Abstract
HLA-DM (DM) is a non-classical major histocompatibility complex II (MHC II) protein that acts as a peptide editor to mediate the exchange of peptides loaded onto MHC II during antigen presentation. Although the ability of DM to promote peptide exchange in vitro and in vivo is well established, the role of DM in epitope selection is still unclear, especially in human response to infectious disease. In this study, we addressed this question in the context of the human CD4 T cell response to vaccinia virus. We measured the IC50, intrinsic dissociation half-life, and DM-mediated dissociation half-life for a large set of peptides derived from the major core protein A10L and other known vaccinia epitopes bound to HLA-DR1, and compared these properties to the presence and magnitude of peptide-specific CD4+ T cell responses. We found that MHC II-peptide complex kinetic stability in the presence of DM distinguishes T cell epitopes from non-recognized peptides in A10L peptides and also in a set of predicted tight binders from the entire vaccinia genome. Taken together, these analyses demonstrate that DM-mediated dissociation half-life is a strong and independent factor governing peptide immunogenicity by favoring the presentation of peptides with greater kinetic stability in the presence of DM.
Introduction
Antigen presentation to CD4 T cells by major histocompatibility complex class II molecules (MHC II)3 is a key process in activation of the adaptive immune system and prevention of infection by pathogens. The process is characterized by the proteolytic cleavage of pathogen-derived proteins and loading of the resultant peptide fragments onto MHC II in specialized endosomal compartment (1–3). The loading of antigenic peptides onto MHC II is catalyzed by HLA-DM (DM), a non-classical MHC II (4). DM also acts as a molecular chaperone to stabilize MHC II in the absence of peptide (5–6). Although DM can catalyze association and dissociation of MHC II-peptide complexes in vitro (7–8) and in vivo (9–10), the role of DM-mediated peptide exchange in epitope selection remains unclear.
There are multiple factors that can influence which peptides are selected as epitopes. Protease cleavage site (11–12), T cell precursor frequency (13–14) and T cell competition (15–16) have been shown to be involved. More recently, the strength of MHC II-peptide interaction during antigen processing and presentation has been demonstrated to be one of the major factors influencing the specificity of T cells (10, 17–20). It has been found that kinetic stability of MHC II-peptide complexes is a key parameter that dictates immunodominance (17). The role of DM in determining peptide immunogenicity has been investigated in an in vitro cell-free epitope selection system (21), using ex vivo antigen presentation and T cell stimulation assays (10, 22–23), and in an animal study (18). DM has been shown to influence epitope selection by favoring the presentation of peptides bound to MHC II molecules with high stabilities (7, 9–10, 18, 23–24). However, there are some immunodominant epitopes identified with low binding affinities and low kinetic stabilities, and, notably, those epitopes all derived from self antigens and were often associated with autoimmune diseases such as multiple sclerosis (MS) (14, 25–26) and rheumatoid arthritis (RA) (27). Despite these advances, a study of DM in epitope selection in the context of a human infectious disease has yet to be done. MHC II-peptide complexes are differentially susceptible to DM-mediated dissociation (7, 28). We hypothesize that DM-mediated half-life is a primary contributory factor that governs peptide immunogenicity in epitope selection and only those peptides bound to MHC II that are less susceptible to DM-mediated dissociation can get presented and selected as epitopes.
In the present study, we used the human CD4 T cell response to vaccinia virus as a test system for evaluating factors contributing to peptide immunogenicity. We evaluated in detail the peptide specificity of the HLA-DR1 restricted response to the A10L major core protein, and we measured MHC II-peptide interaction for a series of overlapping peptides covering the entire A10L sequence. We characterized the MHC-peptide interaction using an equilibrium competition binding assay (IC50), and a kinetic dissociation assay in the absence of DM (intrinsic half-life) and in the presence of DM (DM-mediated half-life). Through statistical analysis between peptide immunogenicity and IC50, intrinsic half-life, and DM-mediated half-life, we found that DM contributes to epitope selection in A10L by favoring the presentation of peptides with higher DM-mediated kinetic stability. In addition, we compared those properties between several epitopes and non-recognized peptides from other vaccinia virus proteins. We found that DM-mediated half-life is a distinguishing feature to separate epitopes from non-recognized peptides, and that MHC II complexes formed by epitope peptides are less susceptible to DM-mediated dissociation.
Materials and Methods
Peptide synthesis and labeling
A10L peptides were synthesized for IFNγ-ELISPOT assay with acetylated N-termini and with N-terminal biotin-PEO (tetrapolyethyleneoxide) for IC50 and dissociation kinetics assays (21st Century Biochemicals, Marlboro, MA). The A10L sequence was obtained from vaccinia virus strain Copenhagen (http://www.ncbi.nlm.nih.gov/protein, GenBank accession number: AAA48129), which shares 99.1% identity with strain MVA (modified vaccinia Ankara), and 98.8% identity with strain Dryvax, the two vaccinia strains to which our donors were exposed. N-terminally acetylated influenza hemaagglutinin analog peptide HA306–318 (Ac-PRFVKQNTLRLAT) was labeled with Alexa-488 tetrafluorophenyl ester (Invitrogen, Eugene, OR) through primary amine of K5 for competition binding studies. Labeled peptides were purified by Jupiter C18 reverse phase chromatography (Phenomenex, Torrance, CA) and subjected to MALDI to confirm the expected molecular weight.
HLA-DR1 and HLA-DM expression and purification
Soluble recombinant HLA-DR1 (HLA DRB1*0101) and HLA-DM were expressed in Drosophila S2 cells and purified by immunoaffinity chromatography followed by Superdex200 (GE Healthcare) size exclusion chromatography as described (29).
Fluorescence Polarization (FP) assay
FP assay was used to measure the IC50 of each peptide bound to HLA-DR1. 25nM Alexa488-labeled HA306–318 as indicator peptide was incubated with 100nM HLA-DR1, along with various dilutions of cold target peptide (five-fold dilutions from 20uM), at 37 °C for 3 days in 200ul pH5.5 binding buffer (100mM sodium citrate, 50mM NaCl, 0.1% octylglucoside, 5mM EDTA, 0.1% NaN3, 1mM DTT, 1mM PMSF) in 96-well non-binding black polystyrene plates (Corning Incorporated, Corning, NY). FP was read using BMG Polarstar plate reader (BMG LABTECH, Cary, NC) at 488nm excitation and 520nm emission. FP values were converted to fractional binding using the values for fully bound (~350 mP) and free (~70 mP) Alexa488-labeled HA306–318. The competition curve was fitted to equation FP=1/(1+[pep]/IC50) in KaleidaGraph (Synergy software, Reading, PA), where FP is the fluorescence polarization value in the presence of unlabeled competitor peptide at concentration [pep] and IC50 is the 50% inhibition concentration.
Peptide dissociation assay
MHC II-peptide complexes were formed by incubating 1uM HLA-DR1 with 10 uM biotin labeled peptides in binding buffer at 37 °C for 3 days. The complexes were purified using Sephadex G-50 Nick columns (GE Healthcare) and incubated at final concentration of 100 nM without or with different concentrations of DM together with 10 uM cold HA306–318 to prevent rebinding of released biotin peptides. 200 ul of dissociation mixture was collected at different time points and mixed with 15 ul 0.5M Tris-HCl (pH 8.0). Samples were frozen immediately after collection and thawed together before assay. The thawed dissociation mixtures were incubated in anti-DR1 antibody LB3.1 pre-coated 96-well Lumitrac 600 white plates (USA Scientific, Ocala, FL) at 4 °C for 3 hours, washed three times with PBS + 0.05% Tween-20, incubated with Europium-streptavidin at 37 °C for 1 hour, washed again, and then mixed with Europium enhancement solution to release EU3+ (PerkinElmer, Shelton, CT). Victor plate reader (PerkinElmer, Shelton, CT) was used to read the time resolved fluorescence of EU3+. The dissociation curve was fitted to single phase exponential decay with constraint 100% bound at time 0 in GraphPad Prism 5 (GraphPad software, San Diego, CA) to determine the off rate koff and half-life t1/2.
Vaccinia virus (VV)-specific CD4 T cell line (TCL)
A HLA-DR1+ donor (donor SL131, DRB1*0101/0407, DRB4*01 DQA1*0101/0301, DQB1*0302/0501, DPA1*0103/01, DPB1*0301/0402) previously immunized against smallpox approximately 35 years earlier and re-immunized with vaccine Dryvax approximately 4 years before this study. The blood collection was done under a protocol approved by the Medical School Institutional Review Board of the University of Massachusetts. HLA class II haplotype was determined by the UMass MHC haplotyping core facility using PCR-based protocols. VV-specific CD4 TCLs were generated by in vitro stimulation. Briefly, peripheral blood mononuclear cells (PBMCs) from this VV-vaccinated HLA-DR1 donor (SL131) were resuspended in cRPMI medium (RPMI1640 supplemented with 10% human serum, 100U/ml penicillin, 100ug/ml streptomycin, 1 mM sodium pyruvate, 2 mM L-glutamine, and 1mM non-essential amino acids from GIBCO) and 1×106 cells in 1 ml were dispensed in a 24-well cell culture plate. Subsequently, 1:100 dilution of heat-inactivated (60 °C for 1hour) vaccinia virus (MVA) -infected CV-1 cell lysate originally containing 1.7×107 pfu/ml, was added to each well. For peptide-specific TCL, use 10 ug/ml peptide as the stimulating antigen. After 3 days, each well was supplemented with 1ml of cRPMI+100U/ml IL-2. Cell expansion proceeded for approximately 17d in cRPMI+100U/ml IL-2 to get enough T cells. TCLs prepared by this method are predominately CD4+ T cells (90–99%) (30). Two non-vaccinated HLA-DR1 donors, SL139 from University of Massachusetts Medical School and 329M from Cellular Technology Limited (CTL Ltd., Shaker Heights, OH), and five additional vaccinia virus-vaccinated HLA-DR1 donors were included in the study. For these additional vaccinated donors, PBMCs from donors 720 and 214B were prepared by CTL Ltd., and PBMCs from donors 2011, 2029 and 2032 before and 45 days after vaccination were prepared by the Saint Louis University Center for Vaccine Development during a study of smallpox vaccines generated by Acambis, Inc. (Cambridge, MA) (31).
IFNγ-ELISPOT
2×104 T cells from the TCL were incubated with 5×104 irradiated autologous PBMC (irPBMC) or HLA-DR1 homozygous human B-lymphoblastoid cells (LG2, DRB1*0101, DQA1*0101, DQB1*050101, DPA1*010301, DPB1*0401) as APCs, using 5 ug/ml peptide as antigen source in 96-well MultiScreen filter plate (Millipore Corporation, Billerica, MA) overnight (~15hr). Number of IFNγ secreting cells was determined using ELISPOT analyzer equipped with ImmunoSpot 5.0.3 software (CTL, Shaker Heights, OH).
