Abstract
Objective: To develop a technique for recognizing critical situations based on laboratory results in settings in which a normal range cannot be defined, because what is “normal” differs widely from patient to patient. To assess the potential of this approach for kidney transplant recipients, where recognition of acute rejections is based on the pattern of changes in serum creatinine.
Design: We developed a case-based reasoning algorithm using dynamic time-warping as the measure of similarity which allows comparison of series of infrequent measurements at irregular intervals for retrieval of the most similar historical cases for the assessment of a new situation.
Measurements: The ability to recognize creatinine courses associated with an acute rejection was tested for a set of cases from a database of transplant patient records and compared with the diagnostic performance of experienced physicians. Tests were performed with case bases of various sizes.
Results: The accuracy of the algorithm increased steadily with the size of the available case base. With the largest case bases, the case-based algorithm reached an accuracy of 78 ± 2%, which is significantly higher than the performance of experienced physicians (69 ± 5.3%) (p < 0.001).
Conclusion: The new case-based reasoning algorithm with dynamic time warping as the measure of similarity allows extension of the use of automatic laboratory alerting systems to conditions in which abnormal laboratory results are the norm and critical states can be detected only by recognition of pathological changes over time.
The use of information technology for the improvement of patient care by detecting and informing clinicians about key clinical events already has a long history with numerous successful examples in various areas of medicine.1,2 Often the success of such systems depends on the feasibility of extracting exact rules from existing comprehensive domain knowledge. Thus, the interpretation of laboratory results is well suited for support by computer systems if the cut-off between normal and critical values is known. Under this condition, the value of automated alerting systems for improving patient care is well proven.3,4
Unfortunately some medical conditions make it impossible to define a normal range for parameters that are essential in monitoring the respective condition. For kidney transplant recipients, a serum creatinine within the “normal” range is not the norm but an exception. Meanwhile, despite ongoing efforts to develop other methods, serum creatinine remains the most important parameter for the assessment of renal graft function.5–7 A rise in serum creatinine corresponds to a deterioration in graft function. The attending physician has to recognize “significant” increases in serum creatinine that warrant further diagnostic measures to exclude or verify an underlying graft rejection, which requires immediate therapy to prevent graft damage or loss. Whether a new measurement constitutes a rise can be determined only in relation to at least one previous measurement. As each patient has an individual range of “usual” creatinine values with an individual size of “usual” changes between consecutive measurements, the decision whether a rise in creatinine is “significant” still requires experience and intuition. Exact rules that define the properties of a “critical” sequence of creatinine values are not available because the pathophysiology of transplant rejections are incompletely understood. Simple algorithms or rule-based expert systems, therefore, are not suitable for the development of diagnostic decision support systems for this or similar problems (e.g., blood cell counts in hematologic disorders, lipase levels in chronic pancreatitis, CD4-leucocyte counts in AIDS8). Instead, a technique that is capable of dealing with sequences (time series) of low-frequency measurements with unequal distances between is required. Because there is only a limited supply of historical cases from which a learning algorithm can extract the inherent information, a method that allows continuous inclusion of new cases as they become available (i.e., a lazy learning approach) seems preferable.
Background
When an incomplete domain theory prohibits the a priori definition of ideal patterns, it is still possible to compare new problems with historical cases. Case-based reasoning (CBR) is a promising approach with existing applications in a number of fields including medicine.9 The idea is to mimic the human technique of problem-solving by analogy. To solve a new problem, the system retrieves similar stored cases and uses the solutions associated with these cases to generate a solution for the new problem.10 Since the task of learning from known examples is delayed until a new case is processed, CBR belongs to the class of lazy learning algorithms.11 Many algorithms that have been successfully used for pattern recognition are eager learning algorithms; that is, they require that the parameters of some sort of a model are learned before the algorithm can be applied. Examples for such algorithms include graphical models such as Bayesian networks or Hidden Markov Models.12 A lazy learning algorithm such as CBR seems to be of advantage in a setting like ours, with cases stemming from a continuously growing electronic patient record. The crucial task in the development of a CBR system is the definition of a similarity measure for the case retrieval. To our knowledge, so far no similarity measure for the comparison of courses of laboratory values by CBR has been established.
