Abstract
More than 60% of intron-containing genes undergo alternative splicing (AS) in plants. This number will increase when AS in different tissues, developmental stages, and environmental conditions are explored. Although the functional impact of AS on protein complexity is still understudied in plants, recent examples demonstrate its importance in regulating plant processes. AS also regulates transcript levels and the link with nonsense-mediated decay and generation of unproductive mRNAs illustrate the need for both transcriptional and AS data in gene expression analyses. AS has influenced the evolution of the complex networks of regulation of gene expression and variation in AS contributed to adaptation of plants to their environment and therefore will impact strategies for improving plant and crop phenotypes.
Frequency and consequences of alternative splicing (AS)
AS (see Glossary) produces multiple mRNAs from the same gene through variable selection of splice sites during pre-mRNA splicing. It plays a key regulatory role in modulating gene expression during development and in response to environmental signals [1–4]. Regulation of AS in different cell types and under different conditions depends on sequence elements in pre-mRNAs and the interactions of RNA-binding proteins which vary in their concentration and activity. The phenotype of a cell is determined by transcriptional, post-transcriptional, and post-translational networks, which include as a key component the regulated AS of thousands of genes. In humans, where >95% of genes are alternatively spliced, extensive protein diversity is largely a result of AS [5]. Next generation sequencing has revolutionized research into AS and global mapping of human splicing regulatory proteins to their target RNA sequences has led to the development of a splicing code that will allow prediction of tissue-dependent AS [6]. AS not only contributes to proteome diversity but also can generate truncated proteins that are potentially regulatory or detrimental to the cell. It also plays a role in gene expression by regulating transcript levels through production of isoforms which are degraded by the nonsense-mediated decay (NMD) pathway.
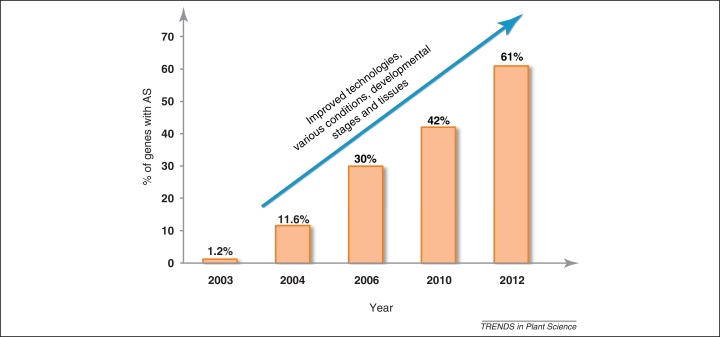
In plants, computational analyses of AS events based on expressed sequence tags and more recently high-throughput transcriptome sequencing have examined the frequency of occurrence of AS in different species [42% and 33% of intron-containing genes in Arabidopsis (Arabidopsis thaliana) and rice (Oryza sativa), respectively] and of different types of AS events [7–10]. In Arabidopsis, the frequency of occurrence has risen significantly over the past 10 years (Figure 1). Recently, an extensive RNA-seq analysis has significantly increased the observed frequency of AS in Arabidopsis to more than 61% of intron-containing genes showing AS. This estimate of 61% of AS is based on analysis of plants grown under normal growth conditions [11] and it is likely that this level will increase further as different tissues at various developmental stages and growth conditions are analyzed [11]. Of the most common types of AS (Figure 2b), intron retention (IR) has been shown to be the most frequent AS event in plants [7–10]. However, some IR events were recently shown to be more likely to represent partially spliced transcripts due to their low abundance [11]. In addition, in the genome-wide analysis above, IR was still the most frequent AS event (40%) but it only occurred in assembled AS transcripts of 23% of the genes providing a more reasonable estimate of the impact of IR to AS plants (Figure 2c) [11]. More importantly, 51% of intron-containing genes utilize alternative 5′ or 3′ splice sites or exon skipping events which can affect the protein coding sequence or generate unproductive mRNAs to affect transcript levels [11]. The widespread occurrence of AS and the range of functional gene groups which it affects supports an essential role for AS in plant development, physiology, metabolism, and responses to environmental conditions and pathogens, all of which have important consequences on plant/crop phenotypes [7,12–18].
