Abstract
A systematic screen of volatile terpene production in the glandular trichomes of 79 accessions of Solanum habrochaites was conducted and revealed the presence of 21 mono- and sesquiterpenes that exhibit a range of qualitative and quantitative variation. Hierarchical clustering identified distinct terpene phenotypic modules with shared patterns of terpene accumulation across accessions. Several terpene modules could be assigned to previously identified terpene synthase (TPS) activities that included members of the TPS-e/f subfamily that utilize the unusual cis-prenyl diphosphate substrates neryl diphosphate and 2z,6z-farnesyl diphosphate. DNA sequencing and in vitro enzyme activity analysis of TPS-e/f members from S. habrochaites identified three previously unassigned enzyme activities that utilize these cisoid substrates. These produce either the monoterpenes α-pinene and limonene, or the sesquiterpene 7-epizingiberene, with the in vitro analyses that recapitulated the trichome chemistry found in planta. Comparison of the distribution of S. habrochaites accessions with terpene content revealed a strong preference for the presence of particular TPS20 alleles at distinct geographic locations. This study reveals that the unusually high intra-specific variation of volatile terpene synthesis in glandular trichomes of S. habrochaites is due at least in part to evolution at the TPS20 locus.
Keywords: specialized metabolism, glandular trichomes, chemical diversity, chemical ecology, terpene synthase, evolution, Solanum habrochaites
Introduction
Plants synthesize an array of specialized metabolites from diverse chemical classes known or hypothesized to provide selective advantage to the host within its ecological niche. These roles include direct and indirect defense against herbivores and insect pests, attraction of pollinators and seed dispersers, UV protection, and facilitation of beneficial symbioses (Li et al., 1993; Landry et al., 1995; Tewksbury and Nabhan, 2001; Wasson et al., 2006; Degenhardt et al., 2009; Unsicker et al., 2009; Zhang et al., 2009; Kang et al., 2010; Klee, 2010; Klahre et al., 2011; Rodriguez et al., 2011; Weinhold and Baldwin, 2011). Many specialized metabolites are synthesized or accumulate in specific tissues or cell types. For example, glandular trichomes are epidermal appendages that occur on the surface of many plant species, which served as a first line of defense against insects and pathogens through the synthesis and storage of specialized metabolites, which included several used commercially as flavors, fragrances, and pharmaceuticals (Turner et al., 1999; Sirikantaramas et al., 2005; Covello et al., 2007; Nagel et al., 2008; Wang et al., 2008; Xie et al., 2008; Olsson et al., 2009).
Glandular trichomes of the Solanaceae are structurally diverse and synthesize several classes of compounds including terpenoids, acyl sugars, phenylpropanoids, alkaloids and methylketones (Luckwill, 1943; van der Hoeven et al., 2000; Kroumova and Wagner, 2003; Fridman et al., 2005; Slocombe et al., 2008; Schilmiller et al., 2010a; Schmidt et al., 2011). These compounds are implicated in plant defense against insect pests and pathogens, acting directly as toxins or repellents or indirectly through tritrophic interactions (Zhang et al., 2008; Alba et al., 2009; Bleeker et al., 2009, 2011a; Weinhold and Baldwin, 2011). The economic importance of tomato, coupled with the availability of wild relatives that synthesize diverse glandular-trichome-derived metabolites has generated interest in utilizing exotic germplasm to improve insect resistance (Lawson et al., 1997; Hartman and St Clair, 1999; Antonious et al., 2005; Antonious and Snyder, 2006; Alba et al., 2009; Lucatti et al., 2010; Maluf et al., 2010). Solanum habrochaites is a wild tomato species native to Southern Ecuador and Peru, which is characterized by an abundance of glandular trichomes that synthesize diverse insect repellent specialized metabolites, including terpenes (Weston et al., 1989; Besser et al., 2009; Bleeker et al., 2009, 2011a; Sallaud et al., 2009; Sifres et al., 2011).
Terpenes are structurally diverse metabolites that are synthesized from the C5 compounds isopentenyl diphosphate (IPP) and dimethylallyl diphosphate (DMAPP) either from the plastid localized 2-C-methyl-d-erythritol 4-phosphate or the mevalonate pathways (Connolly and Hill, 1991; Chen et al., 2011). There is tremendous plasticity in terpene biosynthesis in higher plants with a TPS enzyme frequently synthesizing multiple products from a single substrate (Tholl et al., 2005; Martin et al., 2010; Tholl and Lee, 2011). Furthermore, as plant genome sequences become available, it is increasingly evident that many TPS genes lie within clusters of tandemly duplicated genes that contain both functional and pseudogenes, a feature often indicative of rapid evolution that lead to phenotypic variation (Martin et al., 2010; Chen et al., 2011; Falara et al., 2011).
The genome of the cultivated tomato S. lycopersicum contains approximately 44 TPS genes of which 29 are predicted to be functional and 18 have documented enzyme activity (Falara et al., 2011). Several of these enzymes contribute to the trichome chemical phenotype (van der Hoeven et al., 2000; van Schie et al., 2007; Schilmiller et al., 2009, 2010a). The majority of TPSs utilize the trans substrates GPP and e,e-FPP to synthesize monoterpenes and sesquiterpenes, respectively (Falara et al., 2011). However, three members of the TPS-e/f subfamily, β-phellandrene synthase 1 from S. lycopersicum (SlPHS1) (TPS20), α-phellandrene synthase from S. pennellii (SpPHS1) and santalene/bergamotene synthase from S. habrochaites (ShSBS) were recently identified. These are chloroplast targeted and utilize the atypical cisoid substrates neryl diphosphate (NPP) and 2z,6z-farnesyl diphosphate (2z,6z-FPP) to synthesize monoterpenes and sesquiterpenes, respectively (Sallaud et al., 2009; Schilmiller et al., 2009; Falara et al., 2011). These three activities are encoded by highly conserved genes that reside at a single locus on chromosome 8, which suggested that they may represent orthologous loci (Sallaud et al., 2009; Schilmiller et al., 2009).
The unique features of terpene synthesis in glandular trichomes of tomato and its wild relatives prompted an analysis of the diversity of these isoprenoid compounds within S. habrochaites. The diversity of trichome-derived volatile mono- and sesquiterpenes in 79 accessions of S. habrochaites is described, which revealed qualitative and quantitative variation in the terpene complement. Hierarchical clustering placed the majority of these terpenes within distinct phenotypic modules whose biosynthesis could be traced to individual TPS enzymes. Much of the observed chemical diversity within S. habrochaites can be attributed to sequence variation within TPS20-related enzymes that utilize NPP and 2z,6z-FPP as substrates. These enzymes synthesize several mono- and sesquiterpenes that included three newly identified enzymes that synthesize 7-epizingiberene, limonene and α-pinene. In addition, terpene profiles within S. habrochaites glandular trichomes are highly correlated with geographic distribution of the accessions.
Results
Solanum habrochaites accessions show high variability in their terpene composition
To explore the terpene diversity in the glandular trichomes of S. habrochaites, leaf dips of 79 accessions that cover the known geographic range of the species (Table S1) were screened by gas chromatography–mass spectrometry (GC–MS) (Schilmiller et al., 2010a). A total of 21 analytical signals were identified that corresponded to six monoterpenes, 13 sesquiterpenes and two unknown compounds (Figure 1 and Table S2). These data reveal that some accessions possess a relatively simple terpene composition dominated by a single major compound e.g. LA2109 (group II, with 7-epizingiberene as a major component) and LA1978 (group IV, with δ-elemene) whereas others exhibit more complex profiles, e.g. LA2975 (group I) and LA2107 (group V).
Figure 1.
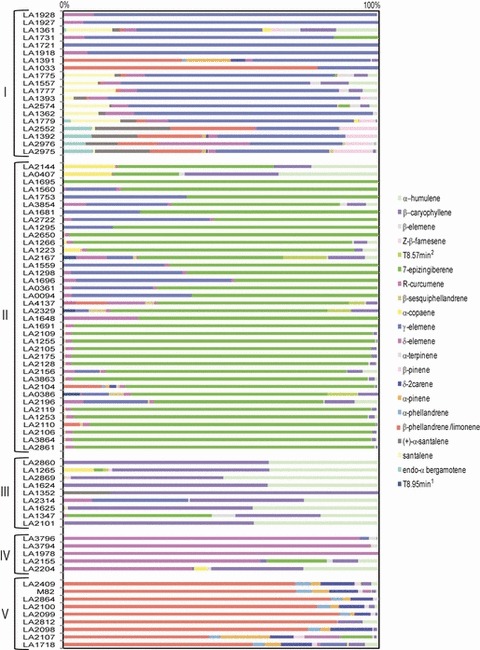
Terpene composition in glandular trichomes of leaves of S. habrochaites. The terpene profile of 79 S. habrochaites accessions and S. lycopersicum cv M82 is shown. Twenty-one analytical signals corresponding to distinct terpenes were identified. The terpene composition was expressed as % of total terpenes. Accessions are grouped according to Figure 2.
