Abstract
Objective
Systemic lupus erythematosus (SLE; OMIM 152700) is a chronic autoimmune disease for which the aetiology includes genetic and environmental factors. ITGAM, integrin αΜ (complement component 3 receptor 3 subunit) encoding a ligand for intracellular adhesion molecule (ICAM) proteins, is an established SLE susceptibility locus. This study aimed to evaluate the independent and joint effects of genetic variations in the genes that encode ITGAM and ICAM.
Methods
The authors examined several markers in the ICAM1–ICAM4–ICAM5 locus on chromosome 19p13 and the single ITGAM polymorphism (rs1143679) using a large-scale case–control study of 17 481 unrelated participants from four ancestry populations. The single marker association and gene–gene interaction were analysed for each ancestry, and a meta-analysis across the four ancestries was performed.
Results
The A-allele of ICAM1–ICAM4–ICAM5 rs3093030, associated with elevated plasma levels of soluble ICAM1, and the A-allele of ITGAM rs1143679 showed the strongest association with increased SLE susceptibility in each of the ancestry populations and the trans-ancestry meta-analysis (ORmeta=1.16, 95% CI 1.11 to 1.22; p=4.88×10−10 and ORmeta=1.67, 95% CI 1.55 to 1.79; p=3.32×10−46, respectively). The effect of the ICAM single-nucleotide polymorphisms (SNPs) was independent of the effect of the ITGAM SNP rs1143679, and carriers of both ICAM rs3093030-AA and ITGAM rs1143679-AA had an OR of 4.08 compared with those with no risk allele in either SNP (95% CI 2.09 to 7.98; p=3.91×10−5).
Conclusion
These findings are the first to suggest that an ICAM–integrin-mediated pathway contributes to susceptibility to SLE.
Systemic lupus erythematosus (SLE; OMIM 152700) is a chronic autoimmune disease for which the aetiology includes diverse genetic and environmental factors. The formation and deposition of immune complexes, which consist of nuclear autoantigens and autoantibodies, in vital organs result in chronic inflammation, organ failure and severe morbidity and mortality in patients with SLE.
Previous genome-wide and candidate-gene association studies reported an association of SLE with a non-synonymous variant in ITGAM, encoding integrin αM (complement component 3 receptor 3 subunit), among European and African descents.1,2 ITGAM is a ligand for intercellular adhesion molecule (ICAM) proteins, suggesting the involvement of an integrin–ICAM-mediated adhesion pathway for SLE susceptibility.
In this candidate gene-association study, we examined the independent and combined effects of common variation in ITGAM and the ICAM1– ICAM4–ICAM5 locus, encoding other components of the integrin–adhesion pathway, with SLE susceptibility using a large multi-ancestry population.
METHODS
Study population
As part of the first phase of this study, we included a total of 14 719 patients with SLE and controls from diverse ancestry backgrounds, recruited from multiple institutions worldwide as part of the Lupus Association Study(LLAS)-2, with approval from the appropriate institutional review boards (online supplementary table S1). The affected had a minimum of four of eleven 1997 American College of Rheumatology revised criteria for the classification of SLE.3 All participants in this study were filtered by principal component analysis to identify population outliers as previously reported.4 The sample exceeding five SDs along any statistically significant principal component were defined as outliers and removed from the study.
For the second phase to demonstrate consistency of results for rs3093030 in this larger Korean population, we constituted an independent cohort consisting of 2762 SLE cases and controls of Korean ancestry. Hospital-based SLE cases and controls were recruited from six hospitals in Korea with approval from the respective institutional review boards (online supplementary table S1).
Genotyping
We genotyped the 12 ICAM variants in phase I (rs5030340, rs5030390, rs5030391, rs5030351, rs5491, rs1799969, rs5498, rs5030400, rs3093032, rs281437, rs3093030, rs2228615), which tagged most of the common single-nucleotide polymorphisms (SNPs) in the ICAM1–ICAM4–ICAM5 locus, in addition to an ITGAM variant (rs1143679), using customised arrays based on the Illumina iSelect platform at the Lupus Genetics Studies Unit of the Oklahoma Medical Research Foundation. For the SLE associated SNP in the ICAM1–ICAM4–ICAM5 locus with the strongest effect (rs3093030), we genotyped this marker for the phase II Korean cohort using the Sequenom iPlex platform at the Korea Advanced Institute of Science and Technology.
SNP association with SLE susceptibility
The statistical assessment for SNP association with SLE susceptibility was performed by the χ2 test with allelic genetic model for each ancestry and meta-analysis across the four ancestries using the PLINK 1.07 http://pngu.mgh.harvard.edu/~purcell/plink/).5 In the meta-analysis, homogeneity of effect sizes was evaluated using the I2 heterogeneity index and Cochrane’s Q statistic.6 For p>0.01 in heterogeneity Q statistic, fixed-effects models were implemented; otherwise random-effects models were used. For meta-analysis, GWAMA 1.4 software (http://www.well.ox.ac.uk/gwama/) was also used to calculate OR and 95% CI.7 Hardy–Weinberg equilibrium (HWE) was examined by comparison between genotype distributions expected and observed in controls using the χ2 test. Pair-wise D’ and r2 values between SNPs were calculated by the Haploview 4.2 program. Statistical power was estimated using the CaTSPower calculator (http://www.sph.umich.edu/csg/abecasis/CaTS/index.html).