Figure 1.
Increasing frequency of occurrence of alternative splicing (AS) in Arabidopsis with time. In 2003, a study using EST (expressed sequence tag) libraries estimated that only 1.2% of the genes in Arabidopsis undergo AS [101]. Subsequently, greater coverage of ESTs and cDNAs libraries allowed the discovery of many more AS events (2004–2006, [7,102–104]). The advent of high-throughput technologies [9,11] has resulted in significant increases in the frequency of AS (almost 60-fold over the past 10 years).
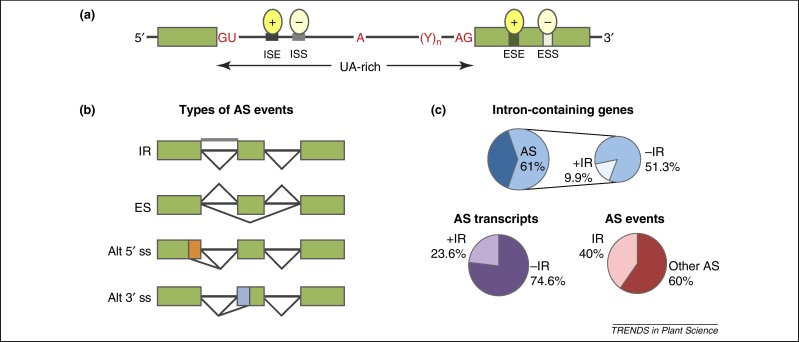
Figure 2.
Main types of alternative splicing (AS) events and frequency in Arabidopsis. (a) Splicing of pre-mRNA is directed by cis elements which include splice sites, branch point, and polypyrimidine tract sequences. Selection of alternative splice sites is affected by trans-acting factors binding to auxiliary exonic and intronic cis elements, termed splicing enhancers and silencers. (b) Types of AS events. (c) Frequency of occurrence of intron retention in Arabidopsis. Intron retention is the most frequent AS event in Arabidopsis (40%) but its contribution to transcript diversity is much lower [11]. Of the 61% of Arabidopsis intron-containing genes with AS, 51% produce AS transcripts which do not involve intron retention (–IR). Among alternatively spliced transcripts, 23.6% contain one or more retained introns (+IR), whereas the rest (74.6%) are produced by other AS events. Colored boxes, exons; lines, introns; GU, 5′ splice site which includes highly conserved GU dinucleotide; AG, 3′ splice site which includes highly conserved AG dinucleotide; A, branch point adenosine; (Y)n, polypyrimidine tract; ovals, positive and negative splicing regulators; carets, splicing events; thick gray line, unspliced (retained) intron. Abbreviations: ESE, exonic splicing enhancers; ESS, exonic splicing silencers; ISE, intronic splicing enhancers; ISS, intronic splicing silencers; Alt 3′ ss, alternative 3′ splice sites; Alt 5′ ss, alternative 5′ splice sites; ES, exon skipping; IR, intron retention.
