Abstract
Natural transformation is the main means of horizontal genetic exchange in the obligate human pathogen Neisseria gonorrhoeae. Neisseria spp. have been shown to preferentially take up and transform their own DNA by recognizing a non-palindromic 10 or 12 nucleotide DNA uptake sequence (DUS10 or DUS12). We investigated the ability of the DUS12 to enhance single-stranded DNA (ssDNA) transformation. Given the non-palindromic nature of the DUS12, we tested whether both strands of the DUS equally enhance transformation. Recombinant single-stranded M13 phage harboring transforming DNA with the Watson DUS12, the Crick DUS12, or no DUS (DUS0) were constructed and circular ssDNA was purified. Southern blots of the purified DNA probed with strand-specific oligonucleotide probes showed greater than 10,000:1 ratio of ssDNA to contaminating dsDNA. The Crick strand of the DUS12 enhanced ssDNA transformation 180–470 fold over DUS0 ssDNA whereas the Watson strand of the DUS only modestly enhanced ssDNA transformation in two strains of N. gonorrhoeae. These data confirm that ssDNA efficiently transforms N. gonorrhoeae but that there is a strand preference, and that part of this strand preference is a greater efficiency of the Crick strand of the DUS12 in enhancing transformation.
Keywords: genetic exchange, pathogen, recombination
INTRODUCTION
Natural transformation is a widespread mechanism for horizontal genetic exchange used by numerous bacterial species (Johnsborg, et al., 2007), and is the main means of horizontal genetic exchange in the obligate human pathogen Neisseria gonorrhoeae and the related pathogen Neisseria meningitidis (Koomey, 1998). With the exception of Helicobacter pylori, all currently identified DNA uptake systems use type IV pili, type II secretion systems, or uptake machinery related to these secretion systems (reviewed in (Chen & Dubnau, 2004)). N. gonorrhoeae use a Type IV pilus for transformation, and are constitutively competent for DNA transformation (Sparling, 1966). The lack of stable clonal lineages indicates that exchange of chromosomal DNA is common between N. gonorrhoeae strains (Smith, et al., 1993). DNA transformation is a multi-step process which includes DNA binding, DNA uptake into the periplasm and cytoplasm, and DNA recombination into the chromosome (reviewed in (Hamilton & Dillard, 2006)).
Neisseria species have been shown to preferentially take up and transform their own DNA by virtue of a non-palindromic Neisseria specific DNA uptake sequence (DUS) (Elkins, et al., 1991). There are two forms of the DUS, DUS10 (5′-GCCGTCTGAA) and DUS12 (5′-ATGCCGTCTGAA), which are necessary for the most efficient transformation into Neisseria, with the DUS12 sequence showing the greatest efficiency (Smith, et al., 1999, Ambur, et al., 2007). Neisseria genomes are enriched for the DUS10 and DUS12 sequences, and many reports have demonstrated increased DNA uptake and transformation with DNA fragments containing one or both DUS sequences (Goodman & Scocca, 1988, Ambur, et al., 2007, Duffin & Seifert, 2010). It appears that the DUS10 and DUS12 sequences function similarly but that the DUS12 provides a small increase in transformation efficiency. The accepted model of DUS action invokes the DUS binding to a putative outer membrane receptor leading to enhanced DNA transport into the periplasm, although the mechanism is uncertain and no receptor has been identified. Recently, we proposed a more complex role for the DUS during transformation which includes undefined roles within the periplasm (Duffin & Seifert, 2010).
Most investigations into transformation of N. gonorrhoeae have used double-stranded DNA (dsDNA) substrates, but a few have utilized single-stranded DNA (ssDNA) substrates to study transformation. Several observations suggest that ssDNA is an important substrate for transformation including: A) single-stranded chromosomal DNA is secreted by the Neisseria type IV secretion system (Salgado-Pabon, et al., 2007) and co-culture experiments show that this secreted DNA transforms recipient cells efficiently (Dillard & Seifert, 2001); B) the secretin PilQ, which is required for DNA uptake, binds ssDNA better than dsDNA (Assalkhou, et al., 2007); and C) ssDNA has been reported to transform at levels similar to dsDNA (Stein, 1991). No reports have investigated the potential role of the two forms of the non-palindromic DUS in ssDNA transformation.