Experience with assessment of time series already exists in other domains with applications ranging from stock prices and meteorological data to electro-cardiograms. Many of these applications work by pattern recognition; that is, a new time series is compared with stored patterns. But many traditional algorithms for comparison of time series, such as Euclidian distance or arithmetic correlation, are based on the assumption of equidistant measurements or equal length of the time series. Blood samples are usually rather infrequent. An approach that is not limited by these assumptions is dynamic time warping (DTW), which has been successfully applied to pattern recognition in time series.13,14 One of its original application domains is speech recognition, in which the matching of spoken words to word templates requires an algorithm that allows for different timing and pronunciation. The result of DTW can be seen as a “warping” of the time axis so that the distance between two time series becomes minimal with respect to a distance function. The cumulative value of this function yields a measure of distance or, since the two concepts are related, similarity.
We performed this study to evaluate whether DTW yields a similarity measure that permits the application of case-based reasoning to time series of laboratory values, thus improving the assessment of creatinine courses in kidney transplant recipients. The diagnostic accuracy of physicians experienced in nephrology was tested as the reference standard.
Method
Data Extraction
All relevant parameters and findings for the patients at the kidney transplant program of the Charité are available through the electronic patient record TBase2®. This system is available from all computers within the Charité intranet that have a web browser installed. Data from other systems (e.g., the laboratory) is imported online via a number of interfaces. TBase2 is based on a relational data model and the stored data can be retrieved via standard structured query language (SQL).15 For this study we extracted laboratory results, information about the transplantation, and information about rejections from the database to analyze courses of creatinine measurements in stable kidney transplant recipients.
A valid creatinine course was defined as having a maximum length of 360 days and starting no earlier than 90 days after transplantation. The second condition was considered necessary to avoid contamination by the regular fall in creatinine during the early posttransplant period. We also required at least four measurements per course to have a sufficient number of measurements to establish a trend.
Each creatinine course was classified as either “critical” (i.e., a graft rejection occurred within two days of the last creatinine measurement) or non-critical (i.e., the patient never had rejection of his current graft). For this study, two classes of 51 courses each were selected by random procedure from all valid courses: The “positive” class was obtained from the critical courses. The “negative” class contains an equal number of noncritical courses. To ensure that during these noncritical courses no clinical situations associated with a rise in creatinine (e.g., infections) occurred, the records of the respective patients were checked manually by a documentation assistant. The 102 courses of these two classes constitute the entire test set. To avoid selection effects, we did not divide the courses of the test set into a case base and a distinct test field. Instead, we repeatedly assembled different case bases from the entire test set. For each assessment, only the course to be classified was excluded from all case bases, and a randomly selected course from the opposite class was also excluded to maintain the same distribution of cases in the case bases (bootstrapping approach).
Algorithm