Antibody inhibition ELISPOT assay
Genetic restriction of peptide-specific TCLs was determined by using LG2 cells and antibodies to MHC molecules. Briefly, IFN-γ ESLISPOT assays were performed as described above using peptide-pulsed LG2 cells as APCs pre-incubated with antibodies to class I (W6/32) or anti-DR (LB3.1). Antibody inhibition is expressed as percentage and indicates the ratio of responses in the presence of antibody to responses without antibody after subtraction of background responses.
HLA-DR1-A10L peptide tetramer staining assay
Tetramer staining was performed as described (32). Briefly, tetramers were produced by stepwise addition of streptavidin-PE (Invitrogen, Eugene, OR) to biotinylated-HLA-DR1-peptide complexes at a final molar ratio of 1:4. T cells were incubated with tetramers for 4 hours at 37°C and then for 20 minutes with anti-CD4-APC antibody (BD Biosciences, La Jolla, CA) on ice before washing with ice-cold buffer. Tetramer and antibody binding was determined using an LSRII Flow cytometer (BD Biosciences, La Jolla, CA).
Epitope prediction algorithms
P9, IEDB and Syfpeithi prediction algorithms were used to predict the peptide immunogenicity of overlapping A10L peptides. The P9 matrix for prediction of peptide binding to HLA-DR1 (including potential 9 amino acids binding motif) was obtained by modification (30) of the virtual DR1 matrix originally described by Sturniolo et al. (33). The IEDB (34) and Syfpeithi (35) predictions were performed using their respective web servers: http://tools.immuneepitope.org/analyze/html/mhc_II_binding.html http://www.syfpeithi.de/Scripts/MHCServer.dll/EpitopePrediction.htm.
Correlation coefficient analysis (CC)
CC analyses relating peptide immunogenicity (shown as spots per well) and IC50, intrinsic half-life (t1/2in), DM-mediated half-life (t1/2DM), DM-susceptibility (DM-sus), P9 prediction, IEDB prediction and Syfpeithi prediction values were done using Graphpad Prism5. DM-susceptibility (DM-sus), the specific rate increase induced by DM was calculated as follows:
koff,in and koff,DM were calculated by fitting the dissociation curve without or with DM to single phase exponential decays as described above; [DM] represents DM concentration.
Receiver Operating Characteristic (ROC) analysis
ROC analyses (36) for the same parameters tested in CC analysis were done using Graphpad Prism5, using the 7 peptides selected in Figure 1 that were CD4 T cell epitopes as positive and the remainder negative. The confidence interval was set as 95%. The area under curve (AUC), and its corresponding p value were obtained based on the ROC curve.
FIGURE 1. Recognition of A10L peptides by a vaccinia virus-specific CD4 T cell line.
(A) The specificity of the vaccinia-specific TCL was tested using IFNγ-ELISPOT. TCL was stimulated with vaccinia virus-infected CV-1 cell lysate (VV, red bar), non-infected CV-1 cell lysate (NI, green bar), and medium only (MED, blue bar), using irradiated autologous PBMC (irPBMC, left panel) or HLA-DR1 matched B-lymphoblastoid cells (LG2,right panel) as antigen presenting cells (APC). The p-value of responses between VV and NI is indicated. This data represent three independent experiments with 4 replicates each. (B, C) The VV-specific CD4 T cell line was stimulated with overlapping A10L peptides to test peptide immunogenicity, shown as spots per well (SPW) in IFNγ-ELISPOT assay using (B) autologous irPBMC (solid blue circle,) or (C) LG2 (solid green circle) as APCs. Medium (MED) was used as control (clear blue circle in (B) and clear green circle in (D) at the very right x-axis). Dashed line represented the threshold we set up for positive peptides (SPW is more than two fold of that of MED, and p value between that peptide and MED is less than 0.01). The positive peptides are labeled with p-value relative to medium control. (B) and (C) represent three independent experiments with at least two replicates each.
Results
Overlapping A10L peptides are recognized differently by a vaccinia-specific T cell line
We selected the vaccinia major core protein A10L, a large and abundant protein, to address the role of DM-mediated peptide exchange on CD4 T cell epitope selection. A10L is one of the five immunodominant proteins that are recognized by CD4 and CD8 T cells from most vaccinia-immunized subjects (37), and also induces significant B cell responses during infection (38). To date, two A10L-derived CD4 epitopes have been identified (30, 39). One of these was sufficiently abundant to be identified among the set of peptides eluted from HLA-DR1 isolated from vaccinia virus-infected B cells (39). Thus A10L is expressed and processed in antigen presenting cells, with competition among the many potential epitopes, making it an ideal target to study factors governing antigen presentation and epitope selection. As for most other antigens, the role of DM in A10L epitope selection has not been addressed.
To identify CD4 T cell epitopes in A10L, we tested a vaccinia virus-specific polyclonal CD4 T cell line (TCL) generated from a vaccinated HLA-DR1 (DR1B*0101) donor (SL131) for reactivity with a set of peptides representing the entire vaccinia A10L sequence. We generated the vaccinia-specific CD4 T cell line by single in vitro expansion of PBMC with heat-inactivated vaccinia virus. The TCL was verified to be specific to vaccinia virus, using either irradiated autologous PBMC (irPBMC) or a HLA-DR1 homozygous lymphoblastoid B cell line (LG2) as antigen presenting cells in an IFNγ-ELISPOT assay (Fig. 1A). For epitope mapping experiments, we synthesized a set of 126 peptides, each 18 residues long, which covers the full length of the A10L protein (supplemental Table I). The peptides were designed with 11-residue overlaps to ensure that any potential epitope (11 residues or shorter) would be present on at least one peptide. Typically class II MHC binding frames are 9 residues long (40), but occasionally the residues immediately flanking the 9-mer binding frame can be recognized by T cells (41–42). T cell recognition of the 126-peptide set was tested in a standard IFNγ-ELISPOT assay using autologous PBMC as antigen presenting cells (Fig 1B). Some of the A10L peptides were recognized by the T cell line, but most were not. Using a cutoff value of SPW (spots per well) above two times background, we identified 7 positive peptides: A10L-42, A10L-43, A10L-50, A10L-51, A10L-75, A10L-121, and A10L-122 (Fig. 1B, and Table I). One potential concern of using TCL is that the epitope reactivity patterns observed may be the result of skewing during the single in vitro expansion. We used virus-specific TCL rather than unstimulated PBMC, because of the low frequency of epitope-specific T cells in PBMC and the limited amount of PBMC available, which prevented screening of individual peptides using PBMC. However, we were able to screen A10L peptides in small pools using PBMC from our original donor (SL131) (Fig. S1A). The highest responses were observed in pools that contained epitope peptides as identified above (pool 5 contains A10L-42, 43, 50; pool 8 contains A10L-75; pool 13 contains A10L-121, 122; Fig. S1A). A non-vaccinated HLA-DR1 donor (SL139) showed no response to these peptide pools (Fig. S1A). Additionally, we confirmed that peptide-specific lines could be expanded from PBMC using each of the epitope peptides, but not a non-epitope peptide (A10L-64) in donor SL131 (Fig. S1B). To verify these positive responses were specific to vaccinia immunization, we also generated a control CD4 TCL from the same non-vaccinia immune HLADR1 donor (SL139). None of these peptides showed positive response to this control TCL (data not shown).
Table I.
Vaccinia A10L peptides containing CD4 T cell epitopes
| Peptide | Sequencea |
|---|---|
| A10L42 | MNFCISMRYQSLIPRLVD |
| A10L43 | RYQSLIPRLVDFFPDIPV |
| A10L50b | RNNKFFINFFNLLAKEQR |
| A10L51 | NFFNLLAKEQRSTAIKVT |
| A10L75 | YQDFIYLLFASMGFKVTT |
| A10L121 | TTDDLVKSYSLIRPKILS |
| A10L122 | SYSLIRPKILSMINYYNE |
The bold underlined 9 amino acids are the predicted binding motifs for HLA-DR1 (see materials and methods for the prediction).
A10L50 has another two potential binding motifs: FFINFFNLL and FINFFNLLA.
Of the seven peptides for which we observed positive ELISPOT responses, six were found as overlapping pairs, A10L-42/43, A10L-50/51, and A10L-121/122. For each of these, the overlapping sequence contained a predicted HLA-DR1 binding motif (underlined in Table I), suggesting that a single shared epitope in the overlap region was recognized. For one of these pairs, we verified the presence of a shared epitope by raising a A10L-43-specific CD4 T cell line, and testing it against both A10L-42 and A10L-43 peptides (Supplemental Fig. S1C). The peptide-specific T cell line showed a dose-dependent response to both peptides A10L-42 and A10L-43, but not to an irrelevant peptide. For the one peptide that was not found as a pair, A10L-75, the predicted binding motif was found in the middle of the peptide. Strikingly, the two peptides with the highest immunogenicity, A10L-42 and A10L-43, contained the epitope previously identified in a study where naturally processed derived peptides were eluted from HLA-DR1 isolated from vaccinia virus-infected cells (39).
Although these T cells were raised from a HLA-DR1 donor, it is possible that some of the peptides might be recognized in the context of other class II MHC alleles, and the biochemical characterizations that we will describe below all utilize HLA-DR1. Thus we wanted to determine which of the responses were restricted by HLA-DR1. We tested the T cell line also using a HLA-DR1 homozygous human B-lymphoblastoid cell line (LG2) as antigen presenting cells (APC), in the same IFNγ-ELISPOT assay (Fig. 1C). The responses were similar (cc>0.87, p<0.0001, data not shown) with the same peptides identified as positive (Fig. 1B and 1C). However, it should be noted that these APCs also share HLA-DQ (DQA1*0101/DQB1*0501). To show that HLA-DR1 is used as the antigen-presenting molecule we generated TCLs specific to peptides A10L-42, A10L-50, A10L-75 and A10L-122, and studied their genetic restriction using anti-MHC antibodies (Fig. S1D). We found that anti-DR antibody inhibited over 75% the responses to peptides A10L-42, A10L-75 and A10L-122 and almost 50% of the response to peptide A10L-50. In sharp contrast inhibitions by anti-class I antibody were below 20% to all but peptide A10L-75. In order to provide additional evidence of the HLA-DR1-resticted response to A10L-75, HLA-DR1 tetramers for this peptide were produced and used to stain the TCL specific to this peptide (Fig. S1E). As shown the frequency of tetramer positive cells with the control HLA-DR1-GAG tetramer is below 0.5% while the frequency of HLA-DR1-A10L-75 tetramer is above 15%. These experiments demonstrate that a substantial fraction of the overall peptide-specific responses that we observed are restricted by HLA-DR1.