We have purified single-stranded transforming DNA carrying the each sequence of the DUS12 sequence. These ssDNA substrates were used to transform two laboratory strains of N. gonorrhoeae to demonstrate that ssDNA transformation is overall less efficient than dsDNA transformation, and that one strand of the DUS12 (Crick strand) strongly enhances ssDNA transformation while the other strand has a modest effect on transformation efficiency. These data suggest that N. gonorrhoeae transformation of ssDNA is largely dependent on the presence of the Crick DUS12.
METHODS AND MATERIALS
Bacterial strains and growth conditions
N. gonorrhoeae was grown on GC Medium Base (GCB) (Difco) plates with Kellogg’s supplements I and II (Kellogg, et al., 1968) and incubated at 37 °C in a 5% CO2 humidified atmosphere. Escherichia coli strain TOP10F′ (Invitrogen) was used to replicate recombinant M13 phage. The F′ episome was maintained in the TOP10F′ cells by addition of tetracycline (15 μg/ml) in the LB or YT media used to grow the E. coli. Transformation was investigated in the laboratory strains FA1090 (Connell, et al., 1988) and MS11 (Meyer, et al., 1982). The concentration of Nalidixic acid (Nal) in GCB was 1 μg/ml for strain FA1090 and 3 μg/ml for strain MS11.
Construction of recombinant DUS0 and DUS12 gyrB1 M13 phages
We have previously constructed plasmids containing DUS0 and DUS12 gyrB1 DNA (plasmids gyrB1 DUS0 and gyrB1 DUS12 (Duffin & Seifert, 2010)); these plasmids were digested with EcoRI and the DNA fragments were cloned into EcoRI digested M13mp18 and M13mp19 replicative form (RF) DNA and positive clones isolated in TOP10F′ cells (Invitrogen) using blue/white screening on Xgal containing media. Recombinant RF DNA was purified from infected TOP10F′ cells and gyrB1 inserts were confirmed by restriction digest analysis and DNA sequencing. M13mp18 and M13mp19, which have opposing orientation of the multiple cloning sites, were utilized so that either the Watson or the Crick strand of the gyrB1 and DUS12 would be expressed from recombinant phage. Recombinant phage harboring both orientations DUS12 gyrB1 DNA and the DUS0 constructs were obtained and used to produce ssDNA. DNA sequencing was carried at the sequencing core of Northwestern University and the program suite VectorNTI (Invitrogen) was used to analyze DNA sequences.
Purification ssDNA and RF dsDNA
TOP10F′ cells were infected with recombinant M13 phage and grown for 5 hours at 37°C with constant agitation. Qiagen M13 and Qiagen miniprep kits were used to purify ssDNA and RF DNA, respectively, from recombinant phage infection following the manufacturer’s instructions. The amount of contaminating dsDNA (from RF DNA) in the ssDNA preps was assessed by Southern blots probed with oligonucleotide probes (see below). Due to variability in the quality of the ssDNA preparations (possibly due to cell lysis during phage infection), each individual ssDNA preparation was measured for ssDNA purity by agarose gel electrophoresis and Southern blot analysis (see below). In order to obtain sufficient ssDNA for the transformation experiments, ssDNA preparations that were deemed pure (less than 1:10,000th contaminating DNA) were pooled together to create ssDNA stocks. The pooled ssDNA stocks were subjected to agarose gel electrophoresis and Southern blot analysis to confirm the purity and then used in transformation experiments.
Southern blots of ssDNA and RF dsDNA
Southern blots probed with DIG-labeled oligonucleotides were used to measure the purity of the ssDNA preparations. Briefly, oligonucleotides gyrBtop2 (5′-GCCATCGACGAAGCACTC) and gyrBbot12 (5′-GGCTTTTTCCAAGGCAAGG) were end labeled with DIG (Roche) following the manufacturer’s instructions. Hybridization, washes, and detection of the Southern blots were performed as per the manufacturer’s instructions (Roche) to determine the relative amounts of ssDNA and RF DNA in the aforementioned preparations.