Imputation
Using the genotype data for the 12 ICAM SNPs, 58 HapMap SNPs located in a chromosome 19 region from position 10 200 kb to 10 320 kb (build 36.3) were imputed using the MACH V.1.0 program (http://www.sph.umich.edu/csg/abecasis/MACH/index.html) with the ancestry-matched template data provided by the International HapMap Project (http://www.hapmap.org) Phase 3 (Genome Browser release 2). Europeans were imputed using the CEU (Utah residents with Northern and Western European ancestry) data, Africans using the ASW (African ancestry in Southwest USA) data, Hispanics using the MEX (Mexican ancestry in Los Angeles, California) data, and Koreans using the CHB (Han Chinese in Beijing, China) data. Among the 58 imputed SNPs, 69.0% in Europeans, 77.6% in Africans, 58.6% in Hispanics and 63.8% in Koreans showed high (≥0.70) average posterior probability for the most likely genotype, although some SNPs were not confidently imputed because of their weak correlations with the 12 genotyped SNPs.
Gene–gene interaction
Epistatic interactions between the seven SLE-associated SNPs in the ICAM1–ICAM4–ICAM5 locus and the non-synonymous SNP rs1143679 in ITGAM were examined using multiplicative logistic regression with the affected status as a dependent factor, two SNPs as main factors and the two-SNP interaction term. The analysis was performed using the PLINK ‘--epistasis’ option.5 8 In addition, case-only analysis using the PLINK ‘--fast-epistasis’ and ‘--case-only’ options was used to examine the dependency or the correlation between the tested SNPs in the groups of SLE cases.5,8 To access combined effects of the risk alleles in rs3093030 and rs1143679, fixed-effects OR and 95% CI were estimated in stratification by the number of risk alleles (0 to 4) as compared with the group having no risk allele, separately among the European, African and Hispanic populations using the Case-control And Tdt Meta-Analysis Package (http://cran.r-project.org/web/packages/catmap/index.html). Two copy carriers homozygous for ICAM or ITGAM risk allele were removed from the two-risk-allele carrier group in order to see the combined effect.
SNP association with gene expression
Using the Genevar 2.0.1 database, which includes SNP genotype and mRNA profiling data for fibroblasts, lymphoblastoid cell lines, and T cells from the 75 GenCord individuals,9,10 the 19 SNPs located in a chromosome 19 region from position 10 200 kb to 10 320 kb were assessed for their associations with mRNA levels of ICAM1, ICAM4 and ICAM5 using a t statistic for Spearman’s rank correlation coefficient (ρ). To minimise the type I error caused by multiple testing, the significance level of the p value was set to α = 0.05/(19 SNPs)/(five probes)/(three cell lines)=1.75×10−4.
RESULTS
ICAM–SLE association study
In phase I, we evaluated 12 common variants directly typed in the ICAM1–ICAM4–ICAM5 locus spanning ~22 kb in a total of 14 719 unrelated SLE cases and controls of European (n=7427; SLE/controls=3936/3491), African (n=3613; 1679/1934), Hispanic (n=2299; 1492/807) and Korean (n=1380; 640/740) ancestries (online supplementary table S1). Proportions of genetic ancestry for the study subjects were filtered using principal component analysis as described in a previous study (online supplementary figure S1).4 All variants were typed with call rates of over 97.9%, and genotype proportions were consistent with expectations of the HWE (p≥0.001) among controls from each of the four ancestries (online supplementary table S2).
Of the 12 ICAM SNPs typed, five were significantly associated with SLE susceptibility under a statistical threshold of p≤4.17×10−3 in the trans-ancestry meta-analysis after Bonferroni correction for multiple comparisons (rs5030390, rs5498, rs281437, rs3093030 and rs2228615; 1.05×10−8 ≤pmeta≤2.80×10−4) (online supplementary table S2). Moreover, the directions of the effect sizes of each SNP were same among the ancestries except for rs5030390, the association of which was considered unreliable (online supplementary table S2).