Besides donor SL131, we also tested A10L peptides in three additional recently vaccinia virus-vaccinated HLA-DR1 donors (donor 2011, 2029 and 2032) (Fig. S2A and S2B), including two with both pre- (day 0) and post-vaccination (day 45) samples (donor 2029 and 2032). We were not able to obtain sufficient samples from these donors for a comprehensive analysis of the entire peptide array, but we were able to analyze reactivity against two epitopes (A10L-43 and A10L-50) and two non-epitope peptides (A10L-7 and A10L-64), as identified above. TCL were raised from PBMC as described for donor SL131 by single in vitro expansion with a vaccinia preparation, and tested for peptide reactivity by ELISPOT, as in the original work. No significant T cell responses against the non-epitope peptides were observed, and no responses to any peptides were observed in the pre-vaccination samples (Fig. S2A). In the post-vaccination samples, significant A10L-43 reactivity was observed for each of the additional donors, whereas significant A10L-50 activity was observed for one of three donors (donor 2029) (Fig. S2A). In each of the three donors, we were able to observe A10L-50 reactivity in TCL raised by in vitro expansion with the peptide (Fig. S2B). Additionally, we raised peptide specific-TCL from another three HLA-DR1 donors (donor 720, 214B and 329M) (Fig. S2C–E). Again, we were able to observe significant and specific responses to epitopes A10L-43 and A10L-50 in vacciniavaccinated donors 720 and 214B (Fig. S2C and S2D), not in non-vaccinated donor 329M (Fig. S2E). No specific response was observed for the non-epitope A10L-64 in all donors (Fig. S2C and S2D).
In summary, we measured the CD4 T cell response to overlapping peptides representing the entire A10L protein sequence and identified seven HLA-DR1 restricted immunodominant peptides in donor SL131. We retested two of the seven positive peptides in several vacciniaimmunized HLA-DR1 donors and confirmed the reactivity to those peptides.
Overlapping A10L peptides have different IC50 to HLA-DR1 and are differentially susceptible to DM-mediated peptide dissociation
For the entire set of A10L peptides we measured several parameters describing the MHC-peptide binding interaction, so that we could evaluate which, if any, were correlated with peptide immunogenicity (measured as peptide-specific CD4 T cell response). We measured peptide IC50 using a competition assay, MHC II-peptide dissociation life-time using a kinetic dissociation assay, and susceptibility to DM using the dissociation assay in the presence of different concentrations of DM. Each of these properties has previously been implicated in epitope selection (10, 17, 22).
For IC50, we used a fluorescence polarization assay (FP) to measure the concentration of peptide required for 50% inhibition of binding of an indicator peptide to HLA-DR1 (IC50) (Fig. 2A, and Table II). The indicator peptide, HA306–318, binds to HLA-DR1 with KD ~ 10nM (43). This assay can measure peptide IC50 from nM to mM. In general HLA-DR-restricted T cell epitopes are thought to have relatively high affinity (IC50 <1000 nM), however more weakly binding epitopes also have been identified (25, 44). For vaccinia virus, all of the identified CD4 T cell epitopes have an IC50 less than 5 uM (30, 45), well within our detection limit of ~1 mM. We found 87 of 126 A10L peptides had measureable binding to HLA-DR1, with a very wide range of IC50 (from 46 nM to 448 uM) (Table II). For example, peptides A10L-42 and A10L-64 exhibited IC50 values of 193±7 nM and 478±25 nM (mean ± standard deviation), respectively (Fig. 2A). No binding was observed for 39 peptides (IC50 > 1mM), including A10L-85 (Fig. 2A).
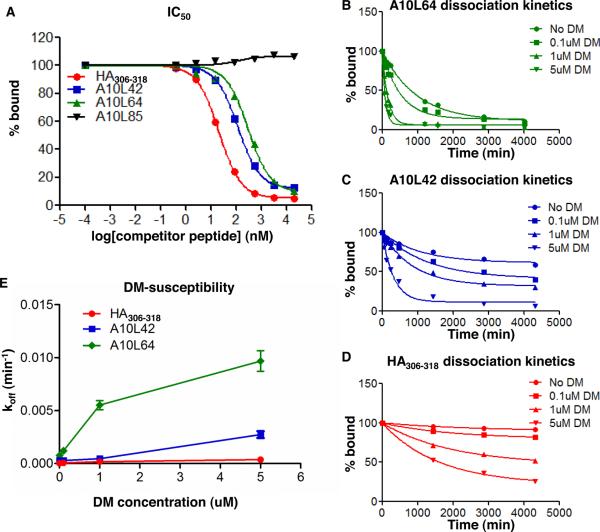
FIGURE 2. Different IC50, dissociation kinetics, and DM-susceptibilities of A10L peptides bound to HLA-DR1.
(A) Representative peptides with different IC50 to HLA-DR1. IC50 is reported as the 50% inhibition concentration calculated from the binding inhibition curve. (B–D) Representative peptides with different dissociation kinetics in the presence of various concentrations of DM. Dissociation half-lives in the absence of DM (intrinsic half-life) and in the presence of DM (DM-mediated half-life) were calculated from these dissociation curves. (E) koff versus DM concentration for representative peptides. The DM susceptibility is calculated as the slope of the koff versus DM concentration curve (using 0–1μM data only in cases where hyperbolic behavior was observed). The IC50 and dissociation kinetics for all A10L peptides were listed in Table II, with fixed DM concentration at 1uM for dissociation kinetics. These data represent three independent experiments with two replicates each.
Table II.
Immunogenicities and HLA-DR1 binding properties of A10L peptides
| Pep#a | TirPBMCb (SPW) | TLG2c (SPW) | IC50d (nM) | t1/2ine (min) | t1/2DMf (min) | DM-susg (10−3min−1 uM−1) | Pep# | TirPBMC (SPW) | TLG2 (SPW) | IC50 (nM) | t1/2in (min) | t1/2DM (min) | DM-sus (10−3min−1 uM−1) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 52 | 55 | n.b. | n.m. | n.m. | n.m. | 64 | 63 | 40 | 478 | 914 | 126 | 4.8 |
| 2 | 47 | 52 | 5080 | 377 | 155 | 2.6 | 65 | 42 | 41 | n.b. | n.m. | n.m. | n.m. |
| 3 | 49 | 31 | 1740 | 286 | 40 | 14.8 | 66 | 58 | 45 | n.b. | n.m. | n.m. | n.m. |
| 4 | 35 | 37 | 448000 | 189 | 66 | 6.9 | 67 | 49 | 51 | 13900 | 112 | 45 | 9.1 |
| 5 | 54 | 47 | 7720 | 4120 | 249 | 2.6 | 68 | 41 | 43 | 818 | 150 | 23 | 25.9 |
| 6 | 60 | 48 | 3490 | 4220 | 290 | 2.2 | 69 | 38 | 36 | 1920 | 88 | 27 | 17.6 |
| 7 | 34 | 33 | 332 | 466 | 46 | 13.7 | 70 | 47 | 38 | n.b. | n.m. | n.m. | n.m. |
| 8 | 35 | 33 | 15100 | 127 | 69 | 4.6 | 71 | 61 | 45 | 21300 | 3000 | 242 | 2.6 |
| 9 | 43 | 35 | 3280 | 404 | 161 | 2.6 | 72 | 49 | 36 | 33500 | 244 | 126 | 2.6 |
| 10 | 34 | 35 | 819 | 752 | 64 | 9.9 | 73 | 52 | 53 | n.b. | n.m. | n.m. | n.m. |
| 11 | 43 | 41 | 7620 | 659 | 139 | 3.9 | 74 | 60 | 62 | 6600 | 1370 | 119 | 5.3 |
| 12 | 58 | 44 | 6500 | 7040 | 325 | 2.0 | 75 | 96 | 90 | 947 | 2860 | 382 | 1.6 |
| 13 | 68 | 57 | 12500 | 499 | 196 | 2.2 | 76 | 60 | 59 | 2960 | 2090 | 204 | 3.1 |
| 14 | 49 | 34 | 213000 | 132 | 58 | 6.6 | 77 | 67 | 41 | 5140 | 1520 | 119 | 5.4 |
| 15 | 54 | 36 | 210000 | 242 | 53 | 10.2 | 78 | 58 | 34 | 176 | 876 | 69 | 9.2 |
| 16 | 34 | 37 | n.b. | n.m. | n.m. | n.m. | 79 | 66 | 51 | 535 | 3450 | 264 | 2.4 |
| 17 | 58 | 37 | 15600 | 378 | 134 | 3.3 | 80 | 65 | 63 | 3650 | 4010 | 282 | 2.3 |
| 18 | 43 | 31 | 7180 | 96 | 86 | 0.8 | 81 | 59 | 33 | n.b. | n.m. | n.m. | n.m. |
| 19 | 33 | 42 | n.b. | n.m. | n.m. | n.m. | 82 | 41 | 37 | 390000 | 878 | 136 | 4.3 |
| 20 | 69 | 53 | 47500 | 7370 | 434 | 1.5 | 83 | 48 | 34 | 6470 | 172 | 43 | 12.0 |
| 21 | 35 | 35 | n.b. | n.m. | n.m. | n.m. | 84 | 35 | 34 | 29000 | 193 | 39 | 14.4 |
| 22 | 32 | 34 | n.b. | n.m. | n.m. | n.m. | 85 | 66 | 59 | n.b. | n.m. | n.m. | n.m. |
| 23 | 38 | 28 | n.b. | n.m. | n.m. | n.m. | 86 | 70 | 42 | 136000 | 715 | 227 | 2.1 |
| 24 | 38 | 26 | 23500 | 93 | 59 | 4.3 | 87 | 53 | 35 | 31100 | 144 | 99 | 2.2 |
| 25 | 58 | 55 | 16300 | 345 | 59 | 9.7 | 88 | 67 | 58 | 60300 | 207 | 116 | 2.6 |
| 26 | 67 | 60 | 54100 | 343 | 143 | 2.8 | 89 | 62 | 50 | n.b. | n.m. | n.m. | n.m. |
| 27 | 62 | 54 | 2970 | 479 | 162 | 2.8 | 90 | 50 | 46 | 87100 | 45 | 28 | 9.5 |
| 28 | 56 | 49 | 15700 | 73 | 48 | 5.0 | 91 | 47 | 42 | 4230 | 282 | 33 | 18.7 |
| 29 | 61 | 51 | 1890 | 523 | 83 | 7.1 | 92 | 52 | 54 | n.b. | n.m. | n.m. | n.m. |
| 30 | 50 | 31 | 8150 | 233 | 33 | 18.1 | 93 | 57 | 60 | n.b. | n.m. | n.m. | n.m. |
| 31 | 37 | 54 | n.b. | n.m. | n.m. | n.m. | 94 | 48 | 45 | n.b. | n.m. | n.m. | n.m. |
| 32 | 70 | 54 | 1280 | 3190 | 508 | 1.2 | 95 | 57 | 30 | 7840 | 282 | 67 | 7.9 |
| 33 | 48 | 52 | 1190 | 298 | 121 | 3.4 | 96 | 44 | 47 | 304000 | 1510 | 182 | 3.4 |
| 34 | 40 | 40 | 12400 | 256 | 109 | 3.7 | 97 | 54 | 45 | 35500 | 164 | 36 | 15.2 |
| 35 | 36 | 31 | 198000 | 77 | 40 | 8.4 | 98 | 56 | 50 | 5920 | 311 | 40 | 15.0 |
| 36 | 59 | 39 | 32500 | 864 | 244 | 2.0 | 99 | 52 | 53 | 12800 | 206 | 58 | 8.7 |