Peak area data normalized to the internal standard and leaf dry weight were used for hierarchical clustering (Figure 2). The presence or absence of analytical signals was used to cluster terpenes to identify distinct phenotypic modules composed of terpenes whose accumulation is correlated across accessions. Four of these modules were assigned to previously identified TPS activities in Solanum trichomes. For example, module A includes endo-α-bergamotene and (+)-α-santalene, products of ShSBS previously characterized from S. habrochaitesLA1777 (Sallaud et al., 2009). Module B consists of six monoterpenes, five of which are the products of SlPHS1 from S. lycopersicum (Schilmiller et al., 2009). Module C includes δ-and ε-elemene, the Cope rearrangement products of germacrene C and B, which are synthesized by TPS9 (Colby et al., 1998). Finally, module D includes the products of TPS12, β-caryophyllene and α-humulene (Schilmiller et al., 2010a; Bleeker et al., 2011b; Falara et al., 2011). Collectively, these observations support the hypothesis that terpenes within a module are produced by the same TPS(s).
Figure 2.
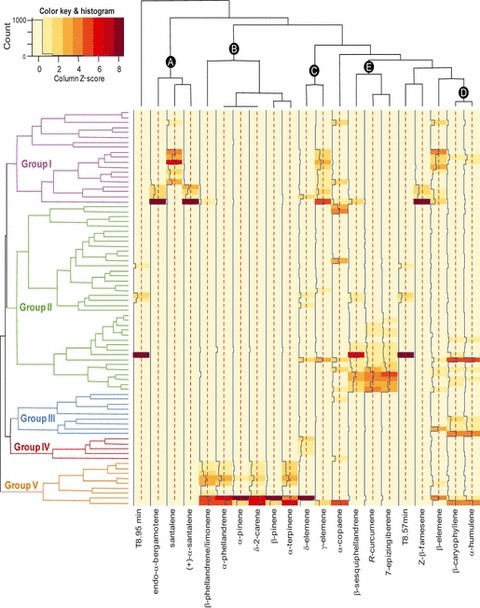
Two-way cluster analysis of normalized peak areas from S. habrochaites accessions. The peak areas normalized to the internal standard and leaf dry weight were calculated for each of the 21 terpene signals in 79 S. habrochaites accessions and M82. The average from two to three replicates was calculated and used for construction of the heatmap. Each row represents the average terpene composition for an accession, and the dendrogram on the left represents the clustering of accessions based on chemical profile. Accessions from the same group (I through V) have similar terpenes. Each column represents the abundance of a particular terpene in all accessions, and the dendrogram representing the clustering of terpenes based on presence/absence in the accessions is shown at the top. Terpenes from the same module (A through E) show similar patterns of accumulation across accessions. Clustering was performed as described in Experimental procedures. The distance of the blue trace line from the center of each cell (red dotted line) is proportional to the abundance of the compound. The peak areas were centered in the column direction. T 8.57 min and T 8.95 min represent unidentified terpenes eluting at 8.57 and 8.95 min, respectively. The order of the accessions in the dendrogram is the same as those presented in Figure 1.
A dendrogram was constructed to cluster accessions according to similarity in terpene content. Five chemical groups (group I through V) that were stable through several iterations of the algorithm were defined, which revealed patterns of terpene distribution across the accessions (Figure 2). Group I, which contains the previously characterized LA1777 (Figures 1 and 2), is defined by high abundance of γ-elemene. Group I could be further divided in two subgroups. Accessions from the first subgroup contain sesquiterpenes synthesized by ShSBS (module A) (e.g. LA1777 and LA1393). The second subgroup also has γ-elemene as the major terpene, but lacks terpenes made by ShSBS (e.g. LA1721). Group II, which constitutes the largest group of 38 accessions is defined by the presence of 7-epizingiberene as the major peak (Bleeker et al., 2011a). The presence of 7-epizingiberene is highly correlated with the presence of R-curcumene and β-sesquiphellandrene, and this chemical phenotype is collectively designated module E. Group V accessions, which also include S. lycopersicum M82, produce a mixture of monoterpenes in which β-phellandrene is the major compound together with smaller amount of limonene, α-pinene, δ-2-carene, α-terpinene and α-phellandrene. β-Caryophyllene and α-humulene are the major compounds identified in nine accessions from group III, whereas δ-elemene is the prominent compound in five accessions from group IV. These compounds are not specific to a single group but are distributed across multiple groups. In contrast, module A is exclusive to group I, module B is mostly present in group V, and module E predominates in group II. Together, results of these analyses simplify the considerable terpene diversity within S. habrochaites trichomes, which suggested common biosynthetic origins that may be reflected in the activity of relatively few TPS activities.
Variation in terpene quantity is associated with terpene composition
A subset of 27 accessions that represent the diversity from the larger dataset were selected and re-grown both to validate the original data derived from 79 accessions and quantify the volatile terpene levels using β-caryophyllene and γ-terpinene standards (see Experimental procedures). The total amount of mono- and sesquiterpenes in this subset of accessions ranged 100-fold from 171 μg g−1 dry weight in LA1265 to 19 200 μg g−1 dry weight in LA2106 (Figure S1). There were differences in terpene quantity between groups (anovaF: 4.753, P-value < 0.01). In addition, a Tukey Honest Significant Differences test revealed that group II means were significantly different from those of group I and III at a P-value < 0.05, due to the high accumulation of 7-epizingiberene in group II accessions. In contrast, the abundance of β-caryophyllene and α-humulene did not vary significantly across the five groups.
Phylogenetic analysis of TPS20-related homologs from S. habrochaites
Three TPS-e/f subfamily enzymes, SlPHS1 (SlTPS20), SpPHS1 and ShSBS, contribute to chemical variation in S. habrochaitesLA1777, S. lycopersicum M82 and S. pennelliiLA0716 trichomes (Sallaud et al., 2009; Schilmiller et al., 2009; Falara et al., 2011). Based on the diverse catalytic activities and the strong level of sequence conservation between these enzymes, we hypothesized that sequence variation within TPS20-related enzymes could contribute to the observed terpene diversity in S. habrochaites. Primers designed from SlPHS1 were used to amplify full-length cDNAs from 23 S. habrochaites accessions that led to the recovery of 34 TPS cDNAs that represented 22 non-redundant sequences. In several accessions distinct cDNAs were recovered that may reflect the presence of separate, closely related loci, but could also be due to heterozygosity which is widespread within S. habrochaites (Rick et al., 1979). The cDNAs encoded predicted proteins of either 777 or 778 amino acids that contain predicted chloroplast targeting sequences together with modified versions of the ‘DDXXD’ (in most of the sequences, the third aspartic acid is replaced by a glutamic acid) and NSE/DTE motifs found in TPSs (Figure S2) (Martin et al., 2010). Phylogenetic studies suggest that all known TPSs are derived from diterpene synthases involved in gibberellin biosynthesis (Chen et al., 2011). While most plant TPSs are shorter than diterpene synthases, the sequences recovered here and others from TPS-e/f have maintained the internal sequence element or ‘γ domain’ (Hillwig et al., 2011).
Phylogenetic analysis of the S. habrochaites amino acid variants and previously characterized enzymes SlPHS1, S. pennellii PHS1 (SpPHS1), and ShSBS revealed separation into two major clusters: M and S (for Monoterpenes and Sesquiterpenes, respectively, see below) (Figure 3 and Figure S3). As expected from the similarity of the chemical profiles, the sequences obtained from group V accessions cluster with SlPHS1 (cluster M in Figure 3). This finding indicates that the S. habrochaites sequences obtained from monoterpene-producing accessions have higher similarity to SlPHS1 than to other S. habrochaites sequences from cluster S. Cluster M is further divided into clade A (more closely related to SlPHS1) and clades B and C. Clade C sequences have an in-frame 3-bp deletion at alanine-538, which is adjacent to the DDXXE motif (Figure S2). This clade includes sequences obtained from groups I and V. However, the accessions that belong to group I, namely LA2976 and LA1391, synthesize monoterpenes such as β-phellandrene and limonene as do other accessions from group V (Figure 1).
Figure 3.
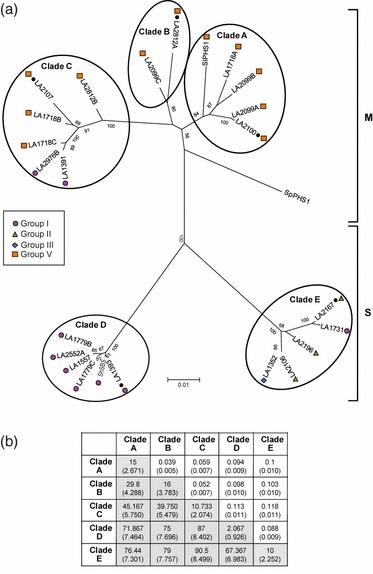
Phylogenetic relationship of TPS20-related sequences from S. habrochaites. (a) An unrooted tree constructed with the ME method using 25 predicted protein sequences, representing a total of 37 sequences from various S. habrochaites accessions (identical sequences at the nucleotide level were eliminated to avoid overcrowding the tree) together with SlPHS1 (FJ797957), SpPHS1 (JN412071), and ShSBS (ACJ38409). Two major groups designated M and S, subdivide into clade A, B and C; and clade D and E, respectively. Bootstrap values above 50 are shown. Colored symbols indicate the chemical group (defined in Figure 2) from which each sequence was isolated. Sequences chosen for codon optimization and expression in E. coli are highlighted (•). (b) Estimates of average evolutionary divergence. Shaded, the number of amino acid differences per sequence from averaging over all sequence pairs within and between each group is shown. White, the p-distance (1-amino acid identity) from averaging over all sequence pairs between groups is shown. Standard error estimates are shown in parentheses. The analysis involved 24 amino acid sequences, corresponding to the five clades shown in (a). Positions containing gaps and missing data were eliminated. Analyses were conducted in MEGA5.
