Abstract
New instrumentation for three-dimensional electron microscopy is facilitating an increase in the throughput of data collection and reconstruction. The increase in throughput creates bottlenecks in the workflow for storing and processing the image data. Here we describe the creation and quantify the throughput of a high-throughput infrastructure supporting collection of three-dimensional data collection.
Keywords: Cryogenic Electron Microscopy, CryoEM, Database, Three-dimensional Electron Microscopy, High-Throughput
Single particle three-dimensional electron microscopy (3DEM) is a powerful technique for determining the structures of biologically relevant macromolecules with several structures now approaching atomic resolution. One of the primary factors that limit resolution of single particle reconstructions is the number of particles that contribute to the reconstruction. So far, the structures that approach atomic resolution have masses of around 1 MDa or greater, and the number of asymmetric units contributing to the structures are from several hundred thousand to several millions of subunits (Cong et al., 2010; Ludtke et al., 2008; Zhang et al., 2008; Zhou, 2011), which agrees well with calculations of the dependence of resolution on numbers of particles (Henderson, 1995; LeBarron et al., 2008; Rosenthal and Henderson, 2003; Stagg et al., 2008). At the same time that the field is approaching atomic resolution for single particle reconstructions, techniques for dealing with heterogeneous data are being developed for tomographic data (Stölken et al., 2011; Winkler, 2007). In the tomographic case, tomographic subvolumes are aligned and classified to sort out the heterogeneity in three-dimensions. Like with single particle data, the quality of subvolume averaging depends on the total number of subvolumes that can be collected. Thus, both single particle and tomographic data collection are driving for an increasing amount of raw data to be able to derive the best possible 3D interpretations. The pressure for more and more data creates bottlenecks in the structure determination pipeline; disk space is required to store all the raw data, increased processing power is required to process the data in a reasonable amount of time, and the disk storage must be able to accommodate reads and writes from many different requests at the same time.
In addition to the techniques requiring more data, new detection devices are coming online such as cameras with large arrays of pixels (Ellisman et al., 2011), hybrid pixel detectors (Faruqi and Henderson, 2007), and monolithic active pixel sensor (MAPS) direct electron detectors (DDD) (Bammes et al., 2012; Milazzo et al., 2011). These developments have the potential to dramatically increase the demands for processing and storage. The commercial MAPS DDDs such as the Direct Electron DE-12, FEI Falcon, and Gatan K2 have fast readout rates with the latter device having a rate of up to 400 frames per second. In the simplest case, many DDD frames are integrated to produce a single EM exposure, and the individual frames contributing to the final exposure are discarded. However there are many potential reasons for storing the contributing frames including dose fractionation and monitoring specimen movement due to beam induced motion (Brilot et al., 2012). Thus, DDDs have the potential to both increase throughput and increase the amount of storage required for the raw data. At the same time that DDDs are being developed, the cameras are getting larger in pixel area (Ellisman et al., 2011). Doubling the linear dimensions of a camera quadruples the storage requirements for an individual image. These technological developments combine to dramatically increase the demands on the processing pipeline and increase the pressure on the previously mentioned bottlenecks.
Dealing with the volume of data coming from EM platforms utilizing new technologies and high-throughput automated data collection requires a nonstandard approach to data storage and processing. Moreover, high-end instruments support many users each with unique data acquisition and storage requirements. The storage and processing facility must be flexible enough to accommodate the different needs of multiple users. This requirement increases the dependence on information technology and computational architecture expertise to acquire the appropriate hardware, software, and support multiple users. Utilizing expertise already in place at a high performance computing (HPC) center facilitates supporting a high-throughput kind of device. However, because high-throughput depends on the robust performance of both the microscope and the processing machines, the considerations described here will be the same even for labs with in-house clusters or that run other automated data collection applications (Mastronarde, 2005; Nickell et al., 2005; Zheng et al., 2007).
Here we describe the throughput and methods for integration of an high performance computing infrastructure with a Titan Krios (FEI Company) equipped with a 4k × 4k pixel CCD camera with automated data collection and processing with Leginon (Suloway et al., 2005) and Appion (Lander et al., 2009). Though we describe our setup using these specific tools, the considerations described are generalizable to any resource running 24 hour-a-day data collection. We describe the considerations for hardware and the tools and methodologies used to ensure seamless integration and ensure dependencies on the processing machines do not adversely impact the availability of the microscope. The setup is scalable and is described with enough detail that our setup can be replicated at other locations by individuals with modest system administration expertise.