In CBR each combination of problem and solution is referred to as a case, and the collection of all available cases is called case base.16 In our setting the problem is time series of creatinine measurements and the solution is the classification are the basis for classifying the course as critical or noncritical. A creatinine course that ends with a rejection is a critical course. The information from the patient record database serves as the gold standard for our tests.
To determine the similarity (i.e., distance) between two creatinine courses, we used DTW as the measure of distance. DTW is a dynamic programming technique that finds a mapping between the measurements from the two time series so that the cumulative value of a given distance function becomes minimal. For our purpose, we require that each measurement of one time series is mapped onto at least one measurement of the other time series and that the mapping preserves the chronological order of the measurements. Therefore the algorithm starts by mapping the first elements of each time series and then successively adds new mappings until the last elements have been included. The mathematical formulation of the algorithm is a special form of the Bellman equation:
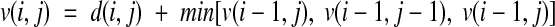 |
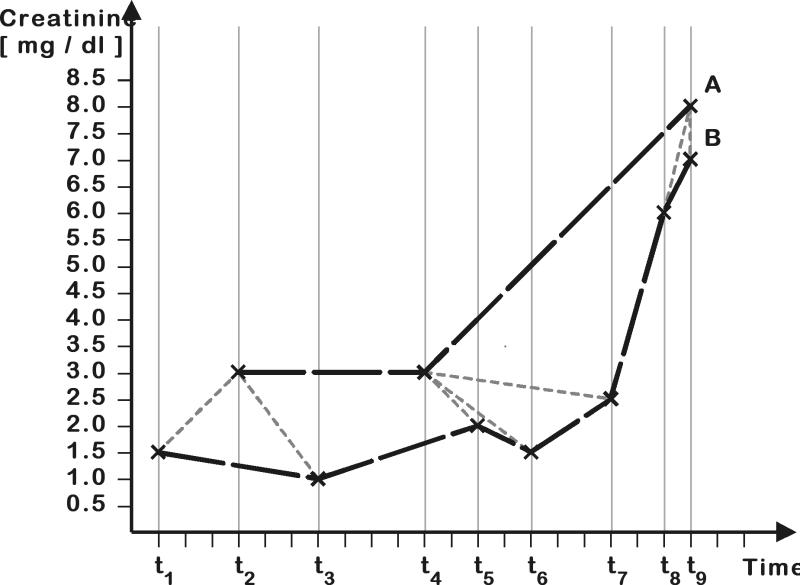
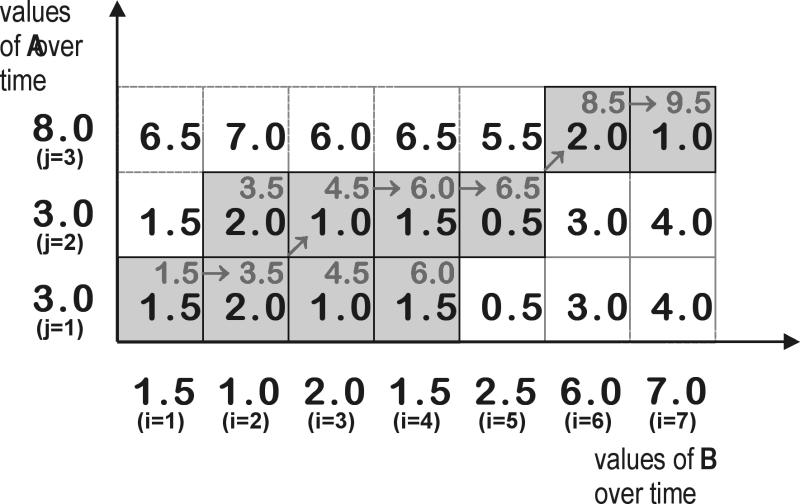
In this equation, v(i,j) is the minimal cumulative distance when mapping the first i values of the first time series to the first j values of the second time series. The function d(i,j) denotes the distance between the i-th value of the first time series and the j-th value of the second time series. Since the chronological order has to be preserved, there are three options to choose the next mapping: (1)move to the next element in the first time series only, (2) move to the next element in the second time series only, or (3) move to the next element in both time series. Thus the value v(i,j) is calculated as the sum of the distance d(i,j) and the minimal cumulative distance for a mapping that ends with (i−1,j), (i−1,j-1), or (i−1,j). We used the absolute value of the difference between two creatinine measurements as local distance function d(i,j). The courses with the lowest cumulative distance to a given course are considered to be most similar. Figure 1▶ shows an example of how DTW maps the measurements of two time series; measurements connected by a gray line are mapped onto each other. Tracing the gray arrows in Figure 2▶, one can see how this mapping was obtained. Note that this is not the only optimal mapping. Since in our example the distance values for two consecutive measurements are the same, both measurements could be used for a mapping.
Figure 1 .
Example for the alignment of measurements by DTW for measuring the distance between two time series. The mapping of the values of time series A onto the values of time series B is depicted with dashed gray lines (e.g., first value of A is mapped onto the first value of B).
Figure 2 .
Graphical example of how DTW obtains a mapping between two time series. The grid shows the distance (absolute difference, d(i,j)) between each pair of measurements from the two time series (black value). For example, the lower left cell contains d(1,1), i.e. |3.0 – 1.5|. The gray arrows mark steps taken to find the optimal mapping shown in Figure 1▶ with the cumulative distance v(i,j) given in the upper right corner of the cell. Shaded cells belong to at least one optimal mapping.
When a new course is presented to our algorithm, it retrieves the five most similar cases from the case base. The new course is classified according to the classification of the majority of these five courses as critical or noncritical. In our setting, the algorithm is not highly sensitive to changes in the size of k, although a very small k (e.g., k = 1) yields problems with outliers and a rather large k leads to a classification based on cases that are not very similar to the given course. To prevent a “draw” situation, an uneven number of cases must be retrieved— in previous tests, retrieval of five cases gave the highest accuracy. With a growing case base, the optimal number of retrieved cases is subject to change.