Regulation of alternative splicing
Assembly of the spliceosome during intron removal and ligation of exons is directed by sequence features of the pre-mRNA. The cis sequences in the pre-mRNA include splice sites, branch point, polypyrimidine tract, enhancer, and suppressor sequences (Figure 2a). In addition, it is well documented in plants that UA-richness of introns contributes to their recognition and is essential for efficient splicing. The splice sites situated at the transition between UA-rich introns and GC-rich exons are preferentially selected for splicing (for review see [19]). The compositional bias for UA-richness has been always considered as a distinguishing feature of plant introns; however, recently it has been shown also for animals that exons have higher GC content than introns [20,21]. Regulation of AS depends on cis signals and their recognition by trans-acting splicing factors. Serine/arginine-rich (SR) and heterogeneous nuclear RNP (hnRNP) proteins function as constitutive and AS splicing factors controlling splice site choice in a concentration-dependent manner [22]. SR proteins are highly conserved in metazoa and plants and, typically, contain one or two RNA recognition motifs (RRMs) and a C-terminal domain (CTD) rich in serine and arginine residues (RS domain). Interestingly, plants possess plant-specific SR proteins and in general they have almost a double number of SR proteins of that in nonphotosynthetic organisms [23]. Many of them have different spatiotemporal expression patterns, implicating diverse target specificities and biological functions [24]. hnRNP proteins, by contrast, constitute a structurally diverse group of RNA-binding proteins associated with nascent pre-mRNA molecules and, in addition to their role in splicing, are involved in a variety of molecular processes [25]. SR and hnRNP proteins bind splicing signals and intronic and exonic enhancer/silencer sequences and through multicomponent interactions with other splicing factors (including cell- and tissue-specific factors) determine splice site selection and where the spliceosome assembles. In general, SR proteins promote splicing and the hnRNP proteins inhibit splice site selection. However, both SRs and hnRNPs can also have opposite functions with, for example, SRSF10 (also known as SRp38) [26] being a negative regulator and polypyrimidine tract-binding protein (PTB; also known as hnRNPI) being a positive regulator [27].
Variation in abundance and activity of splicing factors determines the AS profiles of target genes and therefore their differential expression under different growth conditions and during development [28–31]. Importantly, different growth conditions modulate AS of SR and hnRNP genes causing dynamic changes in the splicing factor profile, further impacting expression of target genes (Figure 3). For example, AS of many Arabidopsis SR genes is affected by temperature, light, salt, hormones, etc. [30–32]. In addition, the activity or localization of SR proteins can be affected by phosphorylation [4,33,34] and protein kinases which phosphorylate plant SR proteins have been identified [35–37]. Not surprisingly therefore, SR protein overexpression and knockout lines show a variety of developmental and growth phenotypes, demonstrating the importance of these proteins to normal growth and development and broad effects on gene expression [28]. The best-studied plant hnRNP proteins to date are the Arabidopsis orthologs of the animal negative splicing regulator, PTB [38], and the glycine-rich RNA-binding proteins, GRP7 and GRP8, components of a slave oscillator coupled to the circadian clock [39–41]. The Arabidopsis PTBs auto- and crossregulate the AS within the family [38]. GRP7 and GRP8 also autoregulate their own AS and crossregulate each other's AS to generate unproductive mRNAs which are targeted by NMD to reduce mRNA and protein levels [39,41]. Finally, the nuclear cap-binding complex (the CBC) consists of two subunits, AtCBP20 and AtCBP80, and AtCBP20 contains a canonical RNA-binding domain (RBD). Mutation of these subunits showed the CBC to be involved in AS of at least some Arabidopsis genes and to preferentially influence AS of the first intron, particularly at the 5′ splice site [42].