Cluster S is composed of sequences that are more similar to ShSBS from LA1777, and are subdivided into clades D and E. Clade D consists of accessions that belong exclusively to group I. Notably, this clade displays the highest level of sequence conservation among members, with a mean amino acid difference between sequence pairs of two, compared with 15, 16, 11 and 10 in clades A, B, C and E, respectively (Figure 3b). Clade E includes accessions from groups I, III, and IV, but predominantly from II.
A genomic rearrangement contributes to TPS-e/f sequence divergence
The sequence divergence within the S. habrochaites TPS-e/f family raises questions about the evolutionary origins of this diversity. To investigate this situation further, a codon alignment was performed that included the sequences of three additional TPS-e/f subfamily members identified within the S. lycopersicum chromosome 8 cluster; TPS18, 19 and 21 (Figure 4) (Falara et al., 2011). These were included because TPS20 (SlPHS1) is 82, 98.5 and 93% identical at the nucleotide level to TPS18, 19 and 21 respectively, which suggested that they arose from a common ancestor by tandem duplication and functional divergence (Falara et al., 2011). Inspection of the alignment reveals that all sequences from group S contain a region of 200 bp (boxed region, Figure 4), which corresponded to exon 4 in SlPHS1, which is more similar to TPS18 than to other sequences from group M. To illustrate this result, phylogenetic analysis of the codon alignments was performed following separation of the coding sequences into three regions, exons 1–3, exon 4, and exons 5–13 (Figure 5a–c). The topology of the tree constructed with exon 4 varied considerably compared with the trees derived from the other regions. Within the exon 4 region, the sequences from group S cluster together with TPS18 from S. lycopersicum, whereas in the other regions, TPS18 forms the out group for all sequences. In exon 4 the average identity of group S to TPS18 is 98% at the nucleotide level, compared with 90 and 79% in the other two regions (Figure 5d). In contrast, S. habrochaites sequences from group M are 84% identical to TPS18 in exon 4, and 89 and 80% identical in the other two regions (Figure 5d). These observations suggest that a form of genomic rearrangement has occurred within exon 4 of S. habrochaites accessions from group S, which introduced a sequence more similar to TPS18 that contributes to sequence divergence within the TPS-e/f subfamily.
Figure 4.
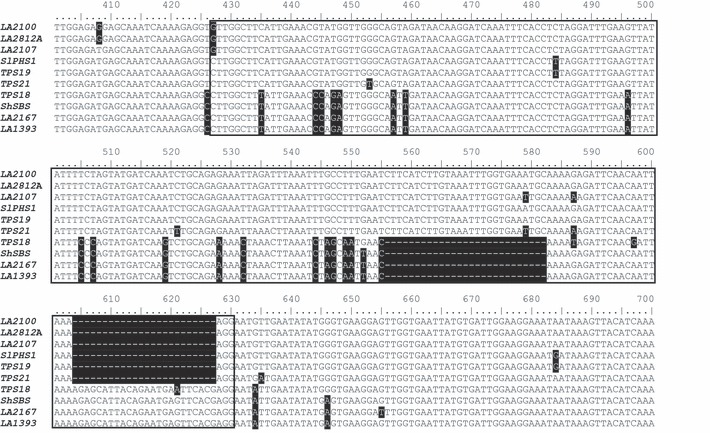
Evidence for a genomic rearrangement within exon 4. (a) Comparison of the nucleotide sequence of the TPS-e/f family members LA2100, LA2812A, LA2107, LA2167, LA1393, and ShSBS, together with TPS18 (JN412088), 19 (JN412072), SlPHS1 (FJ797957) and 21 (JN412087) from S. lycopersicum. Non-identical residues are highlighted in black. Exon 4 is denoted by a box.
Figure 5.
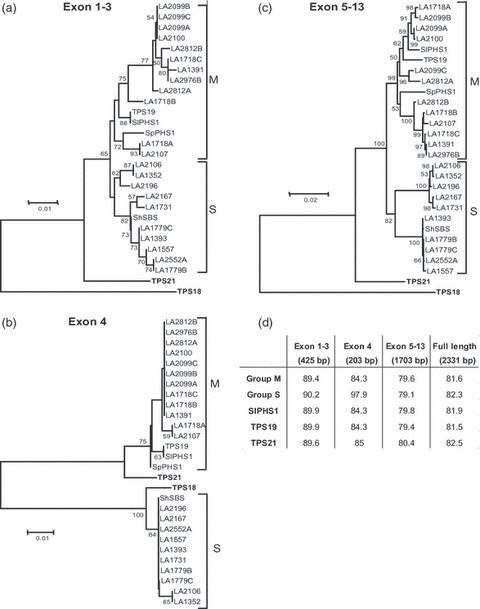
Sequence exchange between TPS20 and TPS18 contributes to TPS diversity. (a–c) Phylogenetic analysis of TPS-e/f nucleotide sequences. A codon alignment performed with MUSCLE was used to reconstruct the phylogeny of the TPS-e/f subfamily with sequences from S. habrochaites together with TPS18 (JN412088), TPS19 (JN412072), TPS21 (JN412087), SlPHS1 (FJ797957), SpPHS1 (JN412071), and ShSBS (FJ194970). The alignment was divided into three segments corresponding to exons 1 through 3, exon 4, and exons 5 through 13 before tree reconstruction with an ME model in MEGA5. (d) Percent nucleotide identity of TPS-e/f sequences compared to TPS18 from S. lycopersicum. A codon alignment was divided into three regions as above. Nucleotide identity between TPS18 from S. lycopersicum and each S. habrochaites sequence was calculated, and the average from each group obtained. For comparison, the nucleotide identity of SlPHS1, TPS19 and TPS21 is shown.
Plasticity in the activity of TPS20-related enzymes from S. habrochaites
SlPHS1 and ShSBS share 91% amino acid sequence identity, yet they use the structurally distinct NPP and 2z,6z-FPP for the synthesis of mono- and sesquiterpenes, respectively (Sallaud et al., 2009; Schilmiller et al., 2009). To explore the hypothesis that sequence diversification has resulted in functional divergence at TPS20 within S. habrochaites, codon optimized versions (Ikemura, 1981) of representatives from each of the five major clades in the phylogenetic tree were expressed in E. coli.
The recombinant proteins were tested with each of the potential substrates: geranyl diphosphate (C10, GPP), e,e-farnesyl diphosphate (C15, e,e-FPP), and the cisoid substrates neryl diphosphate (C10, NPP) and 2z,6z-farnesyl diphosphate (C15, 2z,6z-FPP). The predicted proteins from group M utilized NPP to synthesize a variety of monoterpenes. As predicted based on sequence similarity, the recombinant enzyme from clade A accession LA2100 produced a monoterpene profile indistinguishable from SlPHS1 (Schilmiller et al., 2009), with β-phellandrene as the major product, along with limonene, α-pinene, δ-2-carene, α-phellandrene and γ-terpinene (Figure 6a). The products of this enzyme (designated ShPHS1) with NPP substrate match the terpene profile from trichome extracts of accession LA2100 (Figure S4a). In contrast, the enzyme LA2812A from clade B (one of the sequences recovered from accession LA2812) converted NPP to limonene, which again matches the profile observed in extracts of LA2812 trichomes and is therefore designated ShLMS (Figure 6b, Figures S4b and S5). The enzyme from clade C accession LA2107 converted NPP predominantly to α-pinene together with lower amounts of β-pinene and β-phellandrene and/or limonene (Figure 6c), and therefore is named ShPIS. Trichomes from LA2107 also accumulate the compounds produced by ShPHS1; however the presence of high β-pinene levels in the trichomes can only be explained by the activity of ShPIS (Figure S4c). In general, enzymes from the sesquiterpene-rich clades D and E (from accessions LA1393 and LA2167, respectively) did not produce detectable amounts of monoterpenes from GPP or NPP except for very low amounts of limonene and α-terpineol in assays that contained NPP (Figure 6d, e). However, these compounds were also observed in reactions that contained the empty vector and NPP and, in the case of limonene, are at least two orders of magnitude lower than in reactions that contained ShLMS (Figure 6b,f). Furthermore, α-terpineol was not detected in trichome samples (Figure 1 and Figure S4). Therefore, it is likely that the formation of limonene and α-terpineol can occur either enzymatically or non-enzymatically in the crude E. coli extracts used to perform the enzyme assays.
Figure 6.