Analysis
Each course of the test set was assessed by the algorithm. To study the influence of the size of the case base on the performance of the algorithm, we used case bases of 22, 42, 62, and 82 cases. The case base for each assessment was randomly selected from the test set with stratification for class to maintain an equal distribution. To obtain valid means, we repeated the case base selection and assessment 30 times for each course and each case base size (i.e., 120 assessments per course in the test set).
All results are given as means ± standard deviation. Statistical significance between performance of the experts and the algorithm was established by the Mann-Whitney U-test.
The algorithms have been implemented in SAS (Version 8e, SAS Institute) using the IML programming language. The runtime environment for our test was SAS running on a Windows 95 machine with an AMD Athlon 700 MHz processor and 256 MB RAM.
Quantification of Expert-Performance
To assess the performance of our case-based approach we compared it with the performance of domain experts. Therefore, we designed a test program to investigate how experienced physicians assess the state of renal graft function by means of the creatinine course only. The program is based on web technology and accessible from all workstations within the Department of Nephrology of the Charité. Physicians who take the test see a figure with the creatinine course over time in the same format used in the electronic patient record and assess the course as “rejection is likely” or “rejection is unlikely.” The program successively shows all courses from the test set in random order, and the physicians assessment is stored in a database.
All residents and consultants at the Department of Nephrology of the Charité were invited to participate in this study. All physicians were experienced in care for transplant recipients, although the level of experience differs.
Results
Data Extraction
When data extraction was performed, 1,059,403 laboratory values, 1,143 patients, and 680 rejection episodes were stored in the patient record database. Of the total laboratory values, 43,638 were creatinine measurements. From these, a total of 33,175 different courses with a maximum length of 1 year, measurements from at least four different days, and the first measurement at least 90 days after transplantation were computed.
From a total of 680 rejections, 74 rejections were excluded because of missing creatinine values between transplant and loss of graft. Of the remaining 606 rejections, eight did not include creatinine values at least 90 days after transplantation, and for 360 rejections the date of rejection occurred within the first 90 days after transplantation. Another 28 rejections were excluded because there were no values within the 360 days before the rejection occurred, and for 90 rejections there were no measurements within the last two days before the date of rejection. Finally four of the remaining courses contained fewer than four creatinine measurements and thus were excluded. Thus 116 rejections were associated with a creatinine course without any exclusion criteria. From these the 51 creatinine courses for the “positive” class were chosen by random selection.
Physician Performance
A total of ten physicians took part in the quantification of expert performance by assessing all courses in the test set. The physicians reached an accuracy of 69.2 ± 5.3 % for the whole test set. The sensitivity for recognizing a critical course was 46.7 ± 13.4 % and their specificity was 91.6 ± 4.9 %. The accuracy of the four participating consultants (at least six years of care for transplant patients) was 74.0% (71.6–76.5%) which was higher than the 66.4% (62.7–71.6%) accuracy of the six residents (at least two years of care for transplant patients).
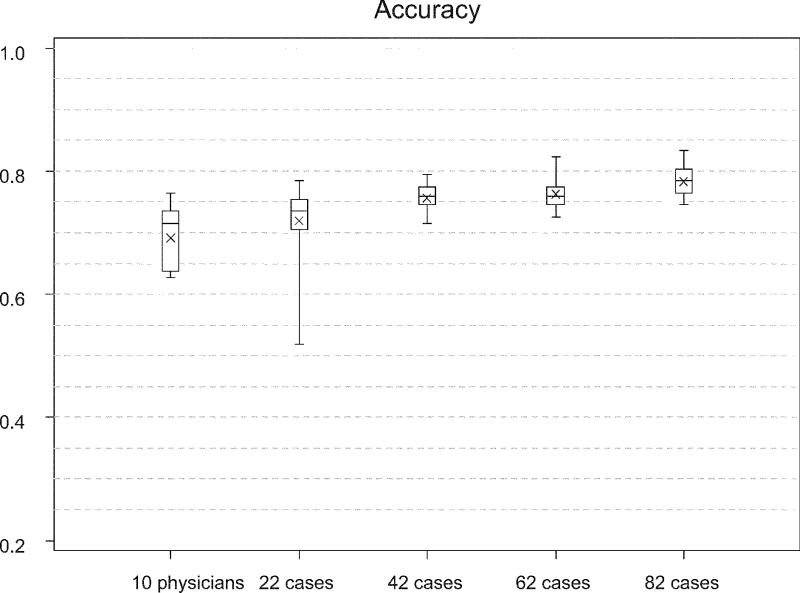
With the smallest case-base size of 22 cases, there was no significant difference between the diagnostic accuracy of the physicians and the algorithm. The accuracy of the algorithm increased with the size of the available case-base (Figure 3▶). With the largest case bases (82 cases), the case-based algorithm reached an accuracy of 78.4 ± 2.3% which is significantly higher than the performance of the experts (p < 0.001).
