;32:593–604. doi: 10.1016/j.immuni.2010.05.007. [DOI] [PubMed] [Google Scholar]
- 66.Munder M, Eichmann K, Modolell M. Alternative metabolic states in murine macrophages reflected by the nitric oxide synthase/arginase balance: competitive regulation by CD4+ T cells correlates with Th1/Th2 phenotype. J Immunol. 1998;160:5347–5354. [PubMed] [Google Scholar]
- 67.Modolell M, Choi BS, Ryan RO, Hancock M, Titus RG, Abebe T, Hailu A, Muller I, Rogers ME, Bangham CR, Munder M, Kropf P. Local suppression of T cell responses by arginase-induced L-arginine depletion in nonhealing leishmaniasis. PLoS Negl Trop Dis. 2009;3:e480. doi: 10.1371/journal.pntd.0000480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Cotter MJ, Muruve DA. Isolation of neutrophils from mouse liver: A novel method to study effector leukocytes during inflammation. J Immunol Methods. 2006;312:68–78. doi: 10.1016/j.jim.2006.02.019. [DOI] [PubMed] [Google Scholar]
- 69.Morrison TE, Simmons JD, Heise MT. Complement receptor 3 promotes severe ross river virus-induced disease. J Virol. 2008;82:11263–11272. doi: 10.1128/JVI.01352-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lidbury BA, Simeonovic C, Maxwell GE, Marshall ID, Hapel AJ. Macrophage-induced muscle pathology results in morbidity and mortality for Ross River virus-infected mice. J Infect Dis. 2000;181:27–34. doi: 10.1086/315164. [DOI] [PubMed] [Google Scholar]
- 71.Hazelton RA, Hughes C, Aaskov JG. The inflammatory response in the synovium of a patient with Ross River arbovirus infection. Aust N Z J Med. 1985;15:336–339. doi: 10.1111/j.1445-5994.1985.tb04048.x. [DOI] [PubMed] [Google Scholar]
- 72.Seay AR, Griffin DE, Johnson RT. Experimental viral polymyositis: age dependency and immune responses to Ross River virus infection in mice. Neurology. 1981;31:656–660. doi: 10.1212/wnl.31.6.656. [DOI] [PubMed] [Google Scholar]
- 73.Wynn TA. Fibrotic disease and the T(H)1/T(H)2 paradigm. Nat Rev Immunol. 2004;4:583–594. doi: 10.1038/nri1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Rulli NE, Guglielmotti A, Mangano G, Rolph MS, Apicella C, Zaid A, Suhrbier A, Mahalingam S. Amelioration of alphavirus-induced arthritis and myositis in a mouse model by treatment with bindarit, an inhibitor of monocyte chemotactic proteins. Arthritis Rheum. 2009;60:2513–2523. doi: 10.1002/art.24682. [DOI] [PubMed] [Google Scholar]
- 75.Rulli NE, Rolph MS, Srikiatkhachorn A, Anantapreecha S, Guglielmotti A, Mahalingam S. Protection from arthritis and myositis in a mouse model of acute chikungunya virus disease by bindarit, an inhibitor of monocyte chemotactic protein-1 synthesis. J Infect Dis. 2011;204:1026–1030. doi: 10.1093/infdis/jir470. [DOI] [PubMed] [Google Scholar]
- 76.Porta C, Rimoldi M, Raes G, Brys L, Ghezzi P, Di Liberto D, Dieli F, Ghisletti S, Natoli G, De Baetselier P, Mantovani A, Sica A. Tolerance and M2 (alternative) macrophage polarization are related processes orchestrated by p50 nuclear factor kappaB. Proc Natl Acad Sci U S A. 2009;106:14978–14983. doi: 10.1073/pnas.0809784106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Sandler NG, Mentink-Kane MM, Cheever AW, Wynn TA. Global gene expression profiles during acute pathogen-induced pulmonary inflammation reveal divergent roles for Th1 and Th2 responses in tissue repair. J Immunol. 2003;171:3655–3667. doi: 10.4049/jimmunol.171.7.3655. [DOI] [PubMed] [Google Scholar]