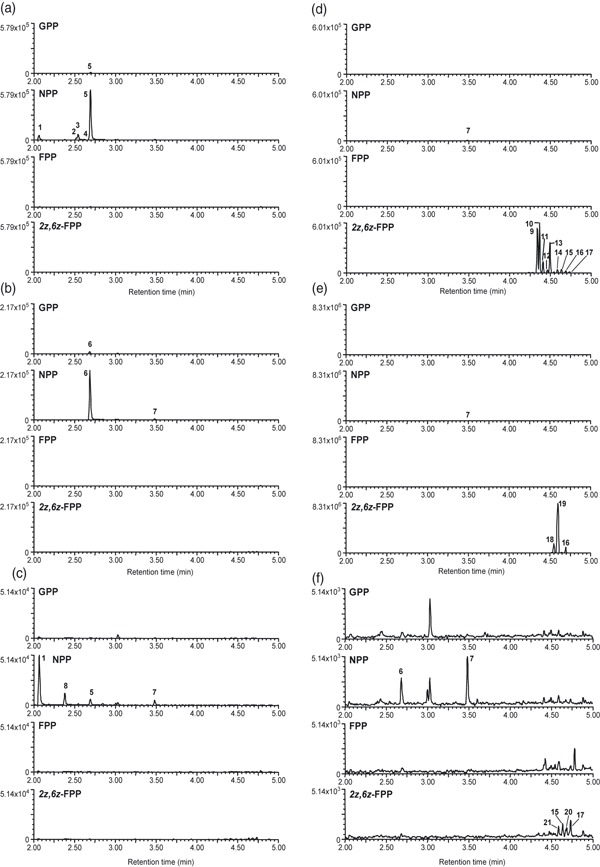
Headspace collection of volatiles produced by recombinant TPS from LA2100 (ShPHS1) (a), LA2812A (ShLMS) (b), LA2107 (ShPIS) (c), LA1393 (ShSBS) (d) and LA2167 (ShZIS) (e) expressed in E. coli and assayed with GPP, NPP, FPP or 2z,6z-FPP as substrates. Reactions with the empty vector are included as a control (f). Extracted ion chromatograms for m/z 93 are shown. 1, α-pinene; 2, δ-2-carene; 3, α-phellandrene; 4, α-terpinene; 5, limonene and β-phellandrene; 6, limonene; 7, α-terpineol; 8, β-pinene; 9, endo-α-bergamotene; 10, (+)-α-santalene; 11, (−)-exo-α-bergamotene; 12, (−)epi-β-santalene; 13, (+)-endo-β-bergamotene; 14, (Z)-β-farnesene; 15, β-bisabolene; 16, β-sesquiphellandrene.; 17, unknown; 18, R-curcumene; 19, 7-epizingiberene; 20, unknown; 21, unknown. A series of chromatograms with improved resolution of low abundance peaks obtained using 2z,6z-FPP as a substrate is provided in Figure S6. Note that peaks 15, 17, 20 and 21 appear to be non-specific products.
The enzyme from clade D produced a sesquiterpene profile identical to that made by the closely related LA1777 ShSBS when supplied with 2z,6z-FPP as substrate (Figure S4d). In contrast, the enzyme from clade E utilized 2z,6z-FPP to synthesize 7-epizingiberene, R-curcumene and β-sesquiphellandrene, the module E terpenes (Figure 2 and Figure S4e). The enzyme therefore is designated ShZIS.
In conclusion, the functional characterization of five different alleles of TPS20 from S. habrochaites revealed five distinct enzyme activities. Enzymes that belong to group M utilize NPP for the production of monoterpenes, whereas enzymes from group S utilize 2z,6z-FPP to synthesize sesquiterpenes. Collectively, these enzymes synthesize many of the major terpenes identified in the trichomes of S. habrochaites.
A geographical-climatic context for terpene diversity
The S. habrochaites accessions used in this study were collected in Peru and Ecuador from 1948 to 1997, and the geographical coordinates of most of the collection sites are available (http://tgrc.ucdavis.edu/). This information was used to examine the terpene variation of S. habrochaites throughout its range of distribution. The accessions were classified according to the five chemical groups described in Figure 2 and mapped using their geographical coordinates (Figure 7a,c). Additionally, annual mean temperature, annual precipitation and elevation were calculated for each group (Figure 7d–f).
Figure 7.
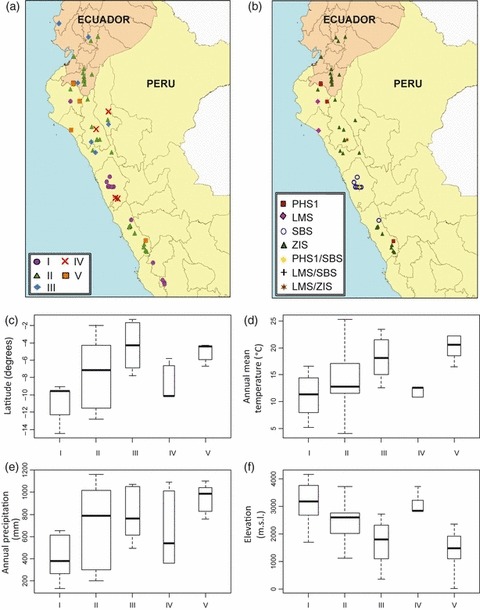
Geographical distribution of S. habrochaites accessions and climatic data. (a, b) Map of Ecuador and Peru showing the collection coordinates of S. habrochaites accessions. (a) Accessions were classified according to the chemical groups determined by hierarchical clustering (Figure 2). (b) Accessions were classified based TPS-e/f derived terpenes. (c–f) Box plots displaying differences between chemical groups in climate data of collection sites. The box plots show lower and upper quartiles, the extreme values that are not outliers and the median. Outliers have been omitted. Boxes are drawn with widths proportional to the square-roots of the number of observations in the group. Maps and climate data were obtained in DIVA-GIS. Box plots were constructed using R.
Co-occurrence of the terpene groups with the geographical distribution of the accessions is observed. Group I accessions were preferentially located in the higher altitude southern range of the distribution, with lower mean temperatures and annual precipitation, whereas groups III and V were located further north, in regions that are at lower elevation, and warmer with a more humid climate. In contrast, group II accessions are present over a wide area of the distribution.
A similar analysis was conducted to classify the accessions based on whether their chemical profile contains the predominant terpenes made by members of the TPS20-related enzymes, namely β-phellandrene, α-pinene and δ-carene for ShPHS1, limonene for ShLMS, santalene/bergamotene for ShSBS, or 7-epizingiberene for ShZIS (Figure 7b and Table S1). The production of santalene and bergamotene is confined to Ancash and Lima in Peru (Figure 7b and Table S1), whereas accessions from group I that lack the products of ShSBS are preferentially located in the southernmost area of the distribution. The narrow distribution of these terpenes is consistent with the low evolutionary divergence found in the sequences recovered from clade D that encode ShSBS enzymes (Figure 3b). A different situation is observed for the β-phellandrene/limonene-producing accessions, which were mainly collected in the border between Ecuador and Peru, yet clade A possessed large sequence divergence (Figure 3b). Meanwhile, the production of 7-epizingiberene is widely distributed and alleles from clade E showed intermediate levels of sequence divergence. Notably, the distribution of accessions that synthesize 7-epizingiberene is not continuous as they are not located in the area where santalene-/bergamotene-producing accessions predominate. Furthermore, no sequence features were detected within clade E that distinguishes the accessions from the southern region of the distribution from those in the northern region that are separated by the santalene- and bergamotene-producing accessions (data not shown).
Discussion
Solanum habrochaites accessions differ in the type and quantity of trichome derived terpenes
The volatile terpene profiles in the trichomes of 79 accessions of S. habrochaites are qualitatively and quantitatively diverse (Figure 1 and Figure S1). Most accessions produce predominantly sesquiterpenes, which included 7-epizingiberene, R-curcumene, endo-α-bergamotene and (+)-α-santalene, which are not present in cultivated S. lycopersicum M82 or within 53 additional S. lycopersicum accessions that we screened (unpublished data). However, a small group of accessions, mainly in group V, resemble cultivated tomato in that they synthesize predominantly monoterpenes with a profile dominated by β-phellandrene (Figure 1 and Figure S1b) (Schilmiller et al., 2009). Moreover, three S. habrochaites TPSs (ShPHS1, ShPIS and ShLMS) convert NPP to monoterpenes; each produces an in vitro terpene profile that matches trichome extracts (Figures 6, 8 and Figure S4). Together, these data suggest the existence of a prenyltransferase in select accessions of S. habrochaites that is similar to the NPP-producing NDPS1 from S. lycopersicum (Schilmiller et al., 2009).
Figure 8.
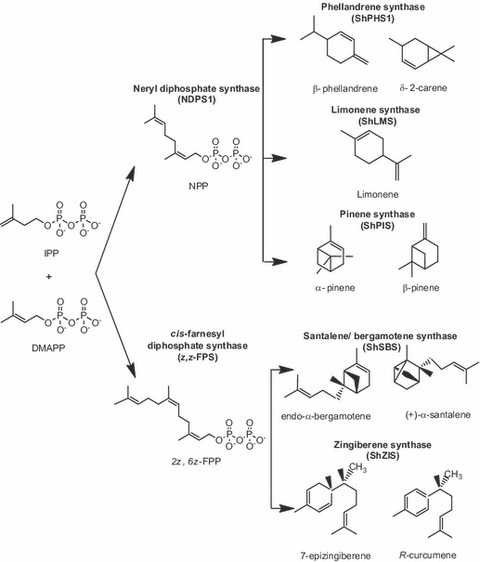
Terpene biosynthesis in S. habrochaites trichomes using the cisoid substrates NPP and 2z,6z-FPP.