Quantitation of throughput
Data collection statistics were acquired for several single particle and tomographic data collection sessions on the Titan Krios equipped with a Gatan 4k × 4k Ultrascan CCD with 4 port readout using automated data collection with Leginon. With single particle data collection, the throughput depends on several factors such as the readout rate of the camera, the stability of the goniometer (drift-rate after a move), and the number of images that can be acquired per target area. We measured the throughput for two data collection sessions with typical Leginon data collection parameters. Dataset 1 was a COPII complex preserved in vitreous ice over a holey carbon film and was collected at 59,000 X magnification (1.5 Å/pix) for final exposures. We were able to collect three images per hole for this session. The overall exposure rate determined as the total number of high magnification exposures over the total session time for this dataset was 95 exposures/hour. Dataset 2 was an adeno-associated virus (AAV) dataset at 120,000 × (0.65 Å/pix), and we collected 93 exposures/hour for this session. For both sessions, the setup time before fully automated data collection was ~3 hours. The diameter of the holes in the support film for both datasets was 2 μm and the diameter of the e− beam was 1.4 μm. This resulted in some beam overlap in the center of the holes, but the area that was overlapping was not imaged by the camera at those magnifications. Given that the beam diameter is required to be greater than 1.3 μm in order to maintain parallel illumination with our imaging conditions, three exposures per hole is the maximum we can attain. The structures associated with these A data are being published elsewhere, but the AAV data reconstructed to 4.5 resolution, which shows that the data collection conditions are sufficient for high-resolution (Lerch et al., 2012). The images are 4k × 4k pixels and are stored as 16 bit signed floats in MRC format that results in an image that takes up 64 MB of disk space. Collecting single particle data in this way for 24 hours requires ~144 GB of disk space.
The situation is similar for tomographic data. In a tomographic data collection session with typical Leginon data collection parameters, we collected 119 images per tilt series and could collect 1.85 series per hour. In 24 hours, we can collect 44 tilt series, which in turn takes up 340 GB of disk space. The Titan Krios can be operated 24 hours-a-day for 6 days-a-week. If we assume 3 days of single particle collection and 3 days of tomographic collection, we would require ~1.5 TB of disk storage per week. These data are summarized in Table 1.
Table 1.
Data collection and throughput statistics
| Collection type | Images/target | Time between exposures (s) | Time between holes (s) | Overall exposure rate | images/day | disk consumption/day |
|---|---|---|---|---|---|---|
| single particle data collection | 3 | 16.5 | 76 | 94 images/hr | 2256 | 144 GB |
| tomographic data collection | 119 | 19.5 | 162 | 1.83 series/hr | 5226 | 340 GB |
Given the throughput afforded by automation and a high-end microscope, it is unfeasible to store the raw data locally on the computer that is driving the data collection. Moreover, processing this much data takes some time, and in a high-throughput scenario, the data is processed immediately after it is acquired. This means that data collection and processing are occurring simultaneously on the same disk volume. Depending on the number of processing jobs, this can be quite taxing on the disk and network that is serving the data. Some of these problems are solved by hosting the data on a distributed file system, but then the limit on the rate of data acquisition becomes dependent on the bandwidth and traffic load of the network. These considerations led us to create a setup where data are staged locally on the computer that runs Leginon and then moved in real time to an off-site secondary data storage system that is connected to the processing computers.