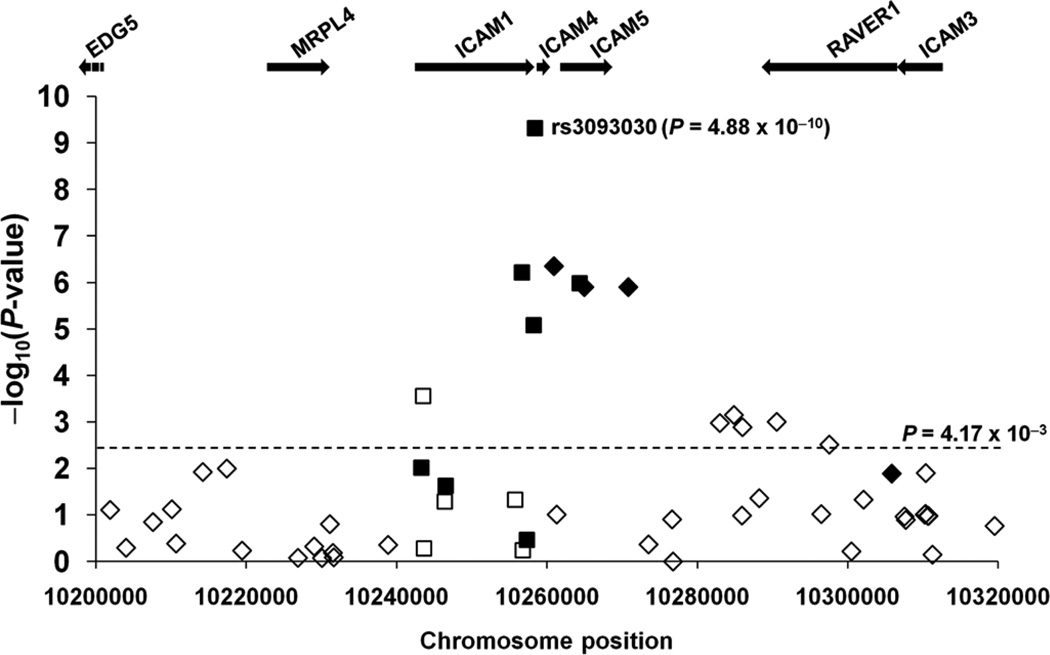
The strongest association with SLE was found with rs3093030 located in the 0.4 kb intergenic region between ICAM1 and ICAM4 (p=2.49×10−8; OR=1.16, 95% CI 1.10 to 1.22). The effect size was similar in each of the four ancestries (online supplementary table S2), with OR=1.12 (95% CI 1.05 to 1.20) in European, OR=1.18 (95% CI 1.01 to 1.37) in African, OR=1.28 (95% CI 1.13 to 1.45) in Hispanic and OR=1.15 (95% CI 0.98 to 1.35) in Korean ancestries (p=0.307 in Q statistic, I2=17.0). In a conditional logistic regression analysis followed by a meta-analysis, we found that the SLE association of rs5498, rs281437 and rs2228615 accounted for that of rs3093030, and they presented a consensus signal of SLE susceptibility locus, possibly because they are in high linkage disequilibrium with rs3093030 (D’ ≥0.84; online supplementary table S3). Although the effect magnitude of rs3093030 among Koreans was similar to those of the other ancestry populations, it did not achieve statistical significance, in contrast with the others (p=0.0841 in Koreans versus p=5.95×10−4 in Europeans, p=4.02×10−2 in Africans and p=7.28×10− 5 in Hispanics), perhaps because of the small sample size of the Korean population relative to the other ancestry populations. With the minor allele frequency of 0.324 in the Korean controls, the statistical power to detect the effect size found in the meta-analysis (fixed-effects OR=1.16) was only 42% in the Korean population. To increase the statistical power for Korean detection, in phase II we recruited an additional independent population of 2762 unrelated Korean participants (online supplementary table S1). Genotyping call rate was 96.9%, and the control subject genotypes were distributed under HWE (p=0.691). Indeed, a similar effect size was detected in the validation population (OR=1.18), and by combining the phase I and phase II populations of Koreans (n=4142), statistical significance (p=1.07×10−3; OR=1.17, 95% CI 1.07 to 1.29) was achieved (online supplementary table S2). Therefore, with the additional Korean validation population, each of the four ancestry populations independently showed a statistically significant association between the ICAM locus SNP rs3093030 and SLE susceptibility, and the meta-analysis across the four ancestral populations revealed a more significant overall relationship (p=4.88×10−10; OR=1.16, 95% CI 1.11 to 1.22; online supplementary table S1). To further characterise this genomic region as an SLE susceptibility locus, we imputed HapMap SNPs around the ICAM gene cluster that included EDG5, MRPL4, ICAM1, ICAM4, ICAM5, RAVER1 and ICAM3. Using the genotype data for the 12 ICAM SNPs, we imputed 58 HapMap SNPs located in a chromosome 19 region from position 10 200 kb to 10 320 kb (Genome build 36.3) using the MACH V.1.0 program, with the ancestry-matched haplotype templates provided by the HapMap3 Genome Browser release 2 of the International HapMap Project.
Three imputed SNPs (rs2569693, rs2569702 and rs892188) with high imputation quality (average posterior probability for the most likely genotype ≥0.97) were significantly associated with SLE susceptibility (table 1) while demonstrating the same direction of SLE-risk effects in each of the SNPs. In addition, all SLE-associated SNPs showed larger effects in Hispanics than in the other ethnic groups. These imputed SLE-associated SNPs were localised to the ICAM1–ICAM4–ICAM5 region and correlated highly with the genotyped SLE-associated SNP rs3093030 (0.81≤r2≤0.96). Trans-ancestry comparisons for the seven SLE associated SNPs (ie, four genotyped and three imputed) supported that the ICAM1–ICAM4–ICAM5 locus alone associated sufficiently with SLE susceptibility (p≤8.28×10−6; figure 1).
Table 1.