Figure 3 .
Accuracy of the case based reasoning algorithm with DTW in detecting creatinine courses associated with a transplant rejection. The diagnostic accuracy is shown for different case base sizes in comparison with the diagnostic accuracy of experienced physicians. The chart shows boxplots giving minimum, 25th percentile, median, 75th percentile and maximum accuracy depicted by the box as well as the mean accuracy shown as x.
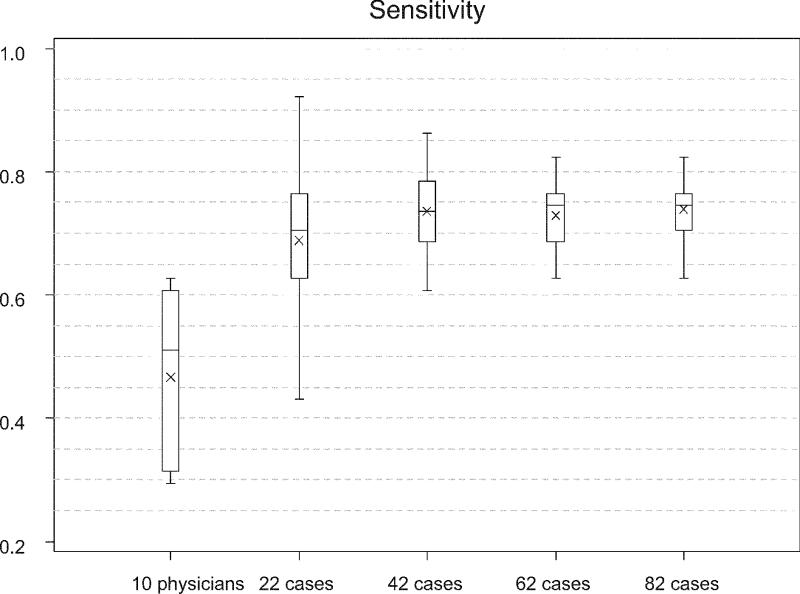
The average specificity of the physicians is significantly higher (p < 0.001) than the specificity of the algorithm for all case-base sizes, despite a trend toward an increase in specificity with the size of the case base. At the same time the mean sensitivity of the algorithm (74.4 ± 5.2% for the largest case base) is significantly higher than the mean sensitivity of the physicians (46.7 ± 13.4; p < 0.001). Again, the sensitivity of the algorithm increases with the size of the case base (Figure 4▶).
Figure 4 .
Sensitivity of the case based reasoning algorithm with DTW in detecting creatinine courses associated with a transplant rejection. The sensitivity is shown for different case base sizes in comparison with the sensitivity of experienced physicians. The chart shows boxplots giving minimum, 25th percentile, median, 75th percentile and maximum sensitivity depicted by the box as well as the mean sensitivity shown as x.
The time required for selection of the five most similar courses from any of the case bases was always well below 1 second. The runtime behavior of different computer systems was not tested.
Discussion
The case-based reasoning algorithm with case retrieval by dynamic time warping demonstrated a higher accuracy than experienced physicians in recognizing creatinine courses associated with a kidney transplant rejection. This is an important application in the care for transplant recipients, because detection of a pathological rise in serum creatinine is still the first step in diagnosing transplant rejection and avoiding graft loss by timely treatment. Moreover, this algorithm might be a solution for similar problems in the care for other chronic conditions, such as the monitoring of CD4-leukocyte counts in AIDS patients, in which a “normal” range can be defined only on an individual basis.8 To our knowledge this is the first report of a working diagnosis support algorithm for the assessment of nonequidistant time series of laboratory results in an area with incomplete domain knowledge.