Hierarchical clustering analysis reveals close association between specific terpenes in different S. habrochaites accessions, leading to the hypothesis that these modules are produced by specific TPSs. In fact, four terpene modules (A–D) correspond to products made by previously characterized Solanum TPSs (Colby et al., 1998; Sallaud et al., 2009; Schilmiller et al., 2009, 2010a; Bleeker et al., 2011b; Falara et al., 2011), and a fifth, characterized by the presence of 7-epizingiberene, are products of ShZIS (Figures 2 and 6). Quantitative variation in terpene levels also correlates with qualitative composition (Figure S1). In general, 7-epizingiberene-producing accessions in group II accumulate higher levels of volatile terpenes than the other groups. In contrast, the products of module A, santalene, (+)-α-santalene and endo-α-bergamotene, which are synthesized by ShSBS, represent minor compounds in group I accessions (Figure S1). This difference in abundance presumably is caused by further metabolism of the ShSBS products to their corresponding acids (Frelichowski and Juvik, 2001, 2005; Besser et al., 2009).
The central role of TPS20-related enzymes in trichome chemical diversity
Previous studies highlighted the importance of TPS20-related enzymes in terpene biosynthesis in the glandular trichomes of tomato and related wild species (Sallaud et al., 2009; Schilmiller et al., 2009; Falara et al., 2011). Furthermore, analysis of nearly isogenic lines that contain genomic regions from LA1777 in a S. lycopersicum background demonstrated that santalene/bergamotene and monoterpene accumulation are controlled by a single locus or tightly linked loci and subsequent identification of SlPHS1 and ShSBS showed that these genes lie within orthologous regions on chromosome 8 (van der Hoeven et al., 2000; Besser et al., 2009; Sallaud et al., 2009; Schilmiller et al., 2009; Falara et al., 2011). These data led us to hypothesize that additional TPS20-related enzymes contribute to the terpene diversity encountered in S. habrochaites trichomes. Homology-based cloning, coupled with phylogenetic analysis and enzyme assays with recombinant proteins, support the hypothesis. Heterologous expression in E. coli revealed that sequences obtained from LA2100 (ShPHS1), LA2812 (ShLMS) and LA2107 (ShPIS) encode monoterpene synthases that convert the C10 substrate NPP to a blend of terpenes that resembles the profiles observed in planta (Figure 8 and Figure S4). Similarly, the TPS20-related enzymes from accessions LA1393 (ShSBS) and LA2167 (ShZIS) synthesize mixtures of sesquiterpenes from 2z,6z-FPP (Figure 8).
The location of genes involved in plant specialized metabolism, which included terpene biosynthesis, within gene clusters is an emerging paradigm and creates the potential for tandem duplications, deletions and rearrangements, possibly generating phenotypic variation (Martin et al., 2010; Chu et al., 2011; Falara et al., 2011). SlPHS1 (TPS20) is part of a gene cluster that contained three additional TPS-e/f subfamily members of unknown function, SlTPS18, SlTPS19 and SlTPS21 (Falara et al., 2011). SlPHS1 (TPS20) is especially closely related to SlTPS19 and these genes likely arose through recent gene duplication (Falara et al., 2011). The TPS20-related sequences identified in this study are more closely related to SlPHS1 and SlTPS19 than to SlTPS18 and SlTPS21 (Figures 4 and 5). However, SlPHS1 and SlTPS19 are more similar to each other than they are to any of the S. habrochaites TPS20-related sequences. Therefore, these relationships render assessment of orthology and paralogy inconclusive at the current level of resolution. Resolution of orthology and paralogy will require analysis of the relative placement of these genes in chemically diverse S. habrochaites accessions to resolve potential tandem gene duplications or additional rearrangements within the gene cluster. Indeed, evidence of a gene rearrangement event was detected that led to incorporation of a sequence derived from SlTPS18 into the clade S genes that encode ShSBS and ShZIS (Figures 4 and 5). The nature of this rearrangement is unknown but may have arisen through a gene conversion event or an alteration in splicing and lead to a change in the exon/intron boundary position (Innan and Kondrashov, 2010). Distinguishing these possibilities will require sequencing of the corresponding genomic loci.
Some of the observed chemical variation in S. habrochaites may be caused by heterozygosity within accessions that result from self-incompatibility and subsequent cross-pollination (Rick et al., 1979). In addition, many of the accessions were collected from more than a single individual, which increased the likelihood that multiple alleles are present within a given seed lot (http://tgrc.ucdavis.edu/). For example, we commonly found plants that produce terpenes characteristic of two TPS20-related enzymes (Table S1 and Figure S7), which suggested either the expression of paralogous genes or heterozygosity within the terpene gene cluster on the top of chromosome 8. Consistent with these diverse chemical phenotypes, more than one TPS20-related sequence was recovered from the same accession on several occasions (Figure 3, accession numbers followed by letters). In addition, many of the accessions were collected from more than a single individual, which increased the likelihood that multiple alleles are present within a given seed lot (http://tgrc.ucdavis.edu/). This situation was observed through the recovery of three TPS20-related alleles from LA1718, LA1779 and LA2099 (designated A through C) (Figure 3 and Figure S3). While the alleles recovered from LA1718 and LA2099 may encode separate monoterpene synthases, the high sequence similarity of the three alleles from LA1779 suggests that all encode a functional SBS (Figure 3 and Figure S3). Furthermore, it is documented (http://tgrc.ucdavis.edu/) that the sample size for this accession was two individuals. Therefore, if three alleles were present in these two individuals, it is possible that they would have been maintained in subsequent seed lots. However, while it is clear that considerable heterogeneity is present within S. habrochaites, it is noteworthy that no accessions were identified that synthesize the products of both ShSBS and ShZIS. The significance of this result remains unclear but may be due to restricted geographic distribution of the ShSBS that contained accessions leading to reproductive isolation (Figure 7b).
Some accessions of S. habrochaites lack terpenes derived from cisoid isoprenoid diphosphate substrates
The glandular trichomes of Solanum spp. possess the ability to utilize both the cytosolic mevalonate and plastidic 2-C-methyl-d-erythritol 4-phosphate pathways to synthesize precursors for terpene biosynthesis (Colby et al., 1998; van der Hoeven et al., 2000; Sallaud et al., 2009; Schilmiller et al., 2009, 2010b). In addition, the use of the plastid localized 2-C-methyl-d-erythritol 4-phosphate pathway to synthesize the cisoid substrates NPP and 2z,6z-FPP has thus far only been reported in Solanum trichomes (Figure 6 and Figure S4) (Sallaud et al., 2009; Schilmiller et al., 2009; Falara et al., 2011). However, most S. habrochaites accessions from groups III and IV are exceptions in that they do not synthesize terpenes from cisoid substrates. In contrast, they accumulate β-caryophyllene/α-humulene and γ-elemene/δ-elemene, the products of SlTPS12 and SlTPS9, respectively that are synthesized in the cytosol from trans e,e-FPP (Figure 1) (Colby et al., 1998; van der Hoeven et al., 2000; Schilmiller et al., 2010b; Falara et al., 2011). Similarly, group I can be divided into a subgroup that contains the sesquiterpene products of ShSBS and a subgroup that does not (Figure 2). Furthermore, accessions from groups III and IV generally accumulate lower levels of volatile terpenes than groups that utilize the cisoid substrates for terpene biosynthesis (Figure S1). Sequestering a proportion of volatile terpene biosynthesis in the chloroplast and utilizing a separate substrate pool creates the opportunity to increase terpene synthesis and generate variation in the terpene profile of glandular trichomes.
Several possibilities exist that could explain the lack of cis substrate-derived terpenes in accessions from groups I, III, and IV. For example, TPS20-related genes may not be expressed in trichomes, are absent, or enzymatically non-functional. Evidence that supported this result derives from failed attempts to clone TPS20-related cDNAs from several of these accessions (e.g. LA2101). Second, the substrate for TPS may be unavailable due to lack of expression or mutation in a cis-prenyltransferase. This finding is supported by the observation that several accessions that do not accumulate 7-epizingiberene (LA2860, LA1625 and LA3794) contain an apparently functional enzyme that is identical to that recovered from 7-epizingiberene-producing accessions (LA2104 and LA2106). Additional characterization of the chromosome 8 TPS cluster from select group I, III and IV accessions will provide insight into these phenomena.