| 37 | 60 | 53 | 20300 | 155 | 67 | 5.9 | 100 | 59 | 59 | n.b. | n.m. | n.m. | n.m. |
| 38 | 53 | 50 | 18200 | 72 | 52 | 3.6 | 101 | 50 | 53 | n.b. | n.m. | n.m. | n.m. |
| 39 | 60 | 56 | 3090 | 468 | 80 | 7.2 | 102 | 56 | 51 | n.b. | n.m. | n.m. | n.m. |
| 40 | 65 | 54 | 18900 | 214 | 97 | 3.9 | 103 | 55 | 57 | n.b. | n.m. | n.m. | n.m. |
| 41 | 55 | 55 | n.b. | n.m. | n.m. | n.m. | 104 | 49 | 50 | 5920 | 1050 | 89 | 7.1 |
| h 42 | 127 | 118 | 193 | 2440 | 1510 | 0.2 | 105 | 58 | 54 | 102000 | 266 | 84 | 5.7 |
| 43 | 131 | 114 | 189 | 4850 | 2420 | 0.1 | 106 | 58 | 47 | 575 | 1060 | 154 | 3.8 |
| 44 | 45 | 52 | 26200 | 256 | 170 | 1.4 | 107 | 37 | 40 | n.b. | n.m. | n.m. | n.m. |
| 45 | 36 | 43 | n.b. | n.m. | n.m. | n.m. | 108 | 48 | 42 | n.b. | n.m. | n.m. | n.m. |
| 46 | 35 | 36 | 6790 | 251 | 67 | 7.6 | 109 | 60 | 57 | 74700 | 163 | 99 | 2.8 |
| 47 | 39 | 39 | n.b. | n.m. | n.m. | n.m. | 110 | 68 | 63 | 1750 | 2620 | 472 | 1.2 |
| 48 | 35 | 30 | 58700 | 206 | 69 | 6.6 | 111 | 65 | 62 | 328 | 1170 | 241 | 2.3 |
| 49 | 53 | 56 | 18000 | 114 | 113 | 0.1 | 112 | 58 | 58 | 824 | 1120 | 164 | 3.6 |
| 50 | 101 | 94 | 98 | 3000 | 416 | 1.4 | 113 | 65 | 64 | n.b. | n.m. | n.m. | n.m. |
| 51 | 94 | 88 | 387 | 2980 | 369 | 1.7 | 114 | 68 | 62 | n.b. | n.m. | n.m. | n.m. |
| 52 | 57 | 32 | n.b. | n.m. | n.m. | n.m. | 115 | 63 | 57 | 3150 | 627 | 184 | 2.7 |
| 53 | 37 | 48 | n.b. | n.m. | n.m. | n.m. | 116 | 37 | 45 | 6150 | 898 | 45 | 14.5 |
| 54 | 45 | 46 | n.b. | n.m. | n.m. | n.m. | 117 | 52 | 44 | 3580 | 2070 | 202 | 3.1 |
| 55 | 49 | 49 | n.b. | n.m. | n.m. | n.m. | 118 | 42 | 43 | 29500 | 265 | 51 | 11.0 |
| 56 | 42 | 35 | 75700 | 246 | 76 | 6.3 | 119 | 36 | 30 | n.b. | n.m. | n.m. | n.m. |
| 57 | 42 | 42 | n.b. | n.m. | n.m. | n.m. | 120 | 46 | 37 | n.b. | n.m. | n.m. | n.m. |
| 58 | 36 | 29 | 2850 | 318 | 74 | 7.3 | 121 | 110 | 107 | 46 | 9030 | 1570 | 0.4 |
| 59 | 39 | 35 | 875 | 483 | 87 | 6.6 | 122 | 108 | 95 | 82 | 5450 | 1580 | 0.3 |
| 60 | 37 | 33 | n.b. | n.m. | n.m. | n.m. | 123 | 53 | 42 | 15000 | 164 | 43 | 11.8 |
| 61 | 58 | 42 | 14700 | 187 | 72 | 6.0 | 124 | 61 | 53 | n.b. | n.m. | n.m. | n.m. |
| 62 | 63 | 37 | 195000 | 333 | 102 | 4.7 | 125 | 58 | 54 | n.b. | n.m. | n.m. | n.m. |
| 63 | 64 | 47 | 575 | 2480 | 210 | 3.0 | 126 | 62 | 48 | n.b. | n.m. | n.m. | n.m. |
Sequential peptide number of 126 A10L overlapping peptides, 18 residues long with 11 residues overlapped. Sequences are shown in supplemental Table I.
CD4 T cell response using irradiated autologous PBMC as antigen presenting cells, shown as spots per well (SPW).
CD4 T cell response using the HLA-DR1 homozygous B-lymphoblastoid cell line LG2 as antigen presenting cells, shown as spots per well (SPW).
50% inhibition concentration, which was measured for all 126 A10L peptides. IC50 values greater than 1mM indicate no binding (n.b.).
Intrinsic dissociation half-life. For those peptides with no binding, t1/2in was not measured (n.m).
DM-mediated dissociation half-life measured in the presence of 1M DM. For those peptides with no binding, t1/2DM was not measured (n.m.). The standard deviation is ~10% for IC50, t1/2in and t1/2DM measurements.
DM-susceptibility. DM-sus was calculated by (koff,DM-koff,in)/[DM], where koff,in=ln2/t1/2in; koff,DM=ln2/t1/2DM; and [DM]=1uM.
The positive peptides identified in Fig. 1 are labeled in all borders.
For measurement of peptide dissociation, we used a streptavidin-based ELISA assay with europium time-delayed fluorescence detection (46). This assay has a high dynamic range and is able to measure dissociation life times from ~20 min to >100 hrs. We fit the dissociation kinetics to single exponential decay curves to determine dissociation half-lives. In the absence of DM these curves represent the intrinsic peptide dissociation, which we characterize as t1/2in values. We were able to measure t1/2in values for the 87 peptides for which binding to HLA-DR1 was observed (Table II). The HLA-DR1-A10L peptide complexes had intrinsic half-lives ranging from 45 min to 150 hr. We also measured the dissociation kinetics in the presence of 1μM DM, which we characterized as t1/2DM. DM-mediated half-lives for the same complexes ranged from 23 minutes to 40 hours. We observed that increased DM concentration accelerates the dissociation of peptides differentially, which we characterized as a DM-susceptibility factor equal to the rate enhancement (koff,DM-koff,in) divided by the DM concentration (1μM). DM-susceptibilities of the various HLA-DR1-A10L complexes varied from 0.08 to 25.88 *10−3 min−1uM−1 (Table II).
The results of the dissociation experiments for three representative peptides are shown in Fig. 2B–2E. HA306–318 is a previously identified influenza epitope with high affinity to HLA-DR1. A10L-64 is a peptide with low IC50 that is not recognized by vaccinia-specific CD4 T cells, while A10L-42 is also a peptide with low IC50 selected as an epitope as noted above. The dissociation half-life and DM-susceptibility of the control influenza hemaagglutinin epitope HA306–318 obtained here are consistent with previous reports (47). A10L-64 and A10L-42 had similar IC50, however with largely different intrinsic half-lives (913 ±59 min, and 2440 ±430 min, respectively) and DM-mediated half-lives (126 ±10 min, and 1506 ±132 min, respectively, at 1uM DM) (Fig. 2B–2D). A10L-64 and A10L-42 also showed different DM-susceptibilities as described by the slope of off rate (koff) versus DM concentration curve (Fig. 2E). Collectively, the HLA-DR1-A10L peptide complexes show a wide range of IC50 and dissociation characteristics.
IC50, intrinsic half-life and DM-mediated half-life are all significant different between epitopes and non-epitopes, however, DM-mediated half life is the most distinguishing feature of epitopes
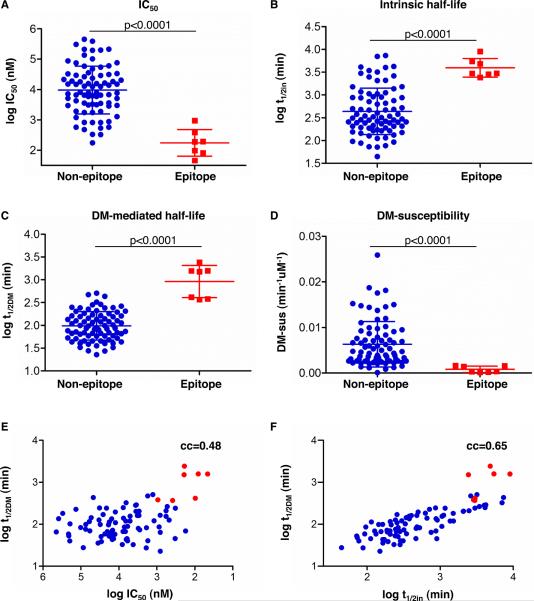
We next explored which of the measured properties of the HLA-DR1-A10L peptide complexes is the most distinguishing feature of peptides with positive CD4 T cell response. IC50, intrinsic half-life, DM-mediated half-life, and DM-susceptibility of epitopes (positive peptides in Fig.1) and non-epitopes (negative peptides in Fig. 1) are shown in Figure 3. We found that IC50 and intrinsic half-life are significantly different between epitopes and non-epitopes (Fig. 3A–3B, p<0.0001). This is consistent with previous reports that the stability of MCH II-peptide complex is important for immunogenicity (17–18). There are some peptides with low IC50 and long intrinsic half-life that were not identified as epitopes, as shown by the overlapping of IC50 and t1/2in between epitopes and non-epitopes (Fig. 3A–3B). Interestingly, DM-mediated half-life and DM-susceptibility are also significantly different for epitopes and non-epitopes, and there is little overlap between epitopes and non-epitopes (Fig. 3C–3D). We also performed a quantitative analysis, evaluating the correlation between peptide-specific CD4 T cell responses and IC50, intrinsic half-life, DM-mediated half-life, and DM-susceptibility values (supplemental Fig. S3). We found that the peptide-specific CD4 T cell response correlated better with DM-mediated half-life (cc=0.82, p<0.0001, Fig. S3C) than for the other parameters (cc=0.46 to 0.68). Significantly, DM-mediated half-life was just weakly correlated with either IC50 (cc=0.48, Fig. 3E) or intrinsic half-life (cc=0.65, Fig. 3F), indicating that DM-mediated half-life is an independent factor affecting peptide immunogenicity. The weak correlation between DM-mediated half-life and intrinsic half-life was also recorded in a previous study for a large set of 36 peptides (cc=0.69) (8). Although there are several models proposed, the mechanism of different MHC II-peptide complexes with different DM-susceptibilities is still unclear (48–51). A previous report observed that DM affect the cryptic and immunodominant fate of CD4 T cell epitopes, however no basic underlying mechanism was found (23). Here we demonstrated that DM can directly and independently influence peptide immunogenicity in epitope selection by favoring the presentation of peptide with longer DM-mediated half-lives.