Many decision-support systems that work with time-related medical data are based on a detailed representation of the domain knowledge. This approach has been formalized by Shahar’s knowledge-based temporal abstraction (KBTA).17A KBTA-system such as RÉSUMÉ17 requires that the domain experts explicitly define abstraction levels and express the temporal relations between events and intervals. For instance, this approach includes what Shahar calls “temporal dynamic knowledge” (e.g., the temporal persistence of the value of a parameter when it is not measured). Although a structured approach such as KBTA seems to help experts express their knowledge, a significant amount of domain specific expert knowledge has to be entered into a KBTA-based system.17–19 The same applies to the approach described by Bellazzi et al., in which time series from diabetic patients are analyzed using temporal abstractions.20 However, in many medical domains (such as the care for kidney transplant recipients), the domain knowledge is still incomplete; therefore, the definition of reliable abstraction rules is not feasible.
A system designed for domains in which curve fitting models are not available (VIE-VENT) has been applied to continuous and discontinuous data from respirators for controlling the ventilation of newborn infants.21,22 This system relies on temporal abstraction for the input data and still requires rules to generate the output. Moreover, VIE-VENT is designed for high-frequency data.
Schmidt et al. describe ICONS,23 a system that is closer to our approach because it uses case-based reasoning and the objective is to recognize renal dysfunction. Still the setting is quite different: the data stems from ICU patients and a number of measured and calculated parameters are available on a daily basis. These parameters are abstracted into a single state of renal function, and this state is followed over a maximum of seven days. For renal graft recipients we consider the definition of a state of renal function problematic, and, as for most chronic conditions, it is virtually impossible to obtain data on a daily basis. A system that has been tested with infrequent and nonequidistant time series is TrenDx.24 Haimowitz et al. assessed growth charts by matching a number of trend templates grouped into a monitor set against the actual chart. The structure of the trend templates was defined in conjunction with pediatric endocrinologists and consists of low-order polynomial regression models. Consequently, the matching is done by regression techniques and the trend templates have to be defined by domain experts. This system again depends on the quality of the knowledge available in the domain.
Recently algorithms based on graphical models have been used successfully in pattern recognition and for the design of decision support systems.12 Hidden Markov Models (HMM) are widely used in speech recognition and could be applied to the classification of clinical time series. We know of no application of HMMs to time series in a setting similar to ours. We preferred the lazy learning nature of CBR. By deferring the learning until a new course needs to be classified, CBR uses all knowledge currently available in the case base. And since only the most similar cases are taken into account in classifying a new course, local groups of similar cases will be recognized without the need of defining an appropriate model prior to learning. The eager learning nature of Hidden Markov Models requires users to relearn the model’s parameters to use new data that are entered in the electronic patient record. Additional research is necessary to compare the performance of our algorithm with that of other algorithms for classification of time series.
The advantage of our case-based approach is that we do not rely on explicit temporal expert knowledge. Instead, we use the knowledge associated with a collection of historical courses of creatinine. Therefore, our algorithm depends on the quality and number of courses in our case base, and results improve as the case base grows. As new cases are continuously entered into the patient record database, the case base can be enhanced by automatic extraction of cases to the case base. An additional advantage of this lazy-learning approach is the automatic adaptation to temporal trends in patient management and outcome. If this change over time is substantial, the age of the observations can be included in the similarity measure.
With a growing case base the runtime of our algorithm might become an issue, since we use linear search for the retrieval and DTW has a runtime of O(nm). In our study the time required for selection of the five most similar courses from any of the case bases was always well below one second. Although this ensures that the system can be used to assess courses on user request, the runtime of DTW can grow rapidly as the number of values per time series increases. The algorithm essentially calculates a matrix with the local distances between all pairs of values. Therefore the runtime depends on the product of n and m, where n is he number of values in the first time series and m is the number of values in the second time series [i.e. the runtime is O(nm)].
A method to accelerate the retrieval of cases is suggested by Schmidt et al. based on a tree of prototypical cases.25 A prototypical case shares most of the attributes of the cases or prototypes for which it stands. The resulting search tree-like structure facilitates faster retrieval. We plan to investigate how such a technique might be used with this kind of data.