Association of directly typed and imputed markers in the ICAM locus and ITGAM with susceptibility to systemic lupus erythematosus
| European (n=7427) | African (n=3613) | Hispanic (n=2299) | Korean (n=1380 or 4142)+ | Meta-analysis (n≤17481)± |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Gene | SNP* | MAF (SLE/controls) | OR | MAF (SLE/controls) | OR | MAF (SLE/controls) | OR | MAF (SLE/controls) | OR | OR | p Value | |
| ICAM1–ICAM4–ICAM5 | ||||||||||||
| rs3093030 | (G>A) | (0.471/0.442) | 1.12 | (0.107/0.092) | 1.18 | (0.580/0.518) | 1.28 | (0.353/0.318) | 1.17 | 1.16 | 4.88×10−10 | |
| rs2569693 | (C>T) | (0.427/0.403) | 1.11 | (0.097/0.085) | 1.16§ | (0.550/0.492) | 1.26 | (0.363/0.332) | 1.14§ | 1.14 | 4.50×10−7 | |
| rs5498 | (A>G) | (0.470/0.445) | 1.11 | (0.188/0.173) | 1.11§ | (0.582/0.524) | 1.26 | (0.414/0.384) | 1.13§ | 1.13 | 6.08×10−7 | |
| rs2228615 | (G>A) | (0.427/0.403) | 1.11 | (0.098/0.087) | 1.14§ | (0.552/0.496) | 1.25 | (0.346/0.319) | 1.13§ | 1.14 | 1.05×10−6 | |
| rs2569702 | (T>C) | (0.431/0.408) | 1.1 | (0.097/0.085) | 1.16§ | (0.552/0.494) | 1.26 | (0.345/0.318) | 1.13§ | 1.14 | 1.27×10−6 | |
| rs892188 | (C>T) | (0.431/0.408) | 1.1 | (0.097/0.085) | 1.16§ | (0.552/0.494) | 1.26 | (0.345/0.318) | 1.13§ | 1.14 | 1.27×10−6 | |
| rs281437 | (G>A) | (0.276/0.288) | 0.94§ | (0.339/0.379) | 0.84§ | (0.175/0.220) | 0.75 | (0.088/0.092) | 0.95§ | 0.89 | 8.28×10−6 | |
| ITGAM | ||||||||||||
| rs1143679 | (G>A) | (0.194/0.126) | 1.67 | (0.157/0.104) | 1.82 | (0.168/0.100) | 1.59 | (0.002/0.000) | NC | 1.67 | 3.32×10−46 | |
Four SNPs (rs3093030, rs5498, rs2228615 and rs281437) were genotyped and the other three SNPs (rs2569693, rs2569702 and rs892188) were imputed. The major allele is more common than the minor allele in the whole control population but not necessarily in each subpopulation.
A total of 4142 Korean participants from phase I and phase II were genotyped for rs3093030, and the separate analyses are shown in online supplementary table S2.
Fixed-effects p value and OR were calculated from the meta-analysis because the effect size of each SNP was not significantly different among the ancestry populations (p>0.01 in Cochran’s Q statistic). For ITGAM rs1143679, the meta-analysis was performed with European, African and Hispanic results.
Allelic associations with SLE were not statistically significant at a threshold of α=0.05. The detailed information is shown in online supplementary table S2.
MAF, minor allele frequency; NC, not calculated; SLE, systemic lupus erythematosus; SNP, single-nucleotide polymorphism.
Figure 1.
A systemic lupus erythematosus association map for the genotyped (squares) and imputed single-nucleotide polymorphisms (SNPs) (diamonds) within and around the ICAM1–ICAM4–ICAM5 locus. Negative logarithms (base 10) of the p values calculated from the meta-analysis (y axis) are plotted against SNP positions on human chromosome 19 (x axis). The direction of effect size was the same in all four ancestries for some SNPs (filled symbols) but not for others (empty symbols). The arrows indicate transcription directions and gene sizes.
Pathway-based ICAM and ITGAM interaction study
Because ICAM1 and ICAM4 are binding partners of an αMβ2 integrin, in which the α subunit is encoded by an SLE-susceptibility gene, ITGAM, we examined the statistical interaction between SLE-risk alleles of the ICAM1–ICAM4–ICAM5 region and the previously reported SLE-risk alleles of the ITGAM SNP rs1143679 to characterise the pathway-based effect on SLE susceptibility.2 For this analysis, all participants of the phase I population were also genotyped for the non-synonymous SNP rs1143679 located in exon 3 of ITGAM. Call rates were ≥99.6% within each ancestry population, and genotype distributions were consistent with expectations of HWE in each of the control groups (online supplementary table S2).
The overall fixed effect of the SLE-risk-associated A allele in the ITGAM SNP was OR=1.67 (95% CI 1.55 to 1.79, p=3.32×10−46), showing similar effect sizes among European, African and Hispanic ancestries (p=0.541 in Q-statistic, I2=0.00%; online supplementary table S2), although the risk-associated allele was too rare (0.07%) to evaluate the interaction among Koreans. The infrequency in Koreans was consistent with the previous report.11 Statistical interaction between ITGAM rs1143679-A and each of the seven SLE-associated SNPs in the ICAM1–ICAM4–ICAM5 locus (p≤4.17×10−3; table 1) were examined among cases and controls using the PLINK ‘-- epistasis’ analysis and among only the cases using the PLINK ‘--fast-epistasis’ analysis.