FIGURE 3. Analysis of IC50, intrinsic half-life, DM-mediated half-life, and DM-susceptibility between epitopes and non-epitopes from A10L.
The positive peptides selected in Fig. 1 were shown as epitopes (red squares), and the remaining negative peptides as non-epitopes (blue circles). Two-tailed unpaired t test between epitope and non-epitope was performed for (A) IC50, (B) intrinsic half-life, (C) DM-mediated half-life, and (D) DM-susceptibility. p-value is indicated in each graph of (A–D). Correlation between DM-mediated half-life and IC50 (E), and intrinsic half-life (F) were shown. The seven positive peptides were highlighted as red dots. Correlation coefficient (cc) is indicated in upper right of (E) and (F). DM-susceptibility =(koff,DM-koff,in)/[DM], where [DM]=1uM.
Taken together, our results suggested a direct and causative relationship between DM-mediated half-life and peptide-specific CD4 T cell response.
DM-mediated half-life is a strong predictor of CD4 T cell epitopes
We also used a qualitative approach to test how well each parameter predicted whether or not a peptide was recognized as an epitope. We performed receiver operating characteristic (ROC) analysis to test which property of HLA-DR1-A10L peptide complex can best predict MHC II-presented CD4 T cell epitopes. The ROC curve is a graphical plot of the true positive prediction rate (sensitivity) versus the true negative prediction rate (specificity), as the discrimination threshold of certain property is varied (36, 52–53). The 7 positive peptides selected in Figure 1 were set as true positive epitopes and the remaining 80 were set as true negative epitopes in our ROC analysis.
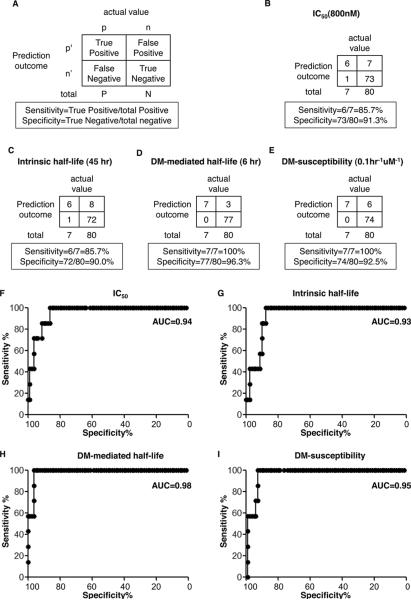
To show how this analysis works, we first arbitrarily selected one threshold for each testing property, and generated 2×2 contingency tables showing the calculated sensitivity and specificity for each property (Fig. 4A–4E). For example, a threshold of IC50 <800nM gives a sensitivity of 86 % (6 predicted of the 7 true positives) and a specificity of 91% (73 predicted of the 80 true negatives). The sensitivity can be increased by lowering the threshold (for example IC50 to 2 uM giving 100% sensitivity) but at a cost to the specificity (78% at 2 uM). The ROC analysis evaluates the tradeoff of specificity and sensitivity throughout the entire range of possible cutoff thresholds (Fig. 4F–4I).
FIGURE 4. ROC analysis shows that DM-mediated half-life is a strong predictor of CD4 T cell epitopes from A10L.
(A) Schematic contingency table showing calculation of true positive rate (sensitivity) and true negative rate (specificity). Sensitivity and specificity were calculated for (B) IC50 (IC50=800nM), (C) intrinsic half-life (t1/2=45hr), (D) DM-mediated half-life (t1/2DM=6hr), and (E) DM-susceptibility (DM-sus=0.1hr−1uM−1), with the selected threshold indicated in parenthesis. The ROC curves with all varying thresholds were plotted for (F) IC50, (G) intrinsic half-life, (H) DM-mediated half-life, and (I) DM-susceptibility. The true positive peptides and true negative peptides set for this analysis were the same ones shown in Fig. 1. Area under curve (AUC) is indicated in upper right of each plot. The IC50 and dissociation kinetics assays were repeated three independent times with two replicates each.
ROC curves for IC50, intrinsic half-life, DM-mediated half-life and DM-susceptibility are shown in Figure 4F–4I. Area under curve (AUC) in ROC analysis is a measure of the probability that the tested property will rank a true positive event higher than a true negative one, with AUC=1.0 corresponding to a perfect predictor and AUC=0.5 a predictor equivalent to a random guess. We found that IC50 (AUC=0.94, p<0.0001) and intrinsic half-life (AUC=0.93, p=0.0002) fairly well predict CD4 T cell epitopes within A10L peptides (Fig. 4F and Fig. 4G). In agreement with this, previous studies have shown that immunodominant epitopes form more stable complexes with MHC II molecules (10, 17–18, 24, 46). However, both IC50 and intrinsic half-life begin to lose sensitivity for specificity greater than ~85%, and both have poor sensitivity (15%) at 100% specificity (Fig. 4F and Fig. 4G). By contrast, DM-mediated half-life predicts the epitopes much better (AUC=0.98, p<0.0001), retaining 100% sensitivity up to 96% specificity, with relatively high 60% sensitivity at 100% specificity (Fig. 4H). Consistently, DM-susceptibility (AUC=0.95, p<0.0001) was a worse predictor than DM-mediated half-life, although substantially better than IC50 and intrinsic half-life (Fig. 4I). The importance of DM in epitope selection has been demonstrated in cell free system and mice (10, 20–22). Here, our ROC analysis indicates that DM-mediated half-life is a strong predictor for CD4 T cell epitopes from a vaccinia virus protein in humans.
DM-mediated half-life is also a distinguishing feature that separates epitopes from nonepitopes from other vaccinia virus proteins
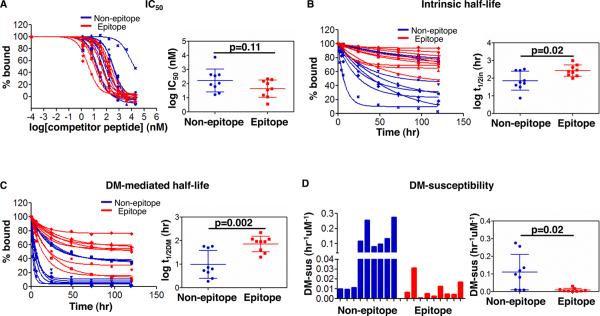
We next looked at whether DM-mediated life can distinguish epitopes from non-recognized peptides in other vaccinia viral proteins. In a previous study of CD4 T cell responses to vaccinia epitopes, we screened 36 peptides scoring highly by a consensus computational prediction algorithm, and identified 25 epitope peptides and 11 high-scoring peptides that were not recognized by any of the donors tested (30). From that set, we selected 9 epitope peptides and 9 non-recognized peptides (non-epitopes) based on their confirmed HLA-DR1 restriction and recognition by the T cell lines from the same donor used in this study (Table III). As for the A10L peptides, we measured IC50, intrinsic half-life, DM-mediated half-life, and DM susceptibility for this set of peptides (Fig. 5). IC50 of epitopes and non-epitopes overlapped (Fig. 5A), indicating that IC50 was not a distinguishing feature of the epitope set. The intrinsic half-lives of the epitopes were longer than those of non-epitopes (Fig 5B), 2.5-fold on average, however, the difference was just slightly significant (p=0.02). The DM-mediated half-lives of the epitopes were consistently longer than those of non-epitopes (Fig 5C), 5-fold on average, and the difference between them was highly significant (p=0.002). Finally, the DM-susceptibility of the epitopes was much lower than that of non-epitopes (Fig. 5D), 10-fold on average, although the difference is not very significant (p=0.02,). In summary, our data indicate that long DM-mediated half-life is a distinguishing feature of epitopes, with MHC II-complexes of epitope peptides less susceptible to DM-mediated dissociation.
Table III.
Binding properties of other vaccinia peptides containing CD4 T cell epitopes and non-epitopes
| Peptide | Sequencea | IC50b (nM) | t1/2inb (hr) | t1/2DMb (hr) | DM-susceptibilityc (*10−3um−1hr−1 |
|---|---|---|---|---|---|
| HLA-DR restricted CD4 T cell epitopesd | |||||
|
| |||||
| A20R (214–228) | KFSFMYIESIKVDRI | 192 | 563 | 91 | 6 |
| A28Ls (15–29) | AVSLLFIQGYSIYEN | 212 | 213 | 20 | 31 |
| A48Rs (41–55) | TTQSMNIMESIPANT | 44 | 136 | 115 | 1 |
| D1R (406–416) | V F R Y M S S E P I I | 12 | 289 | 91 | 5 |
| D6R (156–170) | NKIPFLLLSGSPITN | 4 | 997 | 219 | 3 |
| I1L (8–22) | LVFNSISARALKAYF | 15 | 155 | 41 | 13 |
| F17R(19–31) | GRYLVLKAVKVSD | 37 | 243 | 93 | 5 |
| F1L(202–214) | RE Y LK L I GI T AI M | 99 | 413 | 117 | 4 |
| I7L (193–205) | FDMRF LN S LAI HE | 150 | 99 | 29 | 17 |
|
| |||||
| Non-epitopesd | |||||
|
| |||||
| A38L (219–233) | LI IYYQLAGYILTVL | 431 | 328 | 57 | 10 |
| D5R (315–329) | KVKIVPLDGNKLFNI | 86 | 276 | 58 | 10 |
| I4L (71–85) | HPDYAILAARIAVSN | 62 | 278 | 50 | 11 |
| I8R (243–257) | SLGFKVLDGSPISLR | 17 | 74 | 5 | 119 |
| J6R (924–938) | TP IGI ISAQVLSEKF | 21 | 32 | 3 | 257 |
| A9L (45–55) | W F V V V R A I A S M | 360 | 51 | 7 | 81 |
| D6R (468–480) | VNVYLLAAVYSDF | 425 | 59 | 6 | 98 |
| D11L (386–398) | EPFVNQSGIEILL | 106 | 40 | 5 | 136 |
| L1R (191–205) | IGVI ILAALFMYYAK | 7360 | 8 | 2 | 276 |
Sequence shown are from the vaccinia virus strain modified vaccinia Ankara (MVA).