Other research addresses the runtime of DTW. Keogh and Pazzani describe how piecewise linear segmentation can reduce the runtime of DTW in comparing two time series.26 Before processing, the time series are divided into segments in which the values can be approximated by linear regression with a given error. Depending on the nature of the data, this approach can reduce the number of comparisons considerably. Research by Berndt and Clifford on the way DTW maps values onto each other introduces a “warping window” that restricts how far two compared values can lie apart, limiting the number of necessary calculations.14
Another aspect is that most courses that could be added to the case base are noncritical; critical events are rare. The additional use of many similar courses for the case base seems questionable. It remains to be investigated whether it is more efficient to add only a limited number of noncritical courses and assign different weights to different courses in the case base.
Runtime is especially important if the assessment is initiated by user request as the user will have to wait for the result. This “pull” approach is not necessarily the best solution for this setting. We are considering how to trigger the assessment process automatically each time a new creatinine measurement is entered into the patient record database. The result could be stored directly in the database, eliminating user waiting times. Additionally, this “push” approach would alert the user to new critical results even before the user accesses the file of the respective patient.
With integration of the algorithm into our electronic patient record system, we will be able to evaluate prospectively the actual impact on care of transplant recipients. An upcoming prospective trial will study the system’s ability to determine correctly whether a transplant biopsy (the current gold standard for diagnosing transplant rejection) is indicated will be studied. This can be done either without informing the attending physician about the assessment result and thus measuring the performance of the system alone or by displaying the result and thus measuring the combined human-model performance. In preparation we will assess retrospectively whether the inclusion of other parameters (blood urea nitrogen, C-reactive protein, urinary proteins) is likely to enhance the diagnostic accuracy of the algorithm.
Conclusion
The benefit of automated systems that alert physicians to laboratory results associated with critical situations already has been demonstrated in several studies.3,4,27–30 But so far these systems require predefined limits to describe the normal range and/or the alerting criteria. The case-based reasoning algorithm with dynamic time warping allows extension of the use of alerting systems to conditions in which abnormal laboratory results are the norm and critical states can be detected only by via recognition of a pathological change over time.
Acknowledgments
We are indebted to our colleagues who sacrificed their valuable time to participate in the quantification of expert performance for this study. We are especially grateful to our documentation assistant Tanja Nienkarken RN for her help with data management.
References
- 1.Miller RA. Medical diagnostic decision support systems—past, present, and future: A threaded bibliography and brief commentary. J Am Med Inform Assoc. 1994;1(1):8–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Classen DC. Clinical decision support systems to improve clinical practice and quality of care. JAMA 1998;280(15):1360–1. [DOI] [PubMed] [Google Scholar]
- 3.Raschke RA, Gollihare B, Wunderlich TA, et al. A computer alert system to prevent injury from adverse drug events: development and evaluation in a community teaching hospital. JAMA. 1998;280(15):1317–20. [DOI] [PubMed] [Google Scholar]
- 4.Kuperman GJ, Teich JM, Tanasijevic MJ, et al. Improving response to critical laboratory results with automation: results of a randomized controlled trial. J Am Med Inform Assoc. 1999;6(6):512–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Al-Awwa IA, Hariharan S, First MR. Importance of allograft biopsy in renal transplant recipients: correlation between clinical and histological diagnosis. Am J Kidney Dis. 1998;31(6 Suppl 1):S15–S18. [DOI] [PubMed] [Google Scholar]
- 6.Soulillou JP. Immune monitoring for rejection of kidney transplants. N Engl J Med. 2001;344(13 ):1006–7. [DOI] [PubMed] [Google Scholar]
- 7.Li B, Hartono C, Ding R, et al. Noninvasive diagnosis of renal-allograft rejection by measurement of messenger RNA for perforin and granzyme B in urine. N Engl J Med. 2001;344(13): 947–54. [DOI] [PubMed] [Google Scholar]
- 8.Quinn TC. Acute primary HIV infection. JAMA. 1997; 278(1):58–62. [PubMed] [Google Scholar]
- 9.Gierl L, Bull M, Schmidt R, et al. Case-Based Reasoning Technology: From Foundation to Application. Berlin, Springer, 1998, pp 273–97.
- 10.Kolodner JL. Case-Based Reasoning. San Mateo, CA, Morgan Kauffman, 1993.