Transformation assays
Gonococcal strains were grown for 18 hours on GCB plates and resuspended in liquid transformation media (1.5% protease peptone no. 3 (Difco), 0.1% NaCl, 200 mM HEPES (Sigma), 5mM MgSO4 and Kellogg supplements I and II, pH 7.2) to an optical density at 600nm of approximately 1.5. Thirty μl of the cell suspension was added to tubes containing 0.045 pmol of gyrB1 DNA and 200μl transformation media. DUS12 and DUS0 containing plasmids of gyrB1 (Duffin & Seifert, 2010) were used as transforming dsDNA and purified recombinant phage DNA was used as transforming ssDNA. Following incubation at 37°C for 20 min, transformation mixtures were added to pre-warmed 2ml transformation media and incubated at 37°C in the presence of 5% CO2 for 4 h. The mixtures were serially diluted 10-fold in transformation media lacking MgSO4 and Kellogg supplements and 20 μl serial 10-fold dilutions were spotted on GCB plates in the presence and absence of Nal. Transformation efficiencies are reported as antibiotic resistant CFU divided by total CFU, and are the mean of at least three replicates.
RESULTS
ssDNA preparations contain very little dsDNA
Efficient transformation in N. gonorrhoeae requires the presence of the DUS in the transforming DNA and homology to DNA sequences present within the genome (Ambur, et al., 2007, Duffin & Seifert, 2010). Many N. gonorrhoeae strains harbor a type IV secretions system and thus secrete ssDNA which can serve as substrate for transformation (Dillard & Seifert, 2001, Salgado-Pabon, et al., 2007). No reports have investigated the potential role of the DUS in ssDNA transformation, which may clarify its mechanism of action during transformation. Recombinant M13 phage were used to isolate gyrB1 transforming DNA cloned in both orientations so that the single-stranded DNA would carry either the Watson DUS12 (5′-ATGCCGTCTGAA-3′), the Crick DUS12 (5′-TTCAGACGGCAT-3′), or no DUS (DUS0).
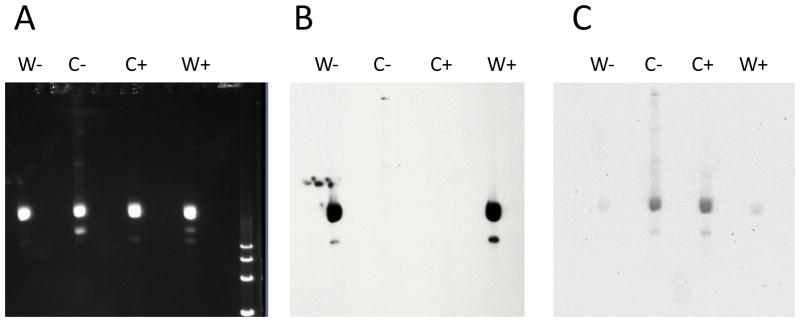
Since dsDNA replicative form DNA (RF DNA) is produced during the course of M13 infection (Sambrook, et al., 2001) and any contaminating dsDNA would transform N. gonorrhoeae, we utilized column purification of the ssDNA following phage isolation (see methods). We then determined the relative amount of dsDNA in the ssDNA preparations using Southern blots with oligonucleotide probes that bind either the Watson or the Crick strand (Fig 1). Southern analysis revealed two distinct species of ssDNA; a major band and a minor smaller band (Fig. 1). The oligonucleotide probe gyrBtop1-DIG hybridized to the Watson ssDNA preparation but not to the Crick ssDNA preparation, indicating low levels of contaminating dsDNA in the Crick ssDNA preparations (Fig. 1B). Similarly, the oligonucleotide probe gyrB2bot-DIG hybridized to the Crick ssDNA preparation, but not to the Watson ssDNA preparation, indicating minimal dsDNA contamination in the Watson ssDNA preparations (Fig. 1C). Analysis of these Southern blot signals shows that the ssDNA preparations contained at most a ca. 10,000:1 ratio of ssDNA to contaminating dsDNA (Fig. 1). However, the supposedly double-stranded RF DNA preparations that were extracted from cells showed considerable ssDNA contamination (data not shown), and thus equal moles of each corresponding plasmid DNA were used for dsDNA controls in transformation experiments.