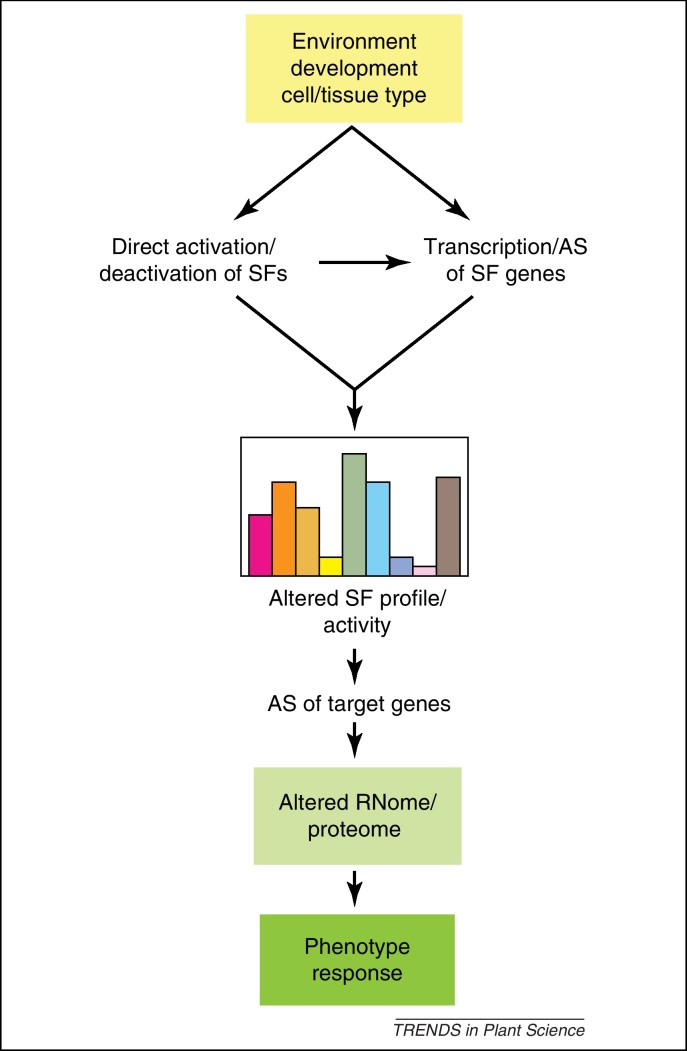
Figure 3.
Dynamic regulation of expression by alternative splicing. Developmental or environmental cues activate signaling pathways which can directly modulate splicing factor activity by post-translational modifications, relocalization, etc. Signaling also directs changes in transcription of splicing factor genes and alternative splicing of these genes changes the abundance, composition, and activity of the splicing factor population. Expression of other target genes including transcription factors is also modulated by alternative splicing responding to the dynamic changes in splicing factor profiles. Changes in the proteome feedback to transcription and alternative splicing (and other post-transcriptional mechanisms) ultimately generating the cellular and organismal phenotype and response. Boxes of colored bars, abundance and/or composition of SFs. Abbreviations: AS, alternative splicing; SF, splicing factor.
Although the abundance and activity of splicing factors determines the AS profiles of downstream genes, very few examples of the biological relevance of AS-derived protein isoforms of splicing factors have been published. Functional diversity of AS isoforms was elegantly demonstrated when two isoforms of SR45 (which differ by only eight amino acids and include putative phosphorylation sites), differentially complemented petal or root developmental phenotypes in the sr45 mutant [43]. Hence, isoforms with very similar sequences can have substantially different morphological outcomes which will reflect a host of gene expression changes in different organs of the plant.
Epigenetic control of alternative splicing
An extensive body of evidence from human and yeast shows that splicing is often coupled to transcription [44–46]. The CTD of RNA polymerase II serves as a landing pad for recruitment of proteins involved in capping, splicing, polyadenylation, and export [47–52]. The rate of transcription elongation by RNA polymerase II may affect splice site choice and thereby AS outcomes [45,46]. A slower rate of transcription tends to favor inclusion of weak upstream exons before the splicing complex is committed to splicing of a stronger downstream exon [45,46]. Importantly, efficiency of the splicing process may also influence the rate of transcription elongation, where, for example, transient pausing of RNAP II at the 3′ splice site of an intron coincides with the appearance of spliced product [53]. The rate of transcript elongation may depend on the chromatin state [50]. For example, nucleosome occupancy varies along a gene with GC-rich exons being relatively nucleosome-rich compared with GC poor introns [20,21,52,54] and transcription through nucleosome-rich regions with compact chromatin tends to be slower [50]. Furthermore, nucleosome occupancy is also lower in alternatively spliced exons compared with constitutively spliced exons [20]. The relations between chromatin state, nucleosome occupancy, RNAP II elongation rates, splicing efficiency, and AS outcomes are key to understanding AS regulation. Indeed, a direct link between histone modifications and AS was demonstrated recently [55]. The splicing of two PTB-dependent mutually exclusive exons in the human fibroblast growth factor receptor 2 (FGFR2) gene depended on histone modifications H3K36me3 and H3K4me1 acting in one direction, and H3K27me3, H3K4me3, and H3K9me1 acting in the opposite direction. The changes in chromatin state are read by the chromatin-binding adapter protein MRG15 which then recruits the splicing factor PTB to the pre-mRNA and affects splicing outcomes [45,46,55]. In plants, the direct link between AS and either chromatin state or RNAP II elongation rates (transcription) has not yet been demonstrated. However, clearly the impact of environmental cues and signaling pathways on chromatin and transcription to generate different AS variants has the potential to further our understanding how plants respond to their ever-changing environment.