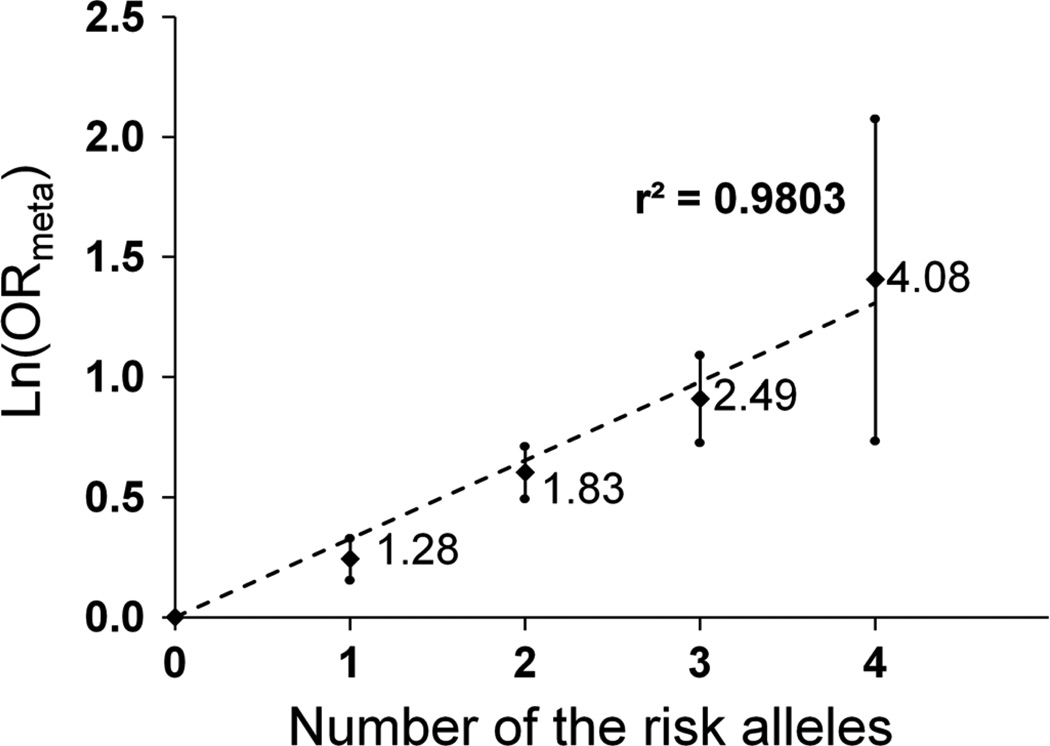
The ITGAM and ICAM SNPs independently affected SLE susceptibility, showing no notable ICAM–ITGAM epistatic interactions (p>0.05; online supplementary table S4). Using five subgroups according to the number of risk-associated alleles (0 to 4) in the ICAM SNP, rs3093030, and the ITGAM SNP, rs1143679, we assessed the combined effects of the SLE risk-associated alleles in the ICAM–integrin pathway. Fixed-effects OR and 95% CI for SLE susceptibility in each subgroup were estimated with reference to the subgroup with no risk allele. The natural logarithm of OR correlated linearly (r2=0.9803) with the number of risk alleles (0 to 4), and four-copy carriers had OR of 4.08 for risk of SLE compared with carriers with 0 copies (figure 2).
Figure 2.
Combined effects of ICAM1–ICAM4–ICAM5 rs3093030-A and ITGAM rs1143679-A alleles on systemic lupus erythematosus (SLE) susceptibility. Natural logarithms of the relative odds of SLE (y axis) are plotted against the number of at-risk alleles (x axis) in the two putative SLE susceptibility single-nucleotide polymorphisms (SNPs), rs3093030 in ICAM1–ICAM4–ICAM5 and rs1143679 in ITGAM. OR values are provided next to the data points, and error bars represent corresponding 95% CI. A linear trend line (dashed) and r2 are shown.
Gene expression
To determine whether the genetic variation has a role in gene expression, we evaluated expression quantitative trait loci (eQTL) associations within the ICAM locus using the Genevar (GENe Expression VARiation) database (http://www.sanger.ac.uk/resources/software/genevar/), which publicly provides data for 75 GenCord individuals on both SNP genotypes and expression profiles from primary fibroblasts, lymphoblastoid cell lines and T cells.9,10 The 19 Genevar SNPs included rs2569693, rs5498, rs2228615 and rs281437 among the SLE associated SNPs. However, none of the evaluated markers were associated with aberrant mRNA levels of ICAM1, ICAM4 or ICAM5 in any cell type (online supplementary figure S2).
DISCUSSION
To our knowledge, this is the first report of a comprehensive approach used to evaluate the effect of the ICAM1–ICAM4–ICAM5 locus on SLE risk. We observed that several SNPs within this locus are associated with susceptibility to SLE in multiple ancestry populations.
ICAM1 is mainly expressed in the vascular endothelium, macrophages and lymphocytes, and plays a role in immunological events including extravasation and T-cell-mediated responses.12 However, a plausible role for ICAM4 and ICAM5 in SLE pathogenesis is less clear because they are preferentially expressed in red blood cells and brain, respectively.13,14
The ICAM polymorphisms were not associated with altered mRNA levels of ICAM1, ICAM4 or ICAM5 in the Genevar database, although the sample size (n=75) was too small to make this null association conclusive. However, soluble ICAM1 levels in plasma have been associated with two SNPs (rs3093030 and rs5498) in the ICAM1–ICAM4–ICAM5 locus by protein QTL analysis in two independent studies involving 6578 European women (p=5.9×10−23 and p=4.8×10−25) and 9813 European individuals (p=3.5×10−23 and p=2.5×10−21).15,16 The alleles that were associated with increased soluble ICAM1 in those studies (A in rs3093030 and G in rs5498) were also associated with increased susceptibility to SLE in this study, providing a plausible mechanism for their effect. In addition, the genotype–phenotype relationships are consistent with several previously reported observations of raised plasma levels of soluble ICAM1 in patients with SLE,17–21 suggesting an underlying aetiological role.