Figure 1.
Characterization of gyrB1 DUS0/12 recombinant M13 ssDNA preparations. A, SYBR green stained agaose gel. B, Southern blot hybridized with oligonucleotide probe, gyrBtop1-DIG. C, Southern blot hybridized with oligonucleotide probe, gyrB2bot-DIG. W indicates Watson strand ssDNA, C indicates Crick strand ssDNA, - indicates DUS0, and + indicates DUS12.
ssDNA transformation is less efficient than dsDNA transformation
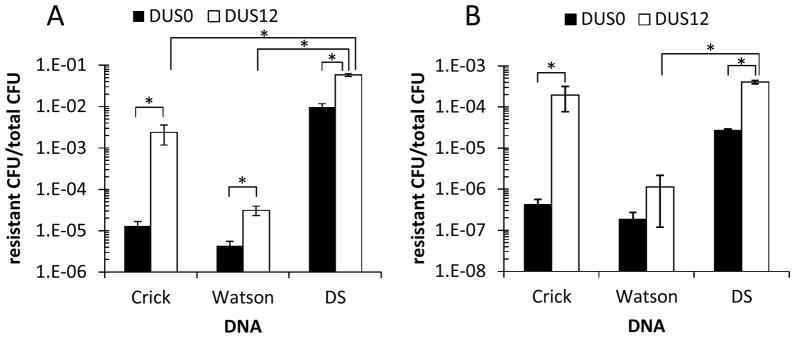
Transformation with equal molar amounts of gyrB1 ssDNA was less efficient in all cases except for Crick DUS12 in MS11 than the identical dsDNA for both strains FA1090 and MS11 (Fig. 2, p<0.05 by Student’s t-test). Watson and Crick DUS0 ssDNA transformation was reduced approximately 740-fold and 2,200-fold, respectively, compared to matched DUS0 dsDNA (Fig. 2A, p<0.05 by Student’s t-test). Similar to DUS0 dsDNA transformation levels, DUS0 ssDNA transformation was less efficient in MS11 than in FA1090 (Fig. 2). Interestingly, Crick DUS0 ssDNA transformation was consistently but not statistically more efficient than Watson DUS0 in ssDNA transformation (p>0.05, 3-fold and 2-fold higher in FA1090 and MS11, respectively).
Figure 2.
ssDNA transformation is less efficient than dsDNA transformation and the Crick strand of DUS12 enhances ssDNA transformation in strain FA1090 (A) and MS11 (B). Strain FA1090 (A) or MS11 (B) were quantitatively transformed with equal moles of DUS0 (black bars) and DUS12 (white bars) Crick ssDNA, Watson ssDNA, and plasmid dsDNA (DS) containing gyrB1. Transformation efficiencies are plotted as resistant CFU/total CFU. *p<0.05. Error bars are SEM.
The Crick strand of DUS12 has a greater effect on ssDNA transformation than the Watson strand
In agreement with previous reports, dsDNA transformation was enhanced by the DUS12 in both FA1090 and MS11, 6 and 16 fold compared to the DUS0 controls, respectively (Fig. 2, p<0.05 by Student’s t-test). Similarly, the Crick DUS12 sequence enhanced transformation of ssDNA in both FA1090 and MS11; however the magnitude of enhancement was much larger than for dsDNA. The Crick DUS12 enhanced ssDNA transformation 182-fold and 467-fold over DUS0 ssDNA in FA1090 and MS11, respectively (Fig. 2, p<0.05 by Student’s t-test). In FA1090, Crick DUS12 ssDNA transformation efficiency was 24-fold lower than dsDNA DUS12 efficiency (p<0.05 by Student’s t-test). However, in MS11 Crick DUS12 ssDNA transformation efficiency was similar to dsDNA DUS12 (2-fold lower, p>0.05) which is consistent with previous findings (Stein, 1991). In contrast, the Watson DUS12 ssDNA only showed a ca. 7-fold increase in transformation enhancement over matched DUS0 ssDNA (Fig. 2, p<0.05 in FA1090, not statistically significant in MS11) and were greatly reduced from dsDNA DUS12 levels (p<0.05, 1871-fold lower and 354-fold lower in FA1090 and MS11, respectively). The results demonstrate that within a ssDNA substrate that the Crick DUS12 sequence show a much greater activity to promote transformation.