Alternative splicing affects protein complexity and transcript levels and stability
The different forms of AS (Figure 2b) and, in particular, alternative 5′ and 3′ splice site selection and exon skipping often lead to changes in protein sequences from the inclusion or removal of a few amino acids (see above for SR45 [43]) to large regions of proteins affecting protein domains, or generating changes in N-terminal or C-terminal regions [56,57]. Different protein isoforms have the potential for differential functions as highlighted by several recent studies. For example, cold-induced sweetening is a serious problem in potatoes where starch is converted to glucose and fructose by vacuolar acid invertase: lines showing resistance to cold-induced sweetening have higher expression of two splice variants (INH2α and INH2β) of the invertase inhibitor gene (INH2) [58]. The flavin-dependent monooxygenase gene, YUCCA4, involved in auxin biosynthesis, undergoes tissue-specific AS to generate isoforms with different intracellular localization. One isoform is expressed in all tissues and is distributed throughout the cytosol, whereas a second is restricted to flowers and is attached to the endoplasmic reticulum [59]. In silico analysis of MADS-box MIKC-type (MADS, Intervening, Keratin-like and C-terminal domain) transcription factors in Arabidopsis predicted protein isoforms which affect dimerization properties or higher order protein complex formation [57]. The potential for AS to influence function was shown by the differential effects on flowering time and floral development of overexpression of isoforms of SHORT VEGETATIVE PHASE, consistent with their different protein–protein interactions [57]. This systematic analysis of a large gene family illustrates the potential of AS to affect key protein domains and function as well as the impact of AS in the evolution of gene families and protein interaction networks.
AS can regulate mRNA levels through the production of AS isoforms containing premature termination codons (PTCs) which are targeted for degradation by NMD [60,61]. Plants possess orthologs of the key eukaryotic NMD proteins, UPF1, UPF2, UPF3, and SMG-7 (but not SMG-1, SMG-5, or SMG-6) and these have been shown to be involved in degrading mRNAs with PTCs [62–68]. Rules for NMD in plants have been established mainly by studying mutations in a small number of model transcripts. The mechanisms of recognition of NMD substrates in plants appear to be fundamentally similar to those in other eukaryotes relying on the distance from a PTC to the 3′ end of the transcript (long 3′UTR) or downstream splice junctions (splicing-dependent) [18,64,69–71]. Recently, by analyzing a large population of endogenous Arabidopsis transcripts, coupled AS, and NMD has been shown to be a widespread mechanism for regulating gene expression with 11–18% of alternatively spliced transcripts being turned over by NMD [18]. This study also showed that transcripts containing PTCs which are NMD substrates are often readily detectable and can contribute significantly to steady-state transcript levels of genes. In addition, some PTC-containing transcripts were not turned over by NMD. For example, some transcripts containing retained introns or parts of introns were unaffected in NMD mutants, suggesting that not all NMD triggering signals or transcript arrangements are understood [18]. Thus, the generation of unproductive AS transcripts can influence the levels of functional mRNAs (full-length protein coding), as has been observed in the regulation of human SR and hnRNP proteins through AS and ultraconserved elements [72–74]. Similarly in plants, SR and PTB genes are regulated by AS [28,38,75] which gives rise to PTC-containing transcripts, suggesting a regulatory function via unproductive mRNAs.