- 78.Satoh T, Takeuchi O, Vandenbon A, Yasuda K, Tanaka Y, Kumagai Y, Miyake T, Matsushita K, Okazaki T, Saitoh T, Honma K, Matsuyama T, Yui K, Tsujimura T, Standley DM, Nakanishi K, Nakai K, Akira S. The Jmjd3-Irf4 axis regulates M2 macrophage polarization and host responses against helminth infection. Nat Immunol. 2010;11:936–944. doi: 10.1038/ni.1920. [DOI] [PubMed] [Google Scholar]
- 79.Qualls JE, Neale G, Smith AM, Koo MS, DeFreitas AA, Zhang H, Kaplan G, Watowich SS, Murray PJ. Arginine usage in mycobacteria-infected macrophages depends on autocrine-paracrine cytokine signaling. Sci Signal. 2010;3:ra62. doi: 10.1126/scisignal.2000955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Sharda DR, Yu S, Ray M, Squadrito ML, De Palma M, Wynn TA, Morris SM, Jr., Hankey PA. Regulation of macrophage arginase expression and tumor growth by the ron receptor tyrosine kinase. J Immunol. 2011;187:2181–2192. doi: 10.4049/jimmunol.1003460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Cuervo H, Guerrero NA, Carbajosa S, Beschin A, De Baetselier P, Girones N, Fresno M. Myeloid-Derived Suppressor Cells Infiltrate the Heart in Acute Trypanosoma cruzi Infection. J Immunol. 2011;187:2656–2665. doi: 10.4049/jimmunol.1002928. [DOI] [PubMed] [Google Scholar]
- 82.Gabrilovich DI, Nagaraj S. Myeloid-derived suppressor cells as regulators of the immune system. Nat Rev Immunol. 2009;9:162–174. doi: 10.1038/nri2506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Binder GK, Griffin DE. Interferon-gamma-mediated site-specific clearance of alphavirus from CNS neurons. Science. 2001;293:303–306. doi: 10.1126/science.1059742. [DOI] [PubMed] [Google Scholar]
- 84.Levine B, Hardwick JM, Trapp BD, Crawford TO, Bollinger RC, Griffin DE. Antibody-mediated clearance of alphavirus infection from neurons. Science. 1991;254:856–860. doi: 10.1126/science.1658936. [DOI] [PubMed] [Google Scholar]
- 85.Levine B, Griffin DE. Persistence of viral RNA in mouse brains after recovery from acute alphavirus encephalitis. J Virol. 1992;66:6429–6435. doi: 10.1128/jvi.66.11.6429-6435.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Brooke CB, Deming DJ, Whitmore AC, White LJ, Johnston RE. T cells facilitate recovery from Venezuelan equine encephalitis virus-induced encephalomyelitis in the absence of antibody. J Virol. 2010;84:4556–4568. doi: 10.1128/JVI.02545-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Rodriguez PC, Quiceno DG, Ochoa AC. L-arginine availability regulates T-lymphocyte cell-cycle progression. Blood. 2007;109:1568–1573. doi: 10.1182/blood-2006-06-031856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Barton ER, Morris L, Kawana M, Bish LT, Toursel T. Systemic administration of L-arginine benefits mdx skeletal muscle function. Muscle Nerve. 2005;32:751–760. doi: 10.1002/mus.20425. [DOI] [PubMed] [Google Scholar]
- 89.Voisin V, Sebrie C, Matecki S, Yu H, Gillet B, Ramonatxo M, Israel M, De la Porte S. L-arginine improves dystrophic phenotype in mdx mice. Neurobiol Dis. 2005;20:123–130. doi: 10.1016/j.nbd.2005.02.010. [DOI] [PubMed] [Google Scholar]
- 90.Das P, Lahiri A, Chakravortty D. Modulation of the arginase pathway in the context of microbial pathogenesis: a metabolic enzyme moonlighting as an immune modulator. PLoS Pathog. 2010;6:e1000899. doi: 10.1371/journal.ppat.1000899. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.