The binding affinity and dissociation kinetics were measured for the interaction to HLA-DR1. The standard deviation is ~10% for IC50, t1/2inand t1/2DM measurements.
DM-susceptibility was measured in the presence of 1uM DM.
These epitopes and non-epitopes were identified in reference(30).
FIGURE 5. DM-mediated half-life distinguishes vaccinia epitopes from non-recognized peptides in a set of predicted HLA-DR1 binders.
Binding properties of a selection of other vaccinia virus peptides predicted to bind to HLA-DR1 and shown to be recognized by a set of vaccinia-immune donors (epitopes, red) or not recognized (non-epitopes, blue). (A) IC50, (B) intrinsic half-lives, (C) DM-mediated half-lives, and (D) DM-susceptibilities of epitopes and non-epitopes were shown with p-value indicated on the right panels. DM concentration was 1uM. The IC50 and dissociation kinetics for each single peptide were listed in Table III. These data represent three independent experiments with two replicates each.
DM-mediated half-life predicts epitopes better than tested prediction algorithms
Epitope prediction algorithms have been widely used to identify epitopes in infectious agents, allergens, and autoantigens, although the accuracy is only approximately 50% to 80% (35, 54–55). However, no algorithm considers the direct effect of DM in the prediction. Therefore, it is interesting to compare DM-mediated half-life and current epitope prediction algorithms in predicting peptide immunogenicity for the 87 A10L peptides with measureable binding considered in this study. We evaluated three different epitope prediction algorithms: P9 and IEDB based on binding affinity of peptides to MHC II molecules (P9 and IEDB) (34, 56), and Syfpeithi, based on sequence analysis of naturally processed peptides found associated with purified MHC II proteins (Syfpeithi) (35). Prediction analyses for these algorithms are shown in Fig. 6. The prediction scores are significant different, although there is substantial overlapping between epitopes and non-epitopes for all three algorithms (Fig. 6A–6C, left panels). ROC analysis revealed relatively weak predictive power (AUC=0.90, p=0.0004 for both P9 and IEDB, and AUC=0.91, p=0.0003 for Syfpeithi; Fig. 6A–6C, right panels). They started to lose specificity when increasing sensitivity to ~20% and had ~70% specificity at 100% sensitivity (Fig. 6, right panels).
FIGURE 6. P9, IEDB and Syfpeithi prediction algorithms on peptide immunogenicity.
Left panels: Two-tailed unpaired t test between epitopes (red squares) and non-epitopes (blue circles) were performed for (A) P9, (B) IEDB and (C) Syfpeithi predicted values. p-value is indicated on each graph. Right panels: ROC analysis for (A) P9, (B) IEDB and (C) Syfpeithi. Area under curve (AUC) is indicated in upper right of each plot. The positive and negative peptides selected for this analysis were the same as described above.
Discussion
In this study, we have evaluated the peptide-specific CD4+ T cell response to 126 overlapping A10L peptides and identified 7 peptides with positive responses. We also measured the IC50, intrinsic dissociation half-life, DM-mediated half-life, and DM-susceptibility of the corresponding HLA-DR1-A10L peptide complexes. Within this data set, DM-mediated half-life is the primary contributory factor of peptide immunogenicity and can predict CD4 T cell epitopes with high accuracy. In another set of peptides from the entire vaccinia virus genome, epitopes again were distinguished by their longer DM-mediated half-lives.
Although T cell response to each individual A10L peptide was only evaluated in donor SL131, we were able to verify some of the peptides in other vaccinia virus-vaccinated or nonvaccinated HLA-DR1 donors (Fig. S2). Moreover, we did not observe any positive responses against A10L peptides in PBMC or CD4 TCL from non-vaccinia-immune HLA-DR1 donors (Fig. S1A, Fig. S2E and data not shown). Whether these positive A10L peptides identified here are in general immunodominant vaccinia epitopes would need to be evaluated in additional donors.
As noted above, other factors beside MHC-peptide interaction may be involved in epitope selection. Endo-peptidases have been demonstrated to either enhance or destroy epitopes available to MHC II molecules (12, 58). A peptide cannot induce immune response if it is degraded by proteases due to the prominent protease cleavage site within the peptide. Protein expression level also must play a role. Besides the interaction between MHC II molecules and peptides, the binding between T cell receptor (TCR) and MHC-peptide complex has also been shown to be important for peptide immunogenicity, both in CD4 (44) and CD8 contexts (59). It is possible that a strong interaction between TCR and MHC II-peptide complex can compensate for the low stability of MHC II-peptide complexes. Finally, the asymmetrical T cell repertoire and TCR frequency would in some case influence peptide immunogenicity and epitope selection (13–14). Despite these caveats, our data still indicate that DM-mediated half-life is a primary factor that governs peptide-specific CD4 T cell responses (Fig. 3–5). Notably, Isamu Hartman and colleagues recently using a cell-free antigen processing system composed of HLA-DR1, DM and cathepsins successfully identified known and novel immunodominant epitopes from various antigens (21), which further demonstrates a role for DM in epitope selection.
Significant progress has been made on understanding the role of MHC II-peptide complex stability in peptide immunogenicity (17–18), which can be utilized to develop more effective epitope prediction algorithms. Most current CD4 T cell epitope prediction algorithms are based on either binding affinity to MHC II molecules, including P9 and IEDB evaluated in this study, or characteristics of endogenously processed peptides associated with MHC II proteins, including Syfpeithi in this study (30, 60). Consistently, we found that the predictive abilities of the three evaluated computational algorithms were similar with those of IC50 and intrinsic half-life. Here, we have shown that DM-mediated half-life predicted CD4 T cell epitopes better than the tested prediction algorithms. The effect of DM on antigen presentation and epitope selection was also observed in previous reports (10, 21–22). Thus, a method to take DM-mediated peptide dissociation into account in epitope prediction algorithms would be likely to increase their accuracy.
DM catalyzes peptide exchange in a specialized compartment for loading peptides onto MHC II molecules (5). The reported half time of MHC II residence in the peptide loading compartment, corresponding to the time for which they exposed to the action of DM, has been measured at ~4 to 6 hrs (61–62). Notably, in our ROC analysis, when the threshold of DM-mediated half-life was set to 6 hrs, the prediction of epitopes was highly efficient with 100% sensitivity and 96 % specificity (Fig. 4D and 4H). Considering this, our observation may indicate that the half-lives of MHC II-peptide complexes in the presence of DM should be longer than the MHC II transit time in order for them to get presented and selected as epitopes.
DM has been shown to be associated with several autoimmune diseases including rheumatoid arthritis (RA), but the mechanism by which DM mediates susceptibility to RA is still unknown (22, 27, 63–65). The expression of DM, as well as the DM:DR ratio, were found to be decreased significantly in RA patients (64). It was also found that the autoantigen type II collagen in RA and autoantigen glutamate decarboxylase in type I diabetes both show DM-dependent presentation in those autoimmune disease models (9, 22). Therefore, it is possible that due to the down regulation or deficiency of DM in antigen presenting cells, self peptides bound to MHC II alleles that are susceptible to DM-mediated dissociation and normally have short DM-mediated half-lives can get presented and cause autoimmune disease. This idea is supported by the observation that the repertoire of autoimmune-encephalitogenic T cells consists primarily of T cells specific for short-lived MHC complexes due to tolerance deletion of T cells that recognize long-lived complexes in an experimental allergic encephalomyelitis study (14). This negative correlation between peptide immunogenicity and strength of HLA-DR binding were also seen in other autoimmune animal studies (66–68), and multiple sclerosis patients study (26), which further support the hypothesis that autoreactive T cells escape negative selection due to the low stability of corresponding MHC-peptide complexes. Thus, the importance of central and peripheral tolerance can likely explain the apparent paradox that foreign pathogenic epitopes form stable and DM-resistant complexes, as seen in this study and previous reports (10, 17, 21), while autoimmune self-epitopes form complexes with low affinity and kinetic stability (14, 25–26).
In conclusion, we have shown that long DM-mediated half-life is a distinguishing feature of CD4 T cell epitopes, and that DM-mediated kinetic stability can effectively predict CD4 T cell epitopes in an example of a human infectious disease.
Supplementary Material
Acknowledgements
We thank Liying Lu for soluble HLA-DR1 and HLA-DM expression and purification, and Loretta Lee for cell lines maintenance and PMBC harvest, and all PBMC donors that participated in this study.
Footnotes
This work was supported by National Institute of Health Grants AI-57319 and AI-48833
Abbreviations used in this paper: VV, vaccinia virus; TCL, T cell line; DM, HLA-DM; FP, fluorescence polarization; koff,in, intrinsic off-rate; koff,DM, DM-mediated off rate, i.e. off-rate in the presence of 1μM DM; t1/2in, intrinsic dissociation half-life; t1/2DM, DM-mediated dissociation half-life, i.e. half-life in the presence of 1μM DM; SPW, spots per well; MHC II, major histocompatibility complex class II alleles; irPBMC, irradiated autologous peripheral blood mononuclear cell; cc, Pearson's correlation coefficient; ROC, receiver operating characteristic; AUC, area under curve.