Alternative splicing in the plant circadian clock
Regulation of expression by AS generating unproductive transcripts has recently been demonstrated for core circadian clock genes in Arabidopsis. Circadian clocks have approximately 24-h rhythms and allow organisms to anticipate the day–night cycle and coordinate their genetic, biochemical, and physiological responses [76–78]. Core clock gene expression is regulated at multiple different levels: transcription, protein degradation, and modification with AS being an emerging theme in regulation of the clock [77,78]. Until recently, examples of AS in clock genes and their functional significance were rare. For example, an IR event in CCA1 was conserved in at least four plant species and levels of IR-containing transcripts increased in high light and decreased in the cold [9]. In addition, a mutant in PROTEIN ARGININE METHYL TRANSFERASE 5 (PRMT5) showed a longer circadian period and dramatic changes in the levels of unproductive AS transcripts of PRR9 [79,80]. Recently, extensive AS in the majority of the core clock genes in Arabidopsis was identified and dynamic changes in AS profiles for many AS events were observed in response to changes in temperature and particularly to lower temperatures [81]. AS events were either induced by low temperatures or increased in abundance to 10–50% of the transcripts; the majority of these events were nonproductive resulting in a reduction of functional mRNAs potentially impacting protein levels [81]. Furthermore, the partially redundant gene pairs, LHY and CCA1, and PRR7 and PRR9 behaved differently with respect to AS, implying functional differences between these related genes. Thus, temperature-associated AS modulating the balance between productive and nonproductive mRNA isoforms is an additional mechanism involved in the operation and control of the plant circadian clock.
Alternative splicing and regulation by small interfering peptides/micro-proteins
The fate of alternatively spliced transcripts containing PTCs is expected to be degradation by the NMD pathway but some PTC-containing transcripts are stable and appear to avoid the NMD machinery [18]. PTC-containing transcripts also have the potential to be translated into truncated proteins or peptides. In plants, intron-containing mRNA transcripts were found associated with polysomes and recently ribosome profiling in mouse has identified novel upstream open reading frames (ORFs) and ORFs in long noncoding RNAs [82,83]. In animal and plant systems, small interfering peptides (siPEPs) or micro-proteins (miPs) named after their analogy with siRNAs and miRNAs have been described [84,85]. Such peptides can have altered functionality by only containing particular domains (e.g., DNA binding, transcriptional activators) and can act as both positive and negative regulators and affect regulatory feedback loops [86]. For example, in animals, an AS isoform generates a miP of the ETS1 transcription factor which regulates growth and development responses, lacks the transactivation domains, and interacts physically with ETS1 blocking ETS1-mediated expression of target genes in a dominant negative manner [86–88]. Similarly, miP AS protein isoforms of the animal transcription factor MEIS2 also interact in a dominant negative manner [89]. Interestingly, one of the splice variants (MEIS2E) is structurally similar to a plant protein ‘KNATM’, which is a member of the TALE homeodomain transcription regulators (controlling meristem formation, organ position and morphogenesis, and some aspects of reproductive phase) [86]. KNATM, however, lacks a homeodomain and by forming nonfunctional heterodimers with the BELL TALE protein regulates leaf pattern [90,91]. It is intriguing that a protein generated via AS in animals appears to exist as a miP equivalent in plants.