The evolution of monoterpene and sesquiterpene biosynthesis is driven by variation in TPS20-related genes in S. habrochaites
The data presented suggest that the diversity observed in terpene production derived from TPS20-related enzymes in S. habrochaites trichomes can be explained by a small number of molecular events. In this model, before the split of S. lycopersicum and S. habrochaites, a member of the TPS-e/f subfamily, likely a diterpene synthase, acquired the ability to use NPP to synthesize monoterpenes. This finding is consistent with the finding that S. pennellii trichomes also utilize NPP to produce monoterpenes (Falara et al., 2011). Additionally, the large sequence divergence within cluster M supports the hypothesis that a monoterpene synthase is the ancestral form of the enzyme (Figure 3). Moreover, monoterpene-producing accessions are preferentially located in the border region between Peru and Ecuador, a hotspot for Solanaceae diversity and the proposed origin of S. habrochaites (Sifres et al., 2011). Subsequently, the appearance of a TPS that could use 2z,6z-FPP instead of NPP could have arisen by introduction of the TPS18 exon four region through gene conversion or other genomic rearrangement (Figures 4 and 5). Although the impact of the exon 4 region from TPS18 on enzymatic activity has not been directly demonstrated, its association with sesquiterpene-producing enzymes is consistent with the hypothesis that it has a role in changing the substrate specificity, allowing the enzyme to accept 2z,6z-FPP as a substrate. This rearrangement occurred prior to the split of ShZIS and ShSBS. Furthermore, it is likely that ShZIS originated before ShSBS as the former has higher evolutionary divergence and a wider geographic distribution (Figures 3 and 7b). It is also possible that the ability to synthesize santalene and bergamotene provides a strong fitness advantage, for example tolerance of insect herbivores or pathogens, which created strong positive selection for ShSBS in central Peru.
Field experiments are required to test the adaptive value of each TPS20-related gene under its native environment. This approach would help establish the biotic factors that influence chemical variation within S. habrochaites. Similarly, the identification of ShZIS provides an opportunity to breed or engineer the biosynthesis of 7-epizingiberene, a known insect repellent (Bleeker et al., 2011a), in trichomes or other tissues of S. lycopersicum and additional crop plants for improved arthropod resistance.
Experimental procedures
Plant material and growth conditions
Seeds from 79 accessions of Solanum habrochaites and Solanum lycopersicum cv. M82 (LA3475) were obtained from the C.M. Rick Tomato Genetics Resource Center (TGRC) (http://tgrc.ucdavis.edu/) (Table S1). Seeds were germinated on filter paper in the dark. After germination, two to three plants per accession were grown on Jiffy-7 Peat Pellets (http://www.hummert.com/) for 3 weeks as previously described (Schilmiller et al., 2009). Plants were transplanted into peat-based compost supplemented with fertilizer in greenhouses equipped with environmental controls and supplemental lighting at Michigan State University, East Lansing, MI, USA.
Metabolite extraction and terpene analysis
Three-week-old plants were used for terpene analysis. Briefly, a leaflet from the second newly emerging leaf was dipped in 1 ml of methyl tert-butyl ether that contained 5 ng μl−1 of tetradecane as an internal standard, and allowed to rock for 1 min. GC–MS analysis was performed as previously described (Schilmiller et al., 2009). The data were exported into MassLynx V4.1 and quantification performed with QuanLynx (http://www.waters.com/). The extracted ion chromatogram was obtained at m/z 93, which is common to all mono- and sesquiterpenes. Peaks with an area >5 in at least one accession were quantified. This criterion narrowed the list of peaks identified; however, minor peaks did not have a reliable mass spectrum for peak identification, nor comparison across multiple samples. The resulting relative abundance for each terpene in a sample was normalized to the internal standard and leaflet dry weight. Terpene identification was based on comparison of mass spectra and retention times of authentic standards when available and with mass spectra in an essential oil library (Adams, 2009). However, in the majority of cases authentic standards were not available and it was therefore not possible to determine the stereochemistry of the identified compounds. Bergamotene, santalene, zingiberene and curcumene stereoisomers were assigned based on previous reports (Sallaud et al., 2009; Bleeker et al., 2011a). Compounds were quantified based on their abundance relative to the internal standard tetradecane and standard curves with β-caryophyllene and γ-terpinene. The average for the two to three replicates was calculated and used for hierarchical clustering applying the Bray–Curtis index, and the distance between samples determined with average linkage with the vegan package for R (Oksanen et al., 2010).
Trichome isolation and gene cloning
Stems and petioles from two to three fully grown plants from the same accession were pooled. Trichomes were collected by fast-freezing the tissue and scraping the frozen trichomes in liquid nitrogen. RNA was extracted using the RNeasy Plant Mini Kit (http://www.qiagen.com/) and cDNA prepared with the Transcriptor First Strand cDNA Synthesis Kit (http://www.roche-applied-science.com) using oligo(dT) as a primer. Amplification of full-length TPS-e/f cDNAs was performed using KOD DNA polymerase (http://www.emdchemicals.com/life-science-research) with conserved primers designed to previously published sequences (Sallaud et al., 2009; Schilmiller et al., 2009) (Table S3). Polymerase chain reaction (PCR) fragments were purified using the Wizard® SV Gel and PCR Clean-Up System (http://www.promega.com/) and cloned into the pCR®4Blunt-TOPO® vector (http://www.invitrogen.com/). Recombinant clones were verified by colony PCR and sequence analysis. Nucleotide sequences were deposited in Genbank under the following accession numbers JN990661–JN990694.
DNA sequence analysis, multiple sequence alignments and phylogenetic analyses
DNA sequences were edited and assembled using Sequencher™ software version 4.8 (http://genecodes.com/). Sequence analysis was performed with MEGA version 5 (Tamura et al., 2011) using MUSCLE to construct the alignments and the Minimum Evolution (ME) method for phylogenetic analysis. A bootstrap test was used to assess the reliability of the tree. Percent identity between nucleotide and predicted amino acid sequences of TPS enzymes was calculated using the alignment made by MUSCLE to calculate distance.
Synthesis of codon optimized genes, recombinant protein expression and activity assays
Codon optimized versions of TPS-e/f enzymes that lacked the chloroplast targeting sequence but containing BamHI and SalI restriction enzyme recognition sequences at the 5′- and 3′-ends respectively, were synthesized by Genscript Corporation (http://www.genscript.com/). The synthetic gene was excised from pUC57 by digestion with BamHI and SalI and ligated into the pHIS8 vector previously linearized with the same enzymes. Recombinant clones were transformed into E. coli BL21 cells. A 5-ml log-phase culture of E. coli BL21 cells containing the expression vector was induced by addition of 0.1 mm IPTG. The induced cells were incubated with agitation at 25°C for 16 h, and then harvested by centrifugation. The cell pellet was resuspended in 1 ml extraction buffer (50 mm HEPES pH 8, 5% glycerol, 100 mm KCl, 7.5 mm MgCl2 containing 1 mm dithiothreitol) prior to sonication. Soluble proteins were harvested after centrifugation at 10 000 g for 20 min and the supernatant utilized for enzyme assays.
For the identification of reaction products, 5 μg of soluble protein were mixed with 10 μm of substrate, either GPP, NPP, e,e-FPP or 2z,6z-FPP (http://www.echelon-inc.com/), in a 2-ml glass vial. Reactions were incubated for 30 min at 30°C, and the headspace was collected for 5 min with a 65-μm polydimethylsiloxane-divinylbenzene solid-phase microextraction fiber (http://www.sigmaaldrich.com/analytical-chromatography.html). For comparison of enzyme products with trichome terpene profiles, stem trichomes were collected from greenhouse-grown plants and an aliquot extracted with hexane containing 1 ng μl−1 of tetradecane as internal standard. Enzyme assays containing 40 μm of substrate and 50 μg of soluble protein in 100 μl were overlaid with two volumes of hexane. After 1 h at 30°C, the vials were vigorously mixed and centrifuged to separate the organic layer. The reactions were frozen at −80°C, and the hexane layer transferred to a new vial. GC–MS analysis was performed as described previously (Schilmiller et al., 2010a).
Analysis of geographical distribution and climate data
When available, latitude and longitude information for the original collection site of S. habrochaites accessions was obtained from the TGRC (Table S1). Locations were mapped using DIVA-GIS 7.5.0.0 software (http://www.diva-gis.org/). Climate and elevation data were obtained from WordClim v1.3 at 5 min resolution (Hijmans et al., 2005). Correlation with volatile terpene profiles was performed using R.
Acknowledgments
This research was supported by NSF DBI-0604336 and IOS-1025636 and a Strategic Partnership Grant from the Michigan State University Foundation. D.E.H. was supported by the Plant Genomics@MSU internship program (http://www.plantgenomics.msu.edu). We thank Amanda Charbonneau for assistance with sample collection, Dr Joe Noel (Salk Institute) for provision of the pHIS8 expression vector, and Dr Sungbeom Lee (MSU) for helpful discussions.
SUPPORTING INFORMATION
Additional Supporting Information may be found in the on-line version of this article:
Figure S1. Terpene abundance and composition in a subset of S. habrochaites accessions.
Figure S2. Alignment of representative TPS-e/f proteins.
Figure S3. Phylogeny of TPS20-related proteins identified in 23 S. habrochaites accessions.
Figure S4. Comparison of recombinant TPS-e/f activities and trichome terpene profiles.
Figure S5. Characterization of major terpenes synthesized by ShPHS1 and ShLMS.
Figure S6. Characterization of minor peaks identified in TPS enzyme activity assays.