Soluble ICAM1 is a cleaved form consisting of the extracellular domain of the membrane-bound full-length ICAM1 that is capable of binding to ligands. Thus soluble and membrane- bound ICAM1 can compete with each other for binding to ligands,12 but the immunological role of either soluble or membrane-bound ICAM1 in SLE pathogenesis has not been characterised. The level of soluble ICAM1 probably correlates with that of membrane-bound ICAM1,12,19 but it is not known whether the increase of soluble ICAM1 in SLE accompanies with the concurrent increase of the membrane form due to increased gene expression or with the reciprocal decrease of the membrane form due to increased proteolytic processing, as the membrane-bound ICAM1 levels have not been measured in patients with SLE. None of the Genevar SNPs in the ICAM1–ICAM4–ICAM5 locus was associated with ICAM1 mRNA expression. Instead, the non-synonymous ICAM1 SNP rs5498 (encoding Lys469Glu) was associated with both increased levels of the soluble form of ICAM1 protein and increased SLE susceptibility. Of note is the fact that it is located at an Ig superfamily domain, which may affect the proteolytic conversion to the soluble ICAM1. However, the level of statistical significance for the SLE association of rs5498 was weaker than that of rs3093030 in all populations, and statistical significance was not reached in the African or Korean ancestry populations. Thus it is possible that rs5498 affects SLE susceptibility in some ancestry populations but not others.
In this study, we also showed the association of ITGAM rs1143679 in European, African and Hispanic descendants, which was also found in previous association studies for systemic sclerosis. 22 This common variant was monomorphic in Koreans, which is consistent with the literature of other Asian populations, 11 perhaps reflecting selective pressures in this population. ITGAM (also known as CD11b, Mac-1 and complement receptor type 3) is a well-characterised molecule in the integrin α chain family that is expressed in a number of myeloid cells including macrophages, monocytes and neutrophils.23–25 ITGAM encodes the α chain of αMβ2 integrin, which regulates neutrophil and monocyte adhesion and migration from the bloodstream via interactions with a wide range of structurally unrelated ligands, including but not limited to ICAM1 and ICAM2.26,27
The previously identified SLE-susceptibility ITGAM SNP, rs1143679, encodes a change in amino acid from arginine at position 77 to histidine (R77H). This conversion of amino acids induces an alteration in the tertiary and quaternary structures of the ligand-binding αMβ2 domain, thereby modifying its overall binding affinity.28 Nath et al suggest that the R77H conversion may ultimately influence the conformation of the αI domain (specific to the ligand-binding domain for ICAM1), with subsequent consequences on αMβ2 ligand binding.2
Indeed, evidence suggests that αMβ2 levels are increased on neutrophils in SLE patients with active disease and may contribute to endothelial injury, consistent with the cumulative organ damage associated with SLE.29 αMβ2 is also involved in immune complex clearance, a process that is impaired in patients with SLE,30 suggesting that the function of αMβ2 may be altered in these people, either directly by structural modification or indirectly by alterations in receptor–ligand binding, possibly resulting from structural modification.2
In addition, this conversion is a target for alloantibodies present in mothers of neonates affected with neonatal autoimmune neutropenia.28 Notably, alloantibodies reactive against the polymorphic αMβ2 molecule are capable of blocking the αMβ2- dependent adhesion of neutrophils and monocytic U937 cells to a number of molecules including ICAM1.28
The functional association of ICAM and ITGAM prompted us to examine the associated SNPs in these genes for epistatic effects. Although a joint role for both ICAM1 and ITGAM in SLE aetiology appears to be plausible, they independently affected SLE susceptibility in this study. This may indicate that the functional role of the ICAM variants is involved in a nonintegrin- mediated pathway or that some factors are affected by ICAM–ITGAM binding through different pathways of ICAM and ITGAM without a common pathway.
In summary, we identified the association of several SNPs within the ICAM1–ICAM4–ICAM5 locus with SLE susceptibility. Although the effect size was modest, the association was consistent across all four ancestries. Our findings suggest that common variations in genes involved in ICAM-mediated or ICAM/integrin- mediated adhesion play a causal role in SLE susceptibility.
Supplementary Material
Acknowledgements
We thank the study participants and physicians who provided samples (Peter K Gregersen, So-Young Bang, BIOLUPUS Network and GENLES Network) and technical support (Taehyeung Kim). The members of the BIOLUPUS Network who provided samples used in this study are Sandra D’Alfonso, RafaellaScorza, Mauro Galleazzi, Gian Domenico Sebastiani, MD Danieli, and Sergio Migliaresi in Italy, Bernard R Lauwerys in Belgium, Emoke Endreffy and László Kovács in Hungary, Carlos asconcelos and Berta Martins da Silva in Portugal, Iñigo Rúa Figueroa, Carmen Gutierrez, Ana Suárez, Norberto Ortego, José Luis Calleja, Enrique de Ramón Garrido, Julio Sanchez Román, and Mario Sabio in Spain, Helle Laustrup and Peter Junker in Denmark, and Marc Bijl and Cees Kallenberg in Holland; Dr Lennart Truedsson provided controls from Southern Sweden (Lund). The members of the GENLES Network who provided samples used in this study are Hugo R Scherbarth, Pilar C Marino, Estela L Motta, Susana Gamron, Cristina Drenkard, Emilia Menso, Alberto Allievi, Guillermo A Tate, Jose L Presas, Simon A Palatnik, Marcelo Abdala, Mariela Bearzotti, Alejandro Alvarellos, Francisco Caeiro, Ana Bertoli, Sergio Paira, Susana Roverano, Cesar E Graf, Estela Bertero, Cesar Caprarulo, Griselda Buchanan, Carolina Guillerón, Sebastian Grimaudo, Jorge Manni, Luis J Catoggio, Enrique R Soriano, Carlos D Santos, Cristina Prigione, Fernando A Ramos, Sandra M Navarro, Guillermo A Berbotto, Marisa Jorfen, Elisa J Romero, Mercedes A Garcia, Juan C Marcos, Ana I Marcos, Carlos E Perandones, Alicia Eimon and Cristina G Battagliotti in Argentina; Eduardo Acevedo and Mariano Cucho in Perú; Ignacio García de la Torre, Mario Cardiel Ríos, José Francisco Moctezuma and Marco Maradiaga Ceceña in Mexico.