Genome-wide analysis of AS in Arabidopsis suggested that 78% of alternative transcripts introduced in-frame PTCs [9], providing a huge potential for production of miPs. Examples of miPs in plants with functional consequences are rare but a recent study showed that the Arabidopsis transcription factor gene, IDD14, produces a splice variant (IDD14β) which lacks the DNA-binding domain but interacts with the functional IDD14α isoform to produce heterodimers. The IDD14α/β heterodimer has reduced binding affinity for the promoter of the Qua-Quine Starch (QQS) gene which regulates starch accumulation by initiating starch degradation [84]. Starch accumulation is one response of plants to cold and as the IDD14β splice variant is only expressed under cold conditions, starch degradation is reduced providing an AS/miP-dependent strategy for maintaining starch reserves at low temperatures. Interestingly, the core clock proteins CCA1 and LHY can both homo- and heterodimerize [92,93], and different combinations have different binding affinity to their target sequences [94]. It is interesting to speculate whether the extensive production of PTC-containing transcripts in core clock genes by AS could add a further level of regulation by miPs. Furthermore, siPEPs/miPs offer another mechanism to modify or knock-down expression of endogenous genes. Artificial siPEPs/miPs encoding dimerization domains could be transformed into plants to reduce the activity of a target gene. As they function at the protein level and depend on homo- or heterodimerization, this should improve specificity and reduce off-target silencing [85].
Alternative splicing diversity in ecotypes and polyploids
Extensive AS occurs under altered growth or stress conditions in plants. Similarly, extensive variation in AS is expected in diverse ecotypes adapted to very different climates thereby achieving environmental and phenotypic plasticity. To address such diversity, genomic and transcriptomic sequencing is being performed on geographically and phenotypically diverse accessions of Arabidopsis. Sequencing of the genomes and transcriptomes of 18 Arabidopsis accessions identified extensive single nucleotide polymorphism and indel variation among the genotypes [95]. When compared with Col-0 (TAIR10) one-third of protein coding genes were disrupted/altered in at least one accession although re-annotation restored coding potential in most cases. Sequence variations affected translation start and stop sites, introduced PTCs, or changed the frame of the coding sequence, or potentially generated protein isoforms in different accessions. Two-thirds of 2572 genes with disrupted splice sites when compared to TAIR10 had new splice sites and a quarter of these sites were close to the splice sites in Col-0 [95]. Clearly, naturally occurring sequence variation can disrupt splice sites and RNA-binding motifs for splicing factors. Such mutations impact protein expression and activity and provide a basis for selection for adaptation of different ecotypes to their environments [7,8,10,57].
Extensive duplication or polyploidization has occurred in the evolutionary history of many plant species [96]. Theoretically, such events could generate immediate and drastic changes in the abundance, composition, and activity of splicing factors which in turn could affect splice site choice among variable analogous splice site sequences. Further mutation, gene loss, or changes in expression or protein functionality provide the basis for selection and continued evolution of the species. Recently, AS patterns have been studied among natural and synthetic polyploids of Brassica napus. Interestingly, two independently synthesized lines showed parallel loss of AS events after polyploidy [97]. This is intriguing because it shows that even in two independent events of polyploidy, using the same species, results in identical pattern of AS loss, pointing towards a non-random response of the so-called ‘genomic shock’ after two genomes physically interact with each other. The same study also showed that 26–30% of the duplicated genes show changes in AS, compared with the parents, with some showing organ specificity or response to abiotic stress [97]. It is likely that AS has played an important role in the evolution and adaptation of cultivated crops to different environmental conditions and niches, because many crop species are polyploids and this whole area will be one of great interest in the future, particularly with the power of high-throughput sequencing.
Evolutionary aspects of alternative splicing factor diversity
The Arabidopsis genome encodes 18 SR proteins which is nearly double the number found in humans. At least 12 of the 18 SR genes are located in duplicated genomic regions and the current data indicate that most of them have different spatiotemporal expression patterns, suggesting functional diversification [24]. In different plant lineages, the number of paralogous SR genes is highly variable. For example, the plant-specific RS subfamily of SR proteins is encoded by four genes in Arabidopsis, two in rice, at least five in Pinus taeda and by one gene both in Physcomitrella patens and Chlamydomonas reinhardtii [98]. Differential or common, redundant functions of SR paralogs in different species remain to be determined. However, it is interesting that several SR paralogs and orthologs are regulated by AS events conserved from unicellular green algae to land plants. These events occur in the analogous long introns situated in the RRM coding regions [98,99] and, moreover, they involve unusually highly conserved sequences around alternative splice sites [98], suggesting an important biological function of such regulation. A recent systematic survey of SR genes in 27 eukaryotic genomes showed that flowering plants on average possess nearly double the number of SR genes than nonphotosynthetic species [23]. Moreover, most of the plant SR genes are under purifying selection, ensuring that paralogous genes which originated due to the duplication events maintain their structure and function, whereas redundancy is reduced via diversification of gene expression [23].