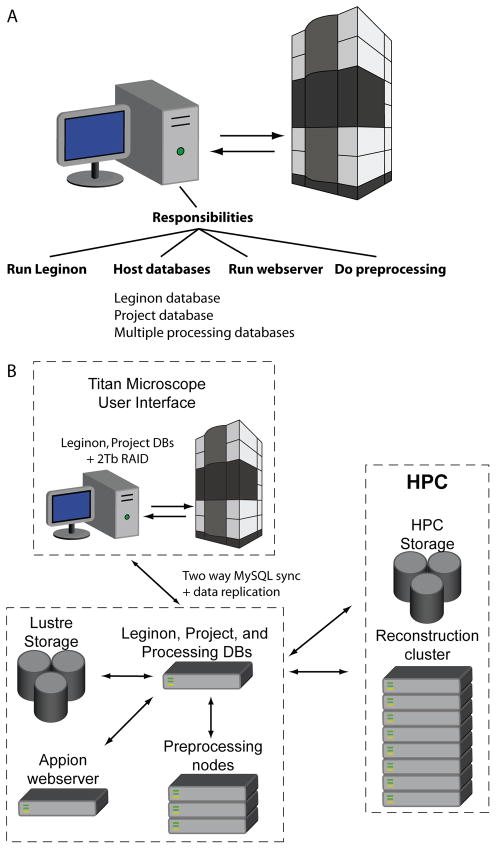
Hosting the data in two physical locations presented a problem for the high-throughput multi-user scenario. The standard setup for data acquisition and processing with the Leginon/Appion software requires a LINUX computer that runs Leginon and is connected to the microscope computer. The Leginon computer also requires network connectivity to a MySQL database to host the metadata and a disk volume to host the image data (Fig 1A). Both the computer driving data collection and the computers doing processing require access to the same image data and database. If we hosted a single database off-site and the network went down, the data collection would go down with it. This problem and the problem of how to store the large volume of image data were solved through the use of a data replication scheme. The computer that is directly connected to the microscope computer hosted a copy of the MySQL database and a small volume of the most recently acquired microscope images, and the image data and metadata database were replicated to a high-performance computing facility with high capacity for disk space and network traffic (Fig. 1B). The duties performed by the HPC computers are split into two functions: 1) pre-processing which includes tasks like particle picking, CTF estimation, and preliminary image classification, and 2) refinement/reconstruction which includes large long-term parallel processing jobs (Fig. 1B). This setup reduces interdependencies of the data collection and data processing tasks and makes data collection more robust and less dependent on network and shared file system availability.
Fig. 1. Schematic of the data collection infrastructure.
A) Schematic showing the responsibilities of a computer running Leginon/Appion in a typical setup. B) Schematic showing the infrastructure that we set up for high-throughput data collection.
Data replication setup
The Appion/Leginon software is supported on the backend by a MySQL database that houses metadata for images produced by the electron microscope. In our replication scheme, this database resides on the Titan Krios desktop as well as a remote pre-processing server housed at the HPC. MySQL includes native support for multi master replication between database servers, and this functionality was used to maintain identical copies of the Appion/Leginon acquisition and project databases on the pre-processing server and the Titan Krios desktop computer. No custom software was required for database replication though a high-bandwidth network connection was required to ensure synchronicity between the databases.
Replication of image data is facilitated through the use of the Linux kernel inotify subsystem which monitors the local file system on the Titan Krios desktop and using inotify-tools and a custom Perl script; an application was developed that copies staged data from the local file system to the HPC Lustre file system via the pre-processing server as it is generated. This script also preserves user and group ownership on the HPC file system, which is necessary to maintain individual and group quotas. We found it necessary to create an hourly batch process (a custom Perl script) that compares image data between the Leginon desktop and the HPC file system. In the event the HPC file system becomes unavailable during data collection and some image data is not moved in real time, a batch process ensures image data is synchronized between the Titan Krios desktop and the HPC file system when it comes back online. This setup ensures that no data is lost during replication.
A secondary function of the remote processing server is to allow end users to monitor the collection process via a web application and perform some pre-processing of collected image data. During normal operations, users rely on the HPC hosted server but in the event HPC facilities are unavailable, users may collect data uninterrupted using the Leginon computer with its associated database and disk volume.
Network and file system considerations
Transfer speeds between the microscope computer and the processing server must be fast to facilitate high-throughput processing. In our case, the computers are connected via a high-speed network with transfers up to 40Gb/second to the storage file system on the HPC. Though we do not approach this is data acquisition rate, it is an important consideration for future equipment enhancements such as high frame rate DDDs. Important considerations for choosing a file system for hosting the data for long-term storage included that it must be robust and that it be easily expandable so that drives could be easily added to the same disk volume. There are several commercial file systems that meet this requirement including PanFS (Panasas, Inc), and IBM’s General Parallel File System. However, the cost of these systems is often outside of the budget for an individual lab. For this reason, the open-source Lustre file system (http://lustre.org) was chosen to host the image data. Lustre is a parallel distributed file system generally used for large scale cluster computing. It is available under the GNU General Public License, and its main advantages over simpler file systems is that it provides scalability in performance and storage capacity.