Funding The work was supported by grants from US NIH (AI063622, AI071651, AI082714, AI083194, AI094377, AI24717, AR042460, AR043274, AR043814, AR044804, AR048940, AR052300, AR053483, AR058554, AR060366, AR43727, AR62277, CA141700-01, GM063483, HD07463, K08-AI083790, K24-AR002138, M01-RR00079, N01-AR-6-227, P01-AR49084, P30-DK42086, P30-AR055385, P60-AR049459 P60-AR053308, P60-2-AR30692, PO1-AR49084, PR094002, R01-AR33062, R21-AI070304, RR015577, RR020143, UL1RR024999, UL1RR025005, UL1RR025741, UL1RR029882 and 5UL1RR025777), the Lupus Foundation of America, the Alliance for Lupus Research, a Kirkland Scholar Award, the US Department of Veterans Affairs, the Korean Healthcare Technology Research and Development Project (A111218-11-GM01), the Korea Research Program for New Drug Target Discovery (20090083335), the National Research Foundation of Korea (2010-0014162), the Lupus Research Institute Novel Research Grant, the Alliance for Lupus Research Target Identification in Lupus Grant, the Arthritis National Research Foundation Eng Tan Scholar Award, the ESF in the framework of the Research Networking Programme European Science Foundation—The Identification of Novel Genes and Biomarkers for Systemic Lupus Erythematosus (BIOLUPUS) 07-RNP-083, the Swedish Research Council, the Swedish Association against Rheumatism, the Swedish International Development Agency, the Gustaf Vth 80th-Jubilee Foundation, the Instituto de Salud Carlos III (PS09/00129) partly financed by the FEDER funds of the European Union, and a grant from the Consejería de Salud de la Junta de Andalucía.
Footnotes
Contributors KK, EEB, JAK, JOO, SKN, BR, AHS, KLM, PMG, JBH, CK, RPK and S-CB designed the study. EEB, C-BC, MEA-R, H-SL, SAB, LAC, GSA, JCE, AMS, COJ, GSG, DLK, BPT, J-MA, JMG, YMK, SCS, C-HS, S-KL, C-SK, JTM, MP, RRG, LMV, TBN, JM, BAP-E, TJV, BIF, KLM, PMG, JDR, JAJ, RHS, JBH, RPK and S-CB recruited and characterised the SLE cases and controls. KK, AA and KMK performed genotyping. SBG, AW, MC and CDL performed quality control analyses. KK and EEB performed statistical analyses. KK, EEB, C-BC, CK, RPK and S-CB prepared the manuscript. All authors approved the final draft.
REFERENCES
- 1.Harley JB, Alarcón-Riquelme ME, Criswell LA, et al. Genome-wide association scan in women with systemic lupus erythematosus identifies susceptibility variants in ITGAM, PXK, KIAA1542 and other loci. Nat Genet. 2008;40:204–210. doi: 10.1038/ng.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nath SK, Han S, Kim-Howard X, et al. A nonsynonymous functional variant in integrin-alpha(M) (encoded by ITGAM) is associated with systemic lupus erythematosus. Nat Genet. 2008;40:152–154. doi: 10.1038/ng.71. [DOI] [PubMed] [Google Scholar]
- 3.Hochberg MC. Updating the American College of Rheumatology revised criteria for the classification of systemic lupus erythematosus. Arthritis Rheum. 1997;40:1725. doi: 10.1002/art.1780400928. [DOI] [PubMed] [Google Scholar]
- 4.Adrianto I, Wen F, Templeton A, et al. Association of a functional variant downstream of TNFAIP3 with systemic lupus erythematosus. Nat Genet. 2011;43:253–258. doi: 10.1038/ng.766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Purcell S, Neale B, Todd-Brown K, et al. PLINK: a tool set for whole genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Huedo-Medina TB, Sánchez-Meca J, Marín-Martínez F, et al. Assessing heterogeneity in meta-analysis: Q statistic or I2 index? Psychol Methods. 2006;11:193–206. doi: 10.1037/1082-989X.11.2.193. [DOI] [PubMed] [Google Scholar]
- 7.Mägi R, Morris AP. GWAMA: software for genome-wide association meta-analysis. BMC Bioinformatics. 2010;11:288. doi: 10.1186/1471-2105-11-288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cordell HJ. Detecting gene-gene interactions that underlie human diseases. Nat Rev Genet. 2009;10:392–404. doi: 10.1038/nrg2579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dimas AS, Deutsch S, Stranger BE, et al. Common regulatory variation impacts gene expression in a cell type-dependent manner. Science. 2009;325:1246–1250. doi: 10.1126/science.1174148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yang TP, Beazley C, Montgomery SB, et al. Genevar: a database and Java application for the analysis and visualization of SNP-gene associations in eQTL studies. Bioinformatics. 2010;26:2474–2476. doi: 10.1093/bioinformatics/btq452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Han S, Kim-Howard X, Deshmukh H, et al. Evaluation of imputation-based association in and around the integrin-alpha-M (ITGAM) gene and replication of robust association between a nonsynonymous functional variant within ITGAM and systemic lupus erythematosus (SLE) Hum Mol Genet. 18:1171–1180. doi: 10.1093/hmg/ddp007. 009; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.van de Stolpe A, van der Saag PT. Intercellular adhesion molecule-1. J Mol Med. 1996;74:13–33. doi: 10.1007/BF00202069. [DOI] [PubMed] [Google Scholar]