Figure S7. Heterogeneity in accession LA2104.
Table S1. S. habrochaites accessions, their geographic distribution and associated climate data.
Table S2. Mono- and sesquiterpenes terpenes identified in S. habrochaites trichomes.
Table S3. Primers used in this study.
Please note: As a service to our authors and readers, this journal provides supporting information supplied by the authors. Such materials are peer-reviewed and may be re-organized for online delivery, but are not copy-edited or typeset. Technical support issues arising from supporting information (other than missing files) should be addressed to the authors.
References
- Adams RP. Identification of Essential Oil Components by Gas Chromatography/Mass Spectrometry. 4th edn. Carik Stream: Allured books; 2009. [Google Scholar]
- Alba JM, Montserrat M, Fernandez-Munoz R. Resistance to the two-spotted spider mite (Tetranychus urticae) by acylsucroses of wild tomato (Solanum pimpinellifolium) trichomes studied in a recombinant inbred line population. Exp. Appl. Acarol. 2009;47:35–47. doi: 10.1007/s10493-008-9192-4. [DOI] [PubMed] [Google Scholar]
- Antonious GF, Snyder JC. Natural products: repellency and toxicity of wild tomato leaf extracts to the two-spotted spider mite, Tetranychus urticae Koch. J. Environ. Sci. Health B. 2006;41:43–55. doi: 10.1080/03601230500234893. [DOI] [PubMed] [Google Scholar]
- Antonious GF, Kochhar TS, Simmons AM. Natural products: seasonal variation in trichome counts and contents in Lycopersicum hirsutum f. glabratum. J. Environ. Sci. Health B. 2005;40:619–631. doi: 10.1081/PFC-200061567. [DOI] [PubMed] [Google Scholar]
- Besser K, Harper A, Welsby N, Schauvinhold I, Slocombe S, Li Y, Dixon RA, Broun P. Divergent regulation of terpenoid metabolism in the trichomes of wild and cultivated tomato species. Plant Physiol. 2009;149:499–514. doi: 10.1104/pp.108.126276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bleeker PM, Diergaarde PJ, Ament K, Guerra J, Weidner M, Schutz S, de Both MTJ, Haring MA, Schuurink RC. The role of specific tomato volatiles in tomato-whitefly interaction. Plant Physiol. 2009;151:925–935. doi: 10.1104/pp.109.142661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bleeker PM, Diergaarde PJ, Ament K, et al. Tomato-produced 7-epizingiberene and R-curcumene act as repellents to whiteflies. Phytochemistry. 2011a;72:68–73. doi: 10.1016/j.phytochem.2010.10.014. [DOI] [PubMed] [Google Scholar]
- Bleeker PM, Spyropoulou EA, Diergaarde PJ, et al. RNA-seq discovery, functional characterization, and comparison of sesquiterpene synthases from Solanum lycopersicum and Solanum habrochaites trichomes. Plant Mol. Biol. 2011b;77:323–336. doi: 10.1007/s11103-011-9813-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen F, Tholl D, Bohlmann J, Pichersky E. The family of terpene synthases in plants: a mid-size family of genes for specialized metabolism that is highly diversified throughout the kingdom. Plant J. 2011;66:212–229. doi: 10.1111/j.1365-313X.2011.04520.x. [DOI] [PubMed] [Google Scholar]
- Chu HY, Wegel E, Osbourn A. From hormones to secondary metabolism: the emergence of metabolic gene clusters in plants. Plant J. 2011;66:66–79. doi: 10.1111/j.1365-313X.2011.04503.x. [DOI] [PubMed] [Google Scholar]
- Colby SM, Crock J, Dowdle-Rizzo B, Lemaux PG, Croteau R. Germacrene C synthase from Lycopersicon esculentum cv. VFNT Cherry tomato: cDNA isolation, characterization, and bacterial expression of the multiple product sesquiterpene cyclase. Proc. Natl Acad. Sci. USA. 1998;95:2216–2221. doi: 10.1073/pnas.95.5.2216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connolly JD, Hill RA. Dictionary of Terpenoids. London: Chapman and Hall; 1991. [Google Scholar]
- Covello PS, Teoh KH, Polichuk DR, Reed DW, Nowak G. Functional genomics and the biosynthesis of artemisinin. Phytochemistry. 2007;68:1864–1871. doi: 10.1016/j.phytochem.2007.02.016. [DOI] [PubMed] [Google Scholar]
- Degenhardt J, Hiltpold I, Kollner TG, Frey M, Gierl A, Gershenzon J, Hibbard BE, Ellersieck MR, Turlings TC. Restoring a maize root signal that attracts insect-killing nematodes to control a major pest. Proc. Nat. Acad. Sci. USA. 2009;106:13213–13218. doi: 10.1073/pnas.0906365106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falara V, Akhtar T, Nguyen TT, et al. The tomato (Solanum lycopersicum) terpene synthase gene family. Plant Physiol. 2011;157:770–789. doi: 10.1104/pp.111.179648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frelichowski JE, Juvik JA. Sesquiterpene carboxylic acids from a wild tomato species affect larval feeding behavior and survival of Helicoverpa zea and Spodoptera exigua (Lepidoptera: Noctuidae) J. Econ. Entomol. 2001;94:1249–1259. doi: 10.1603/0022-0493-94.5.1249. [DOI] [PubMed] [Google Scholar]
- Frelichowski JE, Juvik JA. Inheritance of sesquiterpene carboxylic acid synthesis in crosses of Lycopersicon hirsutum with insect-susceptible tomatoes. Plant Breeding. 2005;124:277–281. [Google Scholar]
- Fridman E, Wang JH, Iijima Y, Froehlich JE, Gang DR, Ohlrogge J, Pichersky E. Metabolic, genomic, and biochemical analyses of glandular trichomes from the wild tomato species Lycopersicon hirsutum identify a key enzyme in the biosynthesis of methylketones. Plant Cell. 2005;17:1252–1267. doi: 10.1105/tpc.104.029736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartman JB, St Clair DA. Variation for aphid resistance and insecticidal acyl sugar expression among and within Lycopersicon pennellii-derived inbred backcross lines of tomato and their F-1 progeny. Plant Breeding. 1999;118:531–536. [Google Scholar]
- Hijmans RJ, Cameron SE, Parra JL, Jones PG, Jarvis A. Very high resolution interpolated climate surfaces for global land areas. Int. J. Climatol. 2005;25:1965–1978. [Google Scholar]
- Hillwig ML, Xu M, Toyomasu T, Tiernan MS, Wei G, Cui G, Huang L, Peters RJ. Domain loss has independently occurred multiple times in plant terpene synthase evolution. Plant J. 2011;68:1051–1060. doi: 10.1111/j.1365-313X.2011.04756.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Hoeven RS, Monforte AJ, Breeden D, Tanksley SD, Steffens JC. Genetic control and evolution of sesquiterpene biosynthesis in Lycopersicon esculentum and L. hirsutum. Plant Cell. 2000;12:2283–2294. doi: 10.1105/tpc.12.11.2283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikemura T. Correlation between the abundance of Escherichia coli transfer RNAs and the occurrence of the respective codons in its protein genes. J. Mol. Biol. 1981;146:1–21. doi: 10.1016/0022-2836(81)90363-6. [DOI] [PubMed] [Google Scholar]
- Innan H, Kondrashov F. The evolution of gene duplications: classifying and distinguishing between models. Nat. Rev. Genet. 2010;11:97–108. doi: 10.1038/nrg2689. [DOI] [PubMed] [Google Scholar]
- Kang JH, Liu GH, Shi F, Jones AD, Beaudry RM, Howe GA. The tomato odorless-2 mutant is defective in trichome-based production of diverse specialized metabolites and broad-spectrum resistance to insect herbivores. Plant Physiol. 2010;154:262–272. doi: 10.1104/pp.110.160192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klahre U, Gurba A, Hermann K, Saxenhofer M, Bossolini E, Guerin PM, Kuhlemeier C. Pollinator choice in Petunia depends on two major genetic loci for floral scent production. Curr. Biol. 2011;21:730–739. doi: 10.1016/j.cub.2011.03.059. [DOI] [PubMed] [Google Scholar]
- Klee HJ. Improving the flavor of fresh fruits: genomics, biochemistry, and biotechnology. New Phytol. 2010;187:44–56. doi: 10.1111/j.1469-8137.2010.03281.x. [DOI] [PubMed] [Google Scholar]
- Kroumova AB, Wagner GJ. Different elongation pathways in the biosynthesis of acyl groups of trichome exudate sugar esters from various solanaceous plants. Planta. 2003;216:1013–1021. doi: 10.1007/s00425-002-0954-7. [DOI] [PubMed] [Google Scholar]
- Landry LG, Chapple CCS, Last RL. Arabidopsis mutants lacking phenolic sunscreens exhibit enhanced ultraviolet-B injury and oxidative damage. Plant Physiol. 1995;109:1159–1166. doi: 10.1104/pp.109.4.1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawson DM, Lunde CF, Mutschler MA. Marker-assisted transfer of acylsugar-mediated pest resistance from the wild tomato, Lycopersicon pennellii, to the cultivated tomato, Lycopersicon esculentum. Mol. Breeding. 1997;3:307–317. [Google Scholar]