Data sharing
The NIH and NSF require a data sharing policy be in place for research that is supported by these agencies. Publishing gigabyte datasets from a web site or identifying jointly accessible disk space can be impractical. Here we made use of methods already developed for sharing large files using the BitTorrent file sharing technology. The FSU HPC allows users to browse HPC file systems (Lustre and panfs) via a custom web application. Using this program, we are able to select specific data sets (folders and/or individual files) and create a metadata file that can be used by any BitTorrent client. We are able to publish a small torrent file (usually less than a few megabytes) on any website. This torrent file can be downloaded and opened on a computer using any freely available BitTorrent client. The BitTorrent client uses data in the torrent file to connect to a server at the HPC which ‘seeds’ or allows the client to download the corresponding dataset directly from the HPC file systems to an end user’s computer.
Toward high-throughput high-resolution cryoEM
We are now in a position where we can ask, how long would it take to collect enough data to determine the near-atomic resolution structure of an asymmetric protein of modest molecular weight (by 3DEM standards). Of course, the ultimate resolution of such a reconstruction will be dependent upon a number of factors including specimen quality, specimen homogeneity, microscope performance, and the detection quantum efficiency (DQE) of the detector (McMullan et al., 2009). In this hypothetical case, we assume that the specimen and microscope are sufficient for a 4.0 Å reconstruction, that we have modest detector performance where the resolution in the images does not extend much beyond 1/2 Nyquist (Booth et al., 2006). With such conditions, for a 4.0 Å reconstruction, we would need a pixel size ≤ 1Å/pix. If we assume that we would need to image 106 particles to reach the target resolution, then a critical parameter for throughput is the average number of particles imaged per micrograph (PPM). In the absence of aggregation, particles will assume random positions in the vitreous ice. We simulated particle positions on micrographs in order to determine the maximum number of usable particles/micrograph for a given particle diameter and pixel size. Positions of particles on a micrograph were simulated assuming random positions with an increasing PPM, a 4k × 4k detector, and a pixel size of 1 Å/pix (Fig. 2). Particles were eliminated if they came within 5% of the particle diameter of each other. To allow for defocus-dependent signal delocalization, particles were also eliminated if they came within 75% of the particle diameter of the image border. For a 100 Å diameter particle, the maximum usable PPM was 167 with 465 total PPM (Fig. 2B). Maximum usable/total PPM values for 200 Å, 300 Å, and 700 Å diameter particles were 39/115, 16/55, and 2/15 respectively (Fig. 2B). This assumes that the particles can be accurately centered and the neighboring particles masked out during alignment and classification. With a more stringent overlap requirement of where the particles are not allowed to come within 1 full diameter of each other, the optimal usable/total PPM values for 100 Å, 200 Å, 300Å, and 700 Å diameter particles were 47/120, 11/31, 5/11, and 1/1 respectively. We estimate that with 100 Å diameter particle, a usable PPM of 167, and a collection rate of 94 images/hour, 106 particles could be collected in ~64 hours. By comparison, it would take just over 11 days to collect 106 particles of a large 200 Å diameter asymmetric particle like a ribosome at a sufficient sampling for near atomic resolution.
Fig. 2. Simulations of the distribution of particles per micrograph.
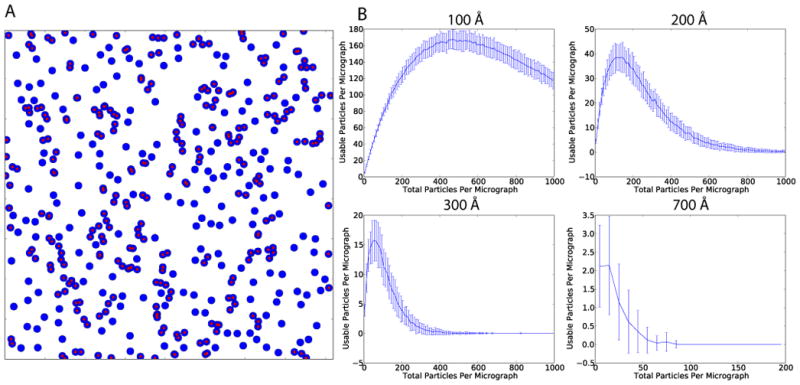
A) Simulated micrograph covering 4096 × 4096 Å with blue circles representing particles of 100 Å diameter. 450 particles are shown in total. Particles that are within 5% of the particle radius from each other and would be excluded from a reconstruction are labeled with a red dot. B) Simulations of usable particles with increasing total number of particles per micrograph. 100 simulations were performed for each total number of particles. Error bars show the standard deviation for each set of simulations. Simulations were performed for particles with 100 Å, 200 Å, 300 Å, and 700 Å diameter particles respectively.