- 11.Mitchell T. Machine Learning. New York, McGraw Hill, 1997.
- 12.Lauritzen SL. Some Modern Applications of Graphical Models. Aalborg Department of Mathematical Sciences, Aalborg University. 2001; R-01–2012 <http//:www.math.auc.dk/~steffen/publications.html>.
- 13.Sakoe H, Chiba S, Waibel A, Lee K-F, (eds). Dynamic programming algorthim optimization for spoken word recognition. In Readings in speech Recognition. San Mateo, CA, Morgan Kauffman, 1990, pp 159–65.
- 14.Berndt DJ, Clifford J, Fayyad UM, et al.(eds). Finding patterns in time series: A dynamic programming approach. Menlo Park, CA, AAAI Press/MIT Press, 1996, pp 229–48.
- 15.Fritsche L, Schroeter K, Lindemann G, et al. A web-based electronic patient record system as a means for collection of clinical data. In Brause RW, Hanisch E (eds): Medical Data Analysis. Berlin, Springer, 2000, pp 198–205.
- 16.Richter MM. Bartsch-Spoerl B, Wess S, Burkhard H-D, Lenz M (eds). Case-Based Reasoning Technology: From Foundation to Application. Berlin, Springer, 1998.
- 17.Shahar Y. A Knowledge-based Method for Temporal Abstraction of Clinical Data. Stanford, CA, Stanford University Press, 1994.
- 18.Shahar Y, Musen MA. Knowledge-based temporal abstraction in clinical domains. Artif Intell Med. 1996;8:267–98. [DOI] [PubMed] [Google Scholar]
- 19.Shahar Y, Musen MA. Evaluation of a temporal-abstraction knowledge acquisition tool. Presented at the 12th Banff Knowledge Acquisition for Knowledge-based Systems Workshop, Banff, Alberta, Canada, 1999.
- 20.Bellazzi R, Larizza C, Magni P, et al. Intelligent Analysis of clinical time series: An application in the diabetes mellitus domain. Artif Intell Med. 2000;20(1):37–57. [DOI] [PubMed] [Google Scholar]
- 21.Miksch S, Horn W, Popow C, et al. Intensive care monitoring and therapy planning for newborns. In: AAAI 1994 Spring Symposium: Artificial Intelligence in Medicine: Interpreting Clinical Data. AAAI Press, 1994, pp 91–4.
- 22.Miksch S, Horn W, Popow C, et al. Therapy planning using qualitative trend descriptions. In Proceedings of the 5th Conference on Artificial Intelligence in Medicine Europe (AIME 95),. 1995.
- 23.Schmidt R, Heindl B, Pollwein B, Gierl L. Multiparametric time course prognoses by means of case-based reasoning and abstractions of data and time. Med Inform (Lond). 1997; 22(3):237–50. [DOI] [PubMed] [Google Scholar]
- 24.Haimowitz IJ, Le PP, Kohane IS. Clinical monitoring using regression-based trend templates. Artif Intell Med. 1995; 7(6):473–96. [DOI] [PubMed] [Google Scholar]
- 25.Schmidt R, Pollwein B, Gierl L. Horn W, et al. (eds). Experiences with case-based reasoning methods and prototypes for medical knowledge-based systems. AIMDM 99. Berlin, Springer, 1999, pp 124–32.
- 26.Keogh EJ, Pazzani MJ. Principles and Practice of Knowledge Discovery in Databases. Berlin, Springer; 1999.
- 27.Warner HRJ, Miller S, Jennings K, et al. Clinical event management using push technology—implementation and evaluation at two health care centers. Proc AMIA Symp. 1998; 106–10. [PMC free article] [PubMed]
- 28.Hripcsak G, Clayton PD, Jenders RA, et al. Design of a clinical event monitor. Comput Biomed Res. 1996;29(3):194–221. [DOI] [PubMed] [Google Scholar]
- 29.Miller JE, Reichley RM, McNamee LA, et al. Notification of real-time clinical alerts generated by pharmacy expert systems. Proc AMIA Symp. 1999;325–9. [PMC free article] [PubMed]
- 30.Kuperman GJ, Teich JM, Bates DW, et al. Detecting alerts, notifying the physician, and offering action items: A comprehensive alerting system. Proc AMIA Annu Fall Symp. 1996;704–8. [PMC free article] [PubMed]