- Li JY, Oulee TM, Raba R, Amundson RG, Last RL. Arabidopsis flavonoid mutants are hypersensitive to UV-B irradiation. Plant Cell. 1993;5:171–179. doi: 10.1105/tpc.5.2.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucatti AF, Alvarez AE, Machado CR, Gilardon E. Resistance of tomato genotypes to the greenhouse whitefly Trialeurodes vaporariorum (West.) (Hemiptera: Aleyrodidae) Neotrop. Entomol. 2010;39:792–798. doi: 10.1590/s1519-566x2010000500019. [DOI] [PubMed] [Google Scholar]
- Luckwill L. The Genus Lycopersicon: A Historical, Biological, and Taxonomic Survey of the Wild and Cultivated Tomatoes. Aberdeen, Scotland: Aberdeen University Press; 1943. [Google Scholar]
- Maluf WR, Silva VD, Cardoso MD, Gomes LAA, Neto ACG, Maciel GM, Nizio DAC. Resistance to the South American tomato pinworm Tuta absoluta in high acylsugar and/or high zingiberene tomato genotypes. Euphytica. 2010;176:113–123. [Google Scholar]
- Martin DM, Aubourg S, Schouwey MB, Daviet L, Schalk M, Toub O, Lund ST, Bohlmann J. Functional annotation, genome organization and phylogeny of the grapevine (Vitis vinifera) terpene synthase gene family based on genome assembly, FLcDNA cloning, and enzyme assays. BMC Plant Biol. 2010;10:226. doi: 10.1186/1471-2229-10-226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagel J, Culley LK, Lu YP, Liu EW, Matthews PD, Stevens JF, Page JE. EST analysis of hop glandular trichomes identifies an O-methyltransferase that catalyzes the biosynthesis of xanthohumol. Plant Cell. 2008;20:186–200. doi: 10.1105/tpc.107.055178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oksanen J, Blanchet FG, Kindt R, Legendre P, O’Hara RG, Simpson GL, Solymos P, Stevens MHH, Wagner H. vegan: Community Ecology Package. 2010. R package version 1.17-1. http://CRAN.R-project.org/package=vegan. [Google Scholar]
- Olsson ME, Olofsson LM, Lindahl AL, Lundgren A, Brodelius M, Brodelius PE. Localization of enzymes of artemisinin biosynthesis to the apical cells of glandular secretory trichomes of Artemisia annua L. Phytochemistry. 2009;70:1123–1128. doi: 10.1016/j.phytochem.2009.07.009. [DOI] [PubMed] [Google Scholar]
- Rick CM, Fobes JF, Tanksley SD. Evolution of mating systems in Lycopersicon hirsutum as deduced from genetic variation in electrophoretic and morphological characters. Plant Syst. Evol. 1979;132:279–298. [Google Scholar]
- Rodriguez A, Andres VS, Cervera M, et al. Terpene down-regulation in orange reveals the role of fruit aromas in mediating interactions with insect herbivores and pathogens. Plant Physiol. 2011;156:793–802. doi: 10.1104/pp.111.176545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sallaud C, Rontein D, Onillon S, et al. A novel pathway for sesquiterpene biosynthesis from Z,Z-farnesyl pyrophosphate in the wild tomato Solanum habrochaites. Plant Cell. 2009;21:301–317. doi: 10.1105/tpc.107.057885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Schie CCN, Haring MA, Schuurink RC. Tomato linalool synthase is induced in trichomes by jasmonic acid. Plant Mol. Biol. 2007;64:251–263. doi: 10.1007/s11103-007-9149-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schilmiller AL, Schauvinhold I, Larson M, Xu R, Charbonneau AL, Schmidt A, Wilkerson C, Last RL, Pichersky E. Monoterpenes in the glandular trichomes of tomato are synthesized from a neryl diphosphate precursor rather than geranyl diphosphate. Proc. Natl Acad. Sci. USA. 2009;106:10865–10870. doi: 10.1073/pnas.0904113106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schilmiller A, Shi F, Kim J, Charbonneau AL, Holmes D, Daniel Jones A, Last RL. Mass spectrometry screening reveals widespread diversity in trichome specialized metabolites of tomato chromosomal substitution lines. Plant J. 2010a;62:391–403. doi: 10.1111/j.1365-313X.2010.04154.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schilmiller AL, Miner DP, Larson M, McDowell E, Gang DR, Wilkerson C, Last RL. Studies of a biochemical factory: tomato trichome deep expressed sequence tag sequencing and proteomics. Plant Physiol. 2010b;153:1212–1223. doi: 10.1104/pp.110.157214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt A, Li C, Shi F, Jones AD, Pichersky E. Polymethylated myricetin in trichomes of the wild tomato species Solanum habrochaites and characterization of trichome-specific 3 ‘/5 ‘- and 7/4 ‘-myricetin O-methyltransferases. Plant Physiol. 2011;155:1999–2009. doi: 10.1104/pp.110.169961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sifres A, Blanca J, Nuez F. Pattern of genetic variability of Solanum habrochaites in its natural area of distribution. Genet. Resour. Crop Evol. 2011;58:347–360. [Google Scholar]
- Sirikantaramas S, Taura F, Tanaka Y, Ishikawa Y, Morimoto S, Shoyama Y. Tetrahydrocannabinolic acid synthase, the enzyme controlling marijuana psychoactivity, is secreted into the storage cavity of the glandular trichomes. Plant Cell Physiol. 2005;46:1578–1582. doi: 10.1093/pcp/pci166. [DOI] [PubMed] [Google Scholar]
- Slocombe SP, Schauvinhold I, McQuinn RP, et al. Transcriptomic and reverse genetic analyses of branched-chain fatty acid and acyl sugar production in Solanum pennellii and Nicotiana benthamiana. Plant Physiol. 2008;148:1830–1846. doi: 10.1104/pp.108.129510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 2011;28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tewksbury JJ, Nabhan GP. Seed dispersal – directed deterrence by capsaicin in chillies. Nature. 2001;412:403–404. doi: 10.1038/35086653. [DOI] [PubMed] [Google Scholar]
- Tholl D, Lee S. Terpene specialized metabolism in Arabidopsis thaliana. Arabidopsis Book. 2011;9:e0143. doi: 10.1199/tab.0143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tholl D, Chen F, Petri J, Gershenzon J, Pichersky E. Two sesquiterpene synthases are responsible for the complex mixture of sesquiterpenes emitted from Arabidopsis flowers. Plant J. 2005;42:757–771. doi: 10.1111/j.1365-313X.2005.02417.x. [DOI] [PubMed] [Google Scholar]
- Turner G, Gershenzon J, Nielson EE, Froehlich JE, Croteau R. Limonene synthase, the enzyme responsible for monoterpene biosynthesis in peppermint, is localized to leucoplasts of oil gland secretory cells. Plant Physiol. 1999;120:879–886. doi: 10.1104/pp.120.3.879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unsicker SB, Kunert G, Gershenzon J. Protective perfumes: the role of vegetative volatiles in plant defense against herbivores. Curr. Opin. Plant Biol. 2009;12:479–485. doi: 10.1016/j.pbi.2009.04.001. [DOI] [PubMed] [Google Scholar]
- Wang GD, Tian L, Aziz N, Broun P, Dai XB, He J, King A, Zhao PX, Dixon RA. Terpene biosynthesis in glandular trichomes of hop. Plant Physiol. 2008;148:1254–1266. doi: 10.1104/pp.108.125187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasson AP, Pellerone FI, Mathesius U. Silencing the flavonoid pathway in Medicago truncatula inhibits root nodule formation and prevents auxin transport regulation by rhizobia. Plant Cell. 2006;18:1617–1629. doi: 10.1105/tpc.105.038232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinhold A, Baldwin IT. Trichome-derived O-acyl sugars are a first meal for caterpillars that tags them for predation. Proc. Natl Acad. Sci. USA. 2011;108:7855–7859. doi: 10.1073/pnas.1101306108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weston PA, Johnson DA, Burton HT, Snyder JC. Trichome secretion composition, trichome densities, and spider-mite resistance of 10 accessions of Lycopersicon hirsutum. J. Am. Soc. Hort. Sci. 1989;114:492–498. [Google Scholar]
- Xie Z, Kapteyn J, Gang DR. A systems biology investigation of the MEP/terpenoid and shikimate/phenylpropanoid pathways points to multiple levels of metabolic control in sweet basil glandular trichomes. Plant J. 2008;54:349–361. doi: 10.1111/j.1365-313X.2008.03429.x. [DOI] [PubMed] [Google Scholar]
- Zhang XM, Thacker RR, Snyder JC. Occurrence of 2,3-dihydrofarnesoic acid, a spidermite repellent, in trichome secretions of Lycopersicon esculentum × L. hirsutum hybrids. Euphytica. 2008;162:1–9. [Google Scholar]
- Zhang J, Subramanian S, Stacey G, Yu O. Flavones and flavonols play distinct critical roles during nodulation of Medicago truncatula by Sinorhizobium meliloti. Plant J. 2009;57:171–183. doi: 10.1111/j.1365-313X.2008.03676.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