There is potential for increasing the data acquisition rate in the future. Currently, the bulk of the time between exposures is spent navigating to new exposure targets, focusing, and waiting for the goniometer to settle after a move. Typically, navigating to new exposure targets involves magnification changes so there is also a time delay for normalizing the lenses to avoid lens hysteresis. Some time could be saved by focusing only once per square, but in Leginon, drift is monitored during focusing, so this would yield only modest time savings. Apparent goniometer settling time is highly dependent on the particular goniometer and sample of interest. On the Titan Krios at FSU, drift is undetectable within a few seconds of a move, so in practice, the goniometer has settled by the time focusing is completed. Careful assessment of goniometer and microscope settling will be necessary to determine exactly how much time must be reserved for settling without adversely effecting image resolution. We anticipate that the largest increase in data acquisition rate will be made by increasing the detective quantum efficiency (DQE) of the detector as is accomplished in the case of direct detection devices (Bammes et al., 2012; Milazzo et al., 2011). With a better DQE, images will not have to be oversampled as much as is required with a CCD camera. This, in-turn, will allow high-resolution data acquisition at lower magnifications, which will yield a greater field of view and more particles per image. Of course, the total amount of data is not the only consideration for achieving high-resolution reconstructions; optimizing specimen preparation and image quality, and dealing with beam induced motion are critical for driving to the highest possible resolution.
It is becoming clear that data collection rates will increase dramatically in the future. Here we have described an infrastructure that will accommodate future improvements in high-throughput high-resolution electron microscopy.
Acknowledgments
We thank Dr. James Wilgenbusch for hosting the resources at the FSU HPC. D.C.S. was supported by a New Florida Award from the FSU Board of Governors. S.M.S. was supported by grants from the AHA (#0835300N) and NIH (R01GM086892)
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Bammes BE, Rochat RH, Jakana J, Chen D-H, Chiu W. Direct electron detection yields cryo-EM reconstructions at resolutions beyond 3/4 Nyquist frequency. J Struct Biol. 2012 doi: 10.1016/j.jsb.2012.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Booth CR, Jakana J, Chiu W. Assessing the capabilities of a 4kx4k CCD camera for electron cryo-microscopy at 300kV. J Struct Biol. 2006;156:556–563. doi: 10.1016/j.jsb.2006.08.019. [DOI] [PubMed] [Google Scholar]
- Brilot AF, Chen JZ, Cheng A, Pan J, Harrison SC, et al. Beam-induced motion of vitrified specimen on holey carbon film. J Struct Biol. 2012;177:630–637. doi: 10.1016/j.jsb.2012.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cong Y, Baker ML, Jakana J, Woolford D, Miller EJ, et al. 4.0-A resolution cryo-EM structure of the mammalian chaperonin TRiC/CCT reveals its unique subunit arrangement. Proc Natl Acad Sci U S A. 2010;107:4967–4972. doi: 10.1073/pnas.0913774107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellisman M, Deerinck T, Bushong E, Bouwer J, Shone T, et al. Advances in Extreme-Scale 3D EM: Specimen Preparation and Recording Systems for Electron Microscopic Tomography and Serial Block Face SEM. Microscopy and Microanalysis. 2011;17:976–977. [Google Scholar]
- Faruqi AR, Henderson R. Electronic detectors for electron microscopy. Curr Opin Struct Biol. 2007;17:549–555. doi: 10.1016/j.sbi.2007.08.014. [DOI] [PubMed] [Google Scholar]
- Henderson R. The potential and limitations of neutrons, electrons and X-rays for atomic resolution microscopy of unstained biological molecules. Q Rev Biophys. 1995;28:171–193. doi: 10.1017/s003358350000305x. [DOI] [PubMed] [Google Scholar]