- 13.Gahmberg CG, Tian L, Ning L, et al. ICAM-5-a novel two-facetted adhesion molecule in the ammalian brain. Immunol Lett. 2008;117:131–135. doi: 10.1016/j.imlet.2008.02.004. [DOI] [PubMed] [Google Scholar]
- 14.Toivanen A, Ihanus E, Mattila M, et al. Importance of molecular studies on major blood groups- intercellular adhesion molecule-4, a blood group antigen involved in multiple cellular interactions. Biochim Biophys Acta. 2008;1780:456–466. doi: 10.1016/j.bbagen.2007.09.003. [DOI] [PubMed] [Google Scholar]
- 15.Paré G, Chasman DI, Kellogg M, et al. Novel association of ABO histo-blood group antigen with soluble ICAM-1: results of a genome-wide association study of 6,578 women. PLoS Genet. 2008;4:e1000118. doi: 10.1371/journal.pgen.1000118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Barbalic M, Dupuis J, Dehghan A, et al. Large-scale genomic studies reveal central role of ABO in sP-selectin and sICAM-1 levels. Hum Mol Genet. 2010;19:1863–1872. doi: 10.1093/hmg/ddq061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Egerer K, Feist E, Rohr U, et al. Increased serum soluble CD14, ICAM-1 and E-selectin correlate with disease activity and prognosis in systemic lupus erythematosus. Lupus. 2000;9:614–621. doi: 10.1191/096120300678828749. [DOI] [PubMed] [Google Scholar]
- 18.Sabry A, Sheashaa H, El-Husseini A, et al. Intercellular adhesion molecules in systemic lupus erythematosus patients with lupus nephritis. Clin Rheumatol. 2007;26:1819–1823. doi: 10.1007/s10067-007-0580-7. [DOI] [PubMed] [Google Scholar]
- 19.Sfikakis PP, Tsokos GC. Clinical use of the measurement of soluble cell adhesion molecules in patients with autoimmune rheumatic diseases. Clin Diagn Lab Immunol. 1997;4:241–246. doi: 10.1128/cdli.4.3.241-246.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tso TK, Huang WN. Elevated soluble intercellular adhesion molecule-1 levels in patients with systemic lupus erythematosus: relation to insulin resistance. J Rheumatol. 2007;34:726–730. [PubMed] [Google Scholar]
- 21.Tulek N, Aydintug O, Ozoran K, et al. Soluble intercellular adhesion molecule-1 (sICAM-1) in patients with systemic lupus erythematosus. Clin Rheumatol. 1996;15:47–50. doi: 10.1007/BF02231684. [DOI] [PubMed] [Google Scholar]
- 22.Anaya JM, Kim-Howard X, Prahalad S, et al. Evaluation of genetic association between an ITGAM non-synonymous SNP (rs1143679) and multiple autoimmune diseases. Autoimmun Rev. 2012;11:276–280. doi: 10.1016/j.autrev.2011.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Abbas AR, Baldwin D, Ma Y, et al. Immune response in silico (IRIS): immune-specific genes identified from a compendium of microarray expression data. Genes Immun. 2005;6:319–331. doi: 10.1038/sj.gene.6364173. [DOI] [PubMed] [Google Scholar]
- 24.Hynes RO. Integrins: versatility, modulation, and signaling in cell adhesion. Cell. 1992;69:11–25. doi: 10.1016/0092-8674(92)90115-s. [DOI] [PubMed] [Google Scholar]
- 25.Lu H, Smith CW, Perrard J, et al. LFA-1 is sufficient in mediating neutrophil emigration in Mac-1-deficient mice. J Clin Invest. 1997;99:1340–1350. doi: 10.1172/JCI119293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fagerholm SC, Varis M, Stefanidakis M, et al. alpha-Chain phosphorylation of the human leukocyte CD11b/CD18 (Mac-1) integrin is pivotal for integrin activation to bind ICAMs and leukocyte extravasation. Blood. 2006;108:3379–3386. doi: 10.1182/blood-2006-03-013557. [DOI] [PubMed] [Google Scholar]
- 27.Li Y, Zhang L. The fourth blade within the beta-propeller is involved specifically in C3bi recognition by integrin alpha M beta 2. J Biol Chem. 2003;278:34395–34402. doi: 10.1074/jbc.M304190200. [DOI] [PubMed] [Google Scholar]
- 28.Sachs UJ, Chavakis T, Fung L, et al. Human alloantibody anti-Mart interferes with Mac-1-dependent leukocyte adhesion. Blood. 2004;104:727–734. doi: 10.1182/blood-2003-11-3809. [DOI] [PubMed] [Google Scholar]
- 29.Buyon JP, Shadick N, Berkman R, et al. Surface expression of Gp 165/95, the complement receptor CR3, as a marker of disease activity in systemic Lupus erythematosus. Clin Immunol Immunopathol. 1988;46:141–149. doi: 10.1016/0090-1229(88)90014-1. [DOI] [PubMed] [Google Scholar]
- 30.Hepburn AL, Mason JC, Wang S, et al. Both Fcgamma and complement receptors mediate transfer of immune complexes from erythrocytes to human macrophages under physiological flow conditions in vitro. Clin Exp Immunol. 2006;146:133–145. doi: 10.1111/j.1365-2249.2006.03174.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.