References
- 1.Germain RN. MHC-dependent antigen processing and peptide presentation: providing ligands for T lymphocyte activation. Cell. 1994;76:287–299. doi: 10.1016/0092-8674(94)90336-0. [DOI] [PubMed] [Google Scholar]
- 2.Bakke O, Dobberstein B. MHC class II-associated invariant chain contains a sorting signal for endosomal compartments. Cell. 1990;63:707–716. doi: 10.1016/0092-8674(90)90137-4. [DOI] [PubMed] [Google Scholar]
- 3.Roche PA, Cresswell P. Invariant chain association with HLA-DR molecules inhibits immunogenic peptide binding. Nature. 1990;345:615–618. doi: 10.1038/345615a0. [DOI] [PubMed] [Google Scholar]
- 4.Sloan VS, Cameron P, Porter G, Gammon M, Amaya M, Mellins E, Zaller DM. Mediation by HLA-DM of dissociation of peptides from HLA-DR. Nature. 1995;375:802–806. doi: 10.1038/375802a0. [DOI] [PubMed] [Google Scholar]
- 5.Denzin LK, Hammond C, Cresswell P. HLA-DM interactions with intermediates in HLA-DR maturation and a role for HLA-DM in stabilizing empty HLADR molecules. J Exp Med. 1996;184:2153–2165. doi: 10.1084/jem.184.6.2153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kropshofer H, Arndt SO, Moldenhauer G, Hammerling GJ, Vogt AB. HLA-DM acts as a molecular chaperone and rescues empty HLA-DR molecules at lysosomal pH. Immunity. 1997;6:293–302. doi: 10.1016/s1074-7613(00)80332-5. [DOI] [PubMed] [Google Scholar]
- 7.Kropshofer H, Vogt AB, Moldenhauer G, Hammer J, Blum JS, Hammerling GJ. Editing of the HLA-DR-peptide repertoire by HLA-DM. EMBO J. 1996;15:6144–6154. [PMC free article] [PubMed] [Google Scholar]
- 8.Belmares MP, Busch R, Wucherpfennig KW, McConnell HM, Mellins ED. Structural factors contributing to DM susceptibility of MHC class II/peptide complexes. J Immunol. 2002;169:5109–5117. doi: 10.4049/jimmunol.169.9.5109. [DOI] [PubMed] [Google Scholar]
- 9.Lich JD, Jayne JA, Zhou D, Elliott JF, Blum JS. Editing of an immunodominant epitope of glutamate decarboxylase by HLA-DM. J Immunol. 2003;171:853–859. doi: 10.4049/jimmunol.171.2.853. [DOI] [PubMed] [Google Scholar]
- 10.Lazarski CA, Chaves FA, Sant AJ. The impact of DM on MHC class II-restricted antigen presentation can be altered by manipulation of MHC-peptide kinetic stability. J Exp Med. 2006;203:1319–1328. doi: 10.1084/jem.20060058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Manoury B, Mazzeo D, Fugger L, Viner N, Ponsford M, Streeter H, Mazza G, Wraith DC, Watts C. Destructive processing by asparagine endopeptidase limits presentation of a dominant T cell epitope in MBP. Nat Immunol. 2002;3:169–174. doi: 10.1038/ni754. [DOI] [PubMed] [Google Scholar]
- 12.Burster T, Beck A, Tolosa E, Marin-Esteban V, Rotzschke O, Falk K, Lautwein A, Reich M, Brandenburg J, Schwarz G, Wiendl H, Melms A, Lehmann R, Stevanovic S, Kalbacher H, Driessen C. Cathepsin G, and not the asparagine-specific endoprotease, controls the processing of myelin basic protein in lysosomes from human B lymphocytes. J Immunol. 2004;172:5495–5503. doi: 10.4049/jimmunol.172.9.5495. [DOI] [PubMed] [Google Scholar]
- 13.Liu GY, Fairchild PJ, Smith RM, Prowle JR, Kioussis D, Wraith DC. Low avidity recognition of self-antigen by T cells permits escape from central tolerance. Immunity. 1995;3:407–415. doi: 10.1016/1074-7613(95)90170-1. [DOI] [PubMed] [Google Scholar]
- 14.Harrington CJ, Paez A, Hunkapiller T, Mannikko V, Brabb T, Ahearn M, Beeson C, Goverman J. Differential tolerance is induced in T cells recognizing distinct epitopes of myelin basic protein. Immunity. 1998;8:571–580. doi: 10.1016/s1074-7613(00)80562-2. [DOI] [PubMed] [Google Scholar]
- 15.Kedl RM, Rees WA, Hildeman DA, Schaefer B, Mitchell T, Kappler J, Marrack P. T cells compete for access to antigen-bearing antigen-presenting cells. J Exp Med. 2000;192:1105–1113. doi: 10.1084/jem.192.8.1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kedl RM, Kappler JW, Marrack P. Epitope dominance, competition and T cell affinity maturation. Curr Opin Immunol. 2003;15:120–127. doi: 10.1016/s0952-7915(02)00009-2. [DOI] [PubMed] [Google Scholar]
- 17.Lazarski CA, Chaves FA, Jenks SA, Wu S, Richards KA, Weaver JM, Sant AJ. The kinetic stability of MHC class II:peptide complexes is a key parameter that dictates immunodominance. Immunity. 2005;23:29–40. doi: 10.1016/j.immuni.2005.05.009. [DOI] [PubMed] [Google Scholar]
- 18.Hall FC, Rabinowitz JD, Busch R, Visconti KC, Belmares M, Patil NS, Cope AP, Patel S, McConnell HM, Mellins ED, Sonderstrup G. Relationship between kinetic stability and immunogenicity of HLA-DR4/peptide complexes. Eur J Immunol. 2002;32:662–670. doi: 10.1002/1521-4141(200203)32:3<662::AID-IMMU662>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- 19.Honey K, Forbush K, Jensen PE, Rudensky AY. Effect of decreasing the affinity of the class II-associated invariant chain peptide on the MHC class II peptide repertoire in the presence or absence of H-2M. J Immunol. 2004;172:4142–4150. doi: 10.4049/jimmunol.172.7.4142. [DOI] [PubMed] [Google Scholar]
- 20.Sant AJ, Chaves FA, Jenks SA, Richards KA, Menges P, Weaver JM, Lazarski CA. The relationship between immunodominance, DM editing, and the kinetic stability of MHC class II:peptide complexes. Immunol Rev. 2005;207:261–278. doi: 10.1111/j.0105-2896.2005.00307.x. [DOI] [PubMed] [Google Scholar]
- 21.Hartman IZ, Kim A, Cotter RJ, Walter K, Dalai SK, Boronina T, Griffith W, Lanar DE, Schwenk R, Krzych U, Cole RN, Sadegh-Nasseri S. A reductionist cell-free major histocompatibility complex class II antigen processing system identifies immunodominant epitopes. Nat Med. 2010;16:1333–1340. doi: 10.1038/nm.2248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Amria S, Hajiaghamohseni LM, Harbeson C, Zhao D, Goldstein O, Blum JS, Haque A. HLA-DM negatively regulates HLA-DR4-restricted collagen pathogenic peptide presentation and T cell recognition. Eur J Immunol. 2008;38:1961–1970. doi: 10.1002/eji.200738100. [DOI] [PubMed] [Google Scholar]
- 23.Nanda NK, Sant AJ. DM determines the cryptic and immunodominant fate of T cell epitopes. J Exp Med. 2000;192:781–788. doi: 10.1084/jem.192.6.781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lovitch SB, Petzold SJ, Unanue ER. Cutting edge: H-2DM is responsible for the large differences in presentation among peptides selected by I-Ak during antigen processing. J Immunol. 2003;171:2183–2186. doi: 10.4049/jimmunol.171.5.2183. [DOI] [PubMed] [Google Scholar]
- 25.Muraro PA, Vergelli M, Kalbus M, Banks DE, Nagle JW, Tranquill LR, Nepom GT, Biddison WE, McFarland HF, Martin R. Immunodominance of a low-affinity major histocompatibility complex-binding myelin basic protein epitope (residues 111–129) in HLA-DR4 (B1*0401) subjects is associated with a restricted T cell receptor repertoire. J Clin Invest. 1997;100:339–349. doi: 10.1172/JCI119539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bielekova B, Sung MH, Kadom N, Simon R, McFarland H, Martin R. Expansion and functional relevance of high-avidity myelin-specific CD4+ T cells in multiple sclerosis. J Immunol. 2004;172:3893–3904. doi: 10.4049/jimmunol.172.6.3893. [DOI] [PubMed] [Google Scholar]
- 27.Patil NS, Pashine A, Belmares MP, Liu W, Kaneshiro B, Rabinowitz J, McConnell H, Mellins ED. Rheumatoid arthritis (RA)-associated HLA-DR alleles form less stable complexes with class II-associated invariant chain peptide than non-RA-associated HLA-DR alleles. J Immunol. 2001;167:7157–7168. doi: 10.4049/jimmunol.167.12.7157. [DOI] [PubMed] [Google Scholar]
- 28.Ferrante A, Gorski J. Cutting edge: HLA-DM-mediated peptide exchange functions normally on MHC class II-peptide complexes that have been weakened by elimination of a conserved hydrogen bond. J Immunol. 2010;184:1153–1158. doi: 10.4049/jimmunol.0902878. [DOI] [PubMed] [Google Scholar]
- 29.Frayser M, Sato AK, Xu L, Stern LJ. Empty and peptide-loaded class II major histocompatibility complex proteins produced by expression in Escherichia coli and folding in vitro. Protein Expr Purif. 1999;15:105–114. doi: 10.1006/prep.1998.0987. [DOI] [PubMed] [Google Scholar]
- 30.Calvo-Calle JM, Strug I, Nastke MD, Baker SP, Stern LJ. Human CD4+ T cell epitopes from vaccinia virus induced by vaccination or infection. PLoS Pathog. 2007;3:1511–1529. doi: 10.1371/journal.ppat.0030144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Frey SE, Newman FK, Kennedy JS, Ennis F, Abate G, Hoft DF, Monath TP. Comparison of the safety and immunogenicity of ACAM1000, ACAM2000 and Dryvax in healthy vaccinia-naive adults. Vaccine. 2009;27:1637–1644. doi: 10.1016/j.vaccine.2008.11.079. [DOI] [PubMed] [Google Scholar]
- 32.Cameron TO, Norris PJ, Patel A, Moulon C, Rosenberg ES, Mellins ED, Wedderburn LR, Stern LJ. Labeling antigen-specific CD4(+) T cells with class II MHC oligomers. J Immunol Methods. 2002;268:51–69. doi: 10.1016/s0022-1759(02)00200-4. [DOI] [PubMed] [Google Scholar]
- 33.Sturniolo T, Bono E, Ding J, Raddrizzani L, Tuereci O, Sahin U, Braxenthaler M, Gallazzi F, Protti MP, Sinigaglia F, Hammer J. Generation of tissue-specific and promiscuous HLA ligand databases using DNA microarrays and virtual HLA class II matrices. Nat Biotechnol. 1999;17:555–561. doi: 10.1038/9858. [DOI] [PubMed] [Google Scholar]