- Lander GC, Stagg SM, Voss NR, Cheng A, Fellmann D, et al. Appion: an integrated, database-driven pipeline to facilitate EM image processing. J Struct Biol. 2009;166:95–102. doi: 10.1016/j.jsb.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeBarron J, Grassucci RA, Shaikh TR, Baxter WT, Sengupta J, Frank J. Exploration of parameters in cryo-EM leading to an improved density map of the E. coli ribosome. J Struct Biol. 2008;164:24–32. doi: 10.1016/j.jsb.2008.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lerch T, O’Donnell J, Meyer N, Xie Q, Taylor K, et al. Structure of AAV-DJ, a Retargeted Gene Therapy Vector: Cryo-Electron Microscopy at 4.5Å Resolution. Structure. 2012 doi: 10.1016/j.str.2012.05.004. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludtke SJ, Baker ML, Chen DH, Song JL, Chuang DT, Chiu W. De novo backbone trace of GroEL from single particle electron cryomicroscopy. Structure. 2008;16:441–448. doi: 10.1016/j.str.2008.02.007. [DOI] [PubMed] [Google Scholar]
- Mastronarde DN. Automated electron microscope tomography using robust prediction of specimen movements. J Struct Biol. 2005;152:36–51. doi: 10.1016/j.jsb.2005.07.007. [DOI] [PubMed] [Google Scholar]
- McMullan G, Chen S, Henderson R, Faruqi AR. Detective quantum efficiency of electron area detectors in electron microscopy. Ultramicroscopy. 2009;109:1126–1143. doi: 10.1016/j.ultramic.2009.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milazzo AC, Cheng A, Moeller A, Lyumkis D, Jacovetty E, et al. Initial evaluation of a direct detection device detector for single particle cryo-electron microscopy. J Struct Biol. 2011;176:404–408. doi: 10.1016/j.jsb.2011.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nickell S, Förster F, Linaroudis A, Net WD, Beck F, et al. TOM software toolbox: acquisition and analysis for electron tomography. J Struct Biol. 2005;149:227–234. doi: 10.1016/j.jsb.2004.10.006. [DOI] [PubMed] [Google Scholar]
- Rosenthal PB, Henderson R. Optimal determination of particle orientation, absolute hand, and contrast loss in single-particle electron cryomicroscopy. Journal of Molecular Biology. 2003;333:721–745. doi: 10.1016/j.jmb.2003.07.013. [DOI] [PubMed] [Google Scholar]
- Stagg SM, Lander GC, Quispe J, Voss NR, Cheng A, et al. A test-bed for optimizing high-resolution single particle reconstructions. J Struct Biol. 2008;163:29–39. doi: 10.1016/j.jsb.2008.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stölken M, Beck F, Haller T, Hegerl R, Gutsche I, et al. Maximum likelihood based classification of electron tomographic data. J Struct Biol. 2011;173:77–85. doi: 10.1016/j.jsb.2010.08.005. [DOI] [PubMed] [Google Scholar]
- Suloway C, Pulokas J, Fellmann D, Cheng A, Guerra F, et al. Automated molecular microscopy: the new Leginon system. J Struct Biol. 2005;151:41–60. doi: 10.1016/j.jsb.2005.03.010. [DOI] [PubMed] [Google Scholar]
- Winkler H. 3D reconstruction and processing of volumetric data in cryo-electron tomography. J Struct Biol. 2007;157:126–137. doi: 10.1016/j.jsb.2006.07.014. [DOI] [PubMed] [Google Scholar]
- Zhang X, Settembre E, Xu C, Dormitzer PR, Bellamy R, et al. Near-atomic resolution using electron cryomicroscopy and single-particle reconstruction. Proc Natl Acad Sci U S A. 2008;105:1867–1872. doi: 10.1073/pnas.0711623105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng SQ, Keszthelyi B, Branlund E, Lyle JM, Braunfeld MB, et al. UCSF tomography: an integrated software suite for real-time electron microscopic tomographic data collection, alignment, and reconstruction. J Struct Biol. 2007;157:138–147. doi: 10.1016/j.jsb.2006.06.005. [DOI] [PubMed] [Google Scholar]
- Zhou ZH. Atomic resolution cryo electron microscopy of macromolecular complexes. Adv Protein Chem Struct Biol. 2011;82:1–35. doi: 10.1016/B978-0-12-386507-6.00001-4. [DOI] [PMC free article] [PubMed] [Google Scholar]