DISCUSSION
Using highly purified ssDNA, we examined the ability of the Watson DUS12 or Crick DUS12 to enhance ssDNA transformation of N. gonorrhoeae. These data show that: A) ssDNA transforms less efficiently than equivalent number of molecules of dsDNA, B) the Crick strand of the DUS12 is more active to enhance ssDNA transformation than the Watson strand of the DUS12, and C) the magnitude of enhancement of the DUS12 is larger for ssDNA than for dsDNA. These data suggest that N. gonorrhoeae transformation by ssDNA is largely dependent on the presence of the Crick DUS12.
A previous study reported efficient ssDNA transformation in N. gonorrhoeae much higher than the levels we measured (Stein, 1991). This study did not report how much contaminating dsDNA was present in the ssDNA preparations and therefore those results are difficult to compare to the results obtained in this study. Our data show that there is significant dsDNA contamination of standard M13 ssDNA preparations and we added a column purification step to enrich for ssDNA molecules. It is possible that the high transformation efficiencies reported previously (Stein, 1991) were due to contaminating double-stranded RF DNA within the recombinant M13 phage preparations.
Our results support the observation of transformation in co-culture experiments with strains secreting ssDNA via the type IV secretion system (Dillard & Seifert, 2001). Interestingly, Crick DUS0 ssDNA transformation was consistently, but not statistically higher than Watson DUS0 ssDNA transformation. We do not presently understand the reason why the Crick strand transforms consistently, but not statistically better without a DUS, but it could be used more efficiently during uptake or recombination into the chromosome, or perhaps is more resistant to nucleases encountered during the transformation process. Although both the Watson and the Crick DUS12 sequences enhanced transformation in both FA1090 and MS11, the magnitude of enhancement was much greater for the Crick DUS12 than the Watson DUS12 (Fig 2). Again, these differences could be mediated at any stage in the transformation process.
The previously accepted model of dsDNA DUS12 action invokes the DUS12 sequence binding to a putative outer membrane receptor leading to increased DNA uptake into the periplasm. We have suggested that the DUS may have more complicated role during the process of transformation (Duffin & Seifert, 2010) which may include a role for the DUS beyond DNA uptake into the periplasm. Many factors are required for the complex process of transformation including DNA binding and DNA uptake into the periplasm and through the inner membrane. Prior reports have shown DUS12 dsDNA uptake is transported into the periplasm, but no reports have shown ssDNA transport. However, since all of the previous studies establish that the dsDUS mediates transport into the periplasm, we do not favor a role for the ssDUS in this step of transformation. A lack of activity in DNA uptake for ssDUS could explain the overall reduction in transformation of ssDNA compared to dsDNA. We therefore propose that the Crick DUS12 may function to enhance transformation efficiency independent of ssDNA uptake into the periplasm.
Acknowledgments
This work was supported by National Institutes of Health grants R37 AI033493 and R01 AI044239 to HSS.