- 34.Peters B, Sidney J, Bourne P, Bui HH, Buus S, Doh G, Fleri W, Kronenberg M, Kubo R, Lund O, Nemazee D, Ponomarenko JV, Sathiamurthy M, Schoenberger S, Stewart S, Surko P, Way S, Wilson S, Sette A. The immune epitope database and analysis resource: from vision to blueprint. PLoS Biol. 2005;3:e91. doi: 10.1371/journal.pbio.0030091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rammensee H, Bachmann J, Emmerich NP, Bachor OA, Stevanovic S. SYFPEITHI: database for MHC ligands and peptide motifs. Immunogenetics. 1999;50:213–219. doi: 10.1007/s002510050595. [DOI] [PubMed] [Google Scholar]
- 36.McNeil BJ, Hanley JA. Statistical approaches to the analysis of receiver operating characteristic (ROC) curves. Med Decis Making. 1984;4:137–150. doi: 10.1177/0272989X8400400203. [DOI] [PubMed] [Google Scholar]
- 37.Jing L, Davies DH, Chong TM, Chun S, McClurkan CL, Huang J, Story BT, Molina DM, Hirst S, Felgner PL, Koelle DM. An extremely diverse CD4 response to vaccinia virus in humans is revealed by proteome-wide T-cell profiling. J Virol. 2008;82:7120–7134. doi: 10.1128/JVI.00453-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Davies DH, Liang X, Hernandez JE, Randall A, Hirst S, Mu Y, Romero KM, Nguyen TT, Kalantari-Dehaghi M, Crotty S, Baldi P, Villarreal LP, Felgner PL. Profiling the humoral immune response to infection by using proteome microarrays: high-throughput vaccine and diagnostic antigen discovery. Proc Natl Acad Sci U S A. 2005;102:547–552. doi: 10.1073/pnas.0408782102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Strug I, Calvo-Calle JM, Green KM, Cruz J, Ennis FA, Evans JE, Stern LJ. Vaccinia peptides eluted from HLA-DR1 isolated from virus-infected cells are recognized by CD4+ T cells from a vaccinated donor. J Proteome Res. 2008;7:2703–2711. doi: 10.1021/pr700780x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stern LJ, Brown JH, Jardetzky TS, Gorga JC, Urban RG, Strominger JL, Wiley DC. Crystal structure of the human class II MHC protein HLA-DR1 complexed with an influenza virus peptide. Nature. 1994;368:215–221. doi: 10.1038/368215a0. [DOI] [PubMed] [Google Scholar]
- 41.Arnold PY, La Gruta NL, Miller T, Vignali KM, Adams PS, Woodland DL, Vignali DA. The majority of immunogenic epitopes generate CD4+ T cells that are dependent on MHC class II-bound peptide-flanking residues. J Immunol. 2002;169:739–749. doi: 10.4049/jimmunol.169.2.739. [DOI] [PubMed] [Google Scholar]
- 42.Zavala-Ruiz Z, Strug I, Walker BD, Norris PJ, Stern LJ. A hairpin turn in a class II MHC-bound peptide orients residues outside the binding groove for T cell recognition. Proc Natl Acad Sci U S A. 2004;101:13279–13284. doi: 10.1073/pnas.0403371101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Roche PA, Cresswell P. High-affinity binding of an influenza hemagglutinin-derived peptide to purified HLA-DR. J Immunol. 1990;144:1849–1856. [PubMed] [Google Scholar]
- 44.Yin Y, Li Y, Kerzic MC, Martin R, Mariuzza RA. Structure of a TCR with high affinity for self-antigen reveals basis for escape from negative selection. EMBO J. 2011;30:1137–1148. doi: 10.1038/emboj.2011.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Moutaftsi M, Bui HH, Peters B, Sidney J, Salek-Ardakani S, Oseroff C, Pasquetto V, Crotty S, Croft M, Lefkowitz EJ, Grey H, Sette A. Vaccinia virus-specific CD4+ T cell responses target a set of antigens largely distinct from those targeted by CD8+ T cell responses. J Immunol. 2007;178:6814–6820. doi: 10.4049/jimmunol.178.11.6814. [DOI] [PubMed] [Google Scholar]
- 46.Baumgartner CK, Ferrante A, Nagaoka M, Gorski J, Malherbe LP. Peptide-MHC class II complex stability governs CD4 T cell clonal selection. J Immunol. 2010;184:573–581. doi: 10.4049/jimmunol.0902107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhou Z, Callaway KA, Weber DA, Jensen PE. Cutting edge: HLA-DM functions through a mechanism that does not require specific conserved hydrogen bonds in class II MHC-peptide complexes. J Immunol. 2009;183:4187–4191. doi: 10.4049/jimmunol.0901663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ferrante A, Anderson MW, Klug CS, Gorski J. HLA-DM mediates epitope selection by a “compare-exchange” mechanism when a potential peptide pool is available. PLoS One. 2008;3:e3722. doi: 10.1371/journal.pone.0003722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Narayan K, Su KW, Chou CL, Khoruzhenko S, Sadegh-Nasseri S. HLADM mediates peptide exchange by interacting transiently and repeatedly with HLA-DR1. Mol Immunol. 2009;46:3157–3162. doi: 10.1016/j.molimm.2009.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Painter CA, Negroni MP, Kellersberger KA, Zavala-Ruiz Z, Evans JE, Stern LJ. Conformational lability in the class II MHC 310 helix and adjacent extended strand dictate HLA-DM susceptibility and peptide exchange. Proc Natl Acad Sci U S A. 2011;108:19329–19334. doi: 10.1073/pnas.1108074108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schulze MS, Wucherpfennig KW. The mechanism of HLA-DM induced peptide exchange in the MHC class II antigen presentation pathway. Curr Opin Immunol. 2012;24:105–111. doi: 10.1016/j.coi.2011.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hanley JA. Receiver operating characteristic (ROC) methodology: the state of the art. Crit Rev Diagn Imaging. 1989;29:307–335. [PubMed] [Google Scholar]
- 53.Hanley JA, McNeil BJ. The meaning and use of the area under a receiver operating characteristic (ROC) curve. Radiology. 1982;143:29–36. doi: 10.1148/radiology.143.1.7063747. [DOI] [PubMed] [Google Scholar]
- 54.Borras-Cuesta F, Golvano J, Garcia-Granero M, Sarobe P, Riezu-Boj J, Huarte E, Lasarte J. Specific and general HLA-DR binding motifs: comparison of algorithms. Hum Immunol. 2000;61:266–278. doi: 10.1016/s0198-8859(99)00153-6. [DOI] [PubMed] [Google Scholar]
- 55.Doolan DL, Southwood S, Freilich DA, Sidney J, Graber NL, Shatney L, Bebris L, Florens L, Dobano C, Witney AA, Appella E, Hoffman SL, Yates JR, 3rd, Carucci DJ, Sette A. Identification of Plasmodium falciparum antigens by antigenic analysis of genomic and proteomic data. Proc Natl Acad Sci U S A. 2003;100:9952–9957. doi: 10.1073/pnas.1633254100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hammer J, Takacs B, Sinigaglia F. Identification of a motif for HLA-DR1 binding peptides using M13 display libraries. J Exp Med. 1992;176:1007–1013. doi: 10.1084/jem.176.4.1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Welsh RM, Che JW, Brehm MA, Selin LK. Heterologous immunity between viruses. Immunol Rev. 2010;235:244–266. doi: 10.1111/j.0105-2896.2010.00897.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Schneider SC, Ohmen J, Fosdick L, Gladstone B, Guo J, Ametani A, Sercarz EE, Deng H. Cutting edge: introduction of an endopeptidase cleavage motif into a determinant flanking region of hen egg lysozyme results in enhanced T cell determinant display. J Immunol. 2000;165:20–23. doi: 10.4049/jimmunol.165.1.20. [DOI] [PubMed] [Google Scholar]
- 59.Aleksic M, Dushek O, Zhang H, Shenderov E, Chen JL, Cerundolo V, Coombs D, van der Merwe PA. Dependence of T cell antigen recognition on T cell receptor-peptide MHC confinement time. Immunity. 2010;32:163–174. doi: 10.1016/j.immuni.2009.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wang P, Sidney J, Kim Y, Sette A, Lund O, Nielsen M, Peters B. Peptide binding predictions for HLA DR, DP and DQ molecules. BMC Bioinformatics. 2010;11:568. doi: 10.1186/1471-2105-11-568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Xu X, Song W, Cho H, Qiu Y, Pierce SK. Intracellular transport of invariant chain-MHC class II complexes to the peptide-loading compartment. J Immunol. 1995;155:2984–2992. [PubMed] [Google Scholar]
- 62.Schafer PH, Green JM, Malapati S, Gu L, Pierce SK. HLA-DM is present in one-fifth the amount of HLA-DR in the class II peptide-loading compartment where it associates with leupeptin-induced peptide (LIP)-HLA-DR complexes. J Immunol. 1996;157:5487–5495. [PubMed] [Google Scholar]
- 63.Pinet V, Combe B, Avinens O, Caillat-Zucman S, Sany J, Clot J, Eliaou JF. Polymorphism of the HLA-DMA and DMB genes in rheumatoid arthritis. Arthritis Rheum. 1997;40:854–858. doi: 10.1002/art.1780400512. [DOI] [PubMed] [Google Scholar]
- 64.Louis-Plence P, Kerlan-Candon S, Morel J, Combe B, Clot J, Pinet V, Eliaou JF. The down-regulation of HLA-DM gene expression in rheumatoid arthritis is not related to their promoter polymorphism. J Immunol. 2000;165:4861–4869. doi: 10.4049/jimmunol.165.9.4861. [DOI] [PubMed] [Google Scholar]
- 65.Morel J, Roch-Bras F, Molinari N, Sany J, Eliaou JF, Combe B. HLA-DMA*0103 and HLA-DMB*0104 alleles as novel prognostic factors in rheumatoid arthritis. Ann Rheum Dis. 2004;63:1581–1586. doi: 10.1136/ard.2003.012294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Vanderlugt CL, Neville KL, Nikcevich KM, Eagar TN, Bluestone JA, Miller SD. Pathologic role and temporal appearance of newly emerging autoepitopes in relapsing experimental autoimmune encephalomyelitis. J Immunol. 2000;164:670–678. doi: 10.4049/jimmunol.164.2.670. [DOI] [PubMed] [Google Scholar]
- 67.Anderton SM, Radu CG, Lowrey PA, Ward ES, Wraith DC. Negative selection during the peripheral immune response to antigen. J Exp Med. 2001;193:1–11. doi: 10.1084/jem.193.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Goverman J. Autoimmune T cell responses in the central nervous system. Nat Rev Immunol. 2009;9:393–407. doi: 10.1038/nri2550. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.