References
- Ambur OH, Frye SA, Tonjum T. New functional identity for the DNA uptake sequence in transformation and its presence in transcriptional terminators. J Bacteriol. 2007;189:2077–2085. doi: 10.1128/JB.01408-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Assalkhou R, Balasingham S, Collins RF, et al. The outer membrane secretin PilQ from Neisseria meningitidis binds DNA. Microbiology. 2007;153:1593–1603. doi: 10.1099/mic.0.2006/004200-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen I, Dubnau D. DNA uptake during bacterial transformation. Nat Rev Microbiol. 2004;2:241–249. doi: 10.1038/nrmicro844. [DOI] [PubMed] [Google Scholar]
- Connell TD, Black WJ, Kawula TH, et al. Recombination among protein II genes of Neisseria gonorrhoeae generates new coding sequences and increases structural variability in the protein II family. Mol Microbiol. 1988;2:227–236. doi: 10.1111/j.1365-2958.1988.tb00024.x. [DOI] [PubMed] [Google Scholar]
- Dillard JP, Seifert HS. A variable genetic island specific for Neisseria gonorrhoeae is involved in providing DNA for natural transformation and is found more often in disseminated infection isolates. Mol Microbiol. 2001;41:263–277. doi: 10.1046/j.1365-2958.2001.02520.x. [DOI] [PubMed] [Google Scholar]
- Duffin PM, Seifert HS. DNA uptake sequence-mediated enhancement of transformation in Neisseria gonorrhoeae is strain dependent. J Bacteriol. 2010;192:4436–4444. doi: 10.1128/JB.00442-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elkins C, Thomas CE, Seifert HS, Sparling PF. Species-specific uptake of DNA by gonococci is mediated by a 10-base-pair sequence. J Bacteriol. 1991;173:3911–3913. doi: 10.1128/jb.173.12.3911-3913.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodman SD, Scocca JJ. Identification and arrangement of the DNA sequence recognized in specific transformation of Neisseria gonorrhoeae. Proc Natl Acad Sci U S A. 1988;85:6982–6986. doi: 10.1073/pnas.85.18.6982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton HL, Dillard JP. Natural transformation of Neisseria gonorrhoeae: from DNA donation to homologous recombination. Mol Microbiol. 2006;59:376–385. doi: 10.1111/j.1365-2958.2005.04964.x. [DOI] [PubMed] [Google Scholar]
- Johnsborg O, Eldholm V, Havarstein LS. Natural genetic transformation: prevalence, mechanisms and function. Res Microbiol. 2007;158:767–778. doi: 10.1016/j.resmic.2007.09.004. [DOI] [PubMed] [Google Scholar]
- Kellogg DS, Jr, Cohen IR, Norins LC, Schroeter AL, Reising G. Neisseria gonorrhoeae. II. Colonial variation and pathogenicity during 35 months in vitro. J Bacteriol. 1968;96:596–605. doi: 10.1128/jb.96.3.596-605.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koomey M. Competence for natural transformation in Neisseria gonorrhoeae: a model system for studies of horizontal gene transfer. APMIS Suppl. 1998;84:56–61. doi: 10.1111/j.1600-0463.1998.tb05649.x. [DOI] [PubMed] [Google Scholar]
- Meyer TF, Mlawer N, So M. Pilus expression in Neisseria gonorrhoeae involves chromosomal rearrangement. Cell. 1982;30:45–52. doi: 10.1016/0092-8674(82)90010-1. [DOI] [PubMed] [Google Scholar]
- Salgado-Pabon W, Jain S, Turner N, van der Does C, Dillard JP. A novel relaxase homologue is involved in chromosomal DNA processing for type IV secretion in Neisseria gonorrhoeae. Mol Microbiol. 2007;66:930–947. doi: 10.1111/j.1365-2958.2007.05966.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Russell DW, Cold Spring Harbor L. Molecular cloning: a laboratory manual/Joseph Sambrook, David W Russell. Cold Spring Harbor Laboratory; Cold Spring Harbor, N.Y: 2001. [Google Scholar]
- Smith HO, Gwinn ML, Salzberg SL. DNA uptake signal sequences in naturally transformable bacteria. Res Microbiol. 1999;150:603–616. doi: 10.1016/s0923-2508(99)00130-8. [DOI] [PubMed] [Google Scholar]
- Smith JM, Smith NH, O’Rourke M, Spratt BG. How clonal are bacteria? Proc Natl Acad Sci U S A. 1993;90:4384–4388. doi: 10.1073/pnas.90.10.4384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sparling PF. Genetic transformation of Neisseria gonorrhoeae to streptomycin resistance. J Bacteriol. 1966;92:1364–1371. doi: 10.1128/jb.92.5.1364-1371.1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein DC. Transformation of Neisseria gonorrhoeae: physical requirements of the transforming DNA. Can J Microbiol. 1991;37:345–349. doi: 10.1139/m91-056. [DOI] [PubMed] [Google Scholar]