Future challenges in alternative splicing research in plants
AS research in plants has made substantial progress in the past 4–5 years. The ever-increasing number of plant genes with AS and the processes in which they are involved point to the importance of understanding the mechanisms and regulation of AS and the functions of AS. The functional impact of AS is one of the most important questions – this is largely due to the relatively small number of examples of AS for which differential functions have been demonstrated for different protein isoforms. However, we draw a parallel with research to discover the extent of AS in plants. Around 10 years ago the first estimate was only 1.2% of plant genes showing AS! With massively improved technologies this number has now grown to more than 61% (Figure 1). Similarly, the increasing number of plant genome sequences and the generation of vast transcriptome data will allow computational analyses to identify conservation of AS events across species and tissue-, developmental-stage and environment-specific regulation of AS providing evidence of functionality. The number of functional examples of AS, whether at the mRNA transcript stability level or protein function, continues to grow and in turn is stimulating wider interest in AS in the plant community. High-throughput sequencing will also address dynamic changes in AS in development and under different environmental conditions and stresses, and how variation in AS patterns in different ecotypes and polyploids contributes to plasticity and adaptation of plant species. Furthermore, we need to understand how signaling pathways affect splicing factor activity directly or via chromatin modification and how transcriptional and AS networks interact [100]. AS is a major mechanism by which plants modulate and fine-tune expression of their genes. The next 5 years will see an explosion of knowledge of the functional significance of AS and understanding its contribution to the complexity of gene expression will offer new opportunities in approaches to modifying plant function for improved phenotypes.
Acknowledgments
This work was supported by the Biotechnology and Biological Sciences Research Council (BBSRC) [BB/G024979/1 (ERA-NET Plant Genomics (PASAS)]; the Scottish Government Rural and Environment Science and Analytical Services Division (RESAS); the Austrian Science Fund (FWF) [SFB 1710, 1711; DK W1207; ERA-NET Plant Genomics (PASAS) I254; SFB RNAreg F43-P10]; the Austria Genomic Program (GENAU III) [ncRNAs]; and the EU FP6 Program Network of Excellence on Alternative Splicing (EURASNET) [LSHG-CT-2005-518238].
Glossary
- Alternative 5′ and 3′ splice sites
use of alternative splice sites at either end of the intron (in the intron or exon sequence) adds or removes sequences.
- Alternative splicing
precursor mRNAs (pre-mRNAs) are spliced differently to generate different mRNA isoforms.
- Exon skipping/inclusion
an exon can be removed in a single splicing event or included by two splicing events. Such exons are called alternative exons or cassette exons.
- Heterogeneous nuclear ribonucleoproteins (hnRNP)
RNA-binding proteins which bind RNA in the cell.
- Intron retention (IR)
one or more introns is/are not removed from a pre-mRNA. It is often difficult to determine whether intron retention is due to DNA contamination or partial splicing of pre-mRNAs at the time of RNA extraction or are actively retained.
- Micro-protein (miP)
micro-proteins are usually generated by translation of a transcript containing a premature termination codon, lack one or more functional domains, and have fewer than 100 amino acids [82].
- Nonsense-mediated decay (NMD)
a cellular quality control mechanism that recognizes mRNA transcripts containing PTCs and targets them for degradation.
- Polypyrimidine tract-binding protein (PTB)
an hnRNP protein which binds pyrimidine-rich sequences in RNA to regulate alternative splicing and other mRNA biogenesis processes.
- Precursor messenger RNA (pre-mRNA)
the primary transcript of a gene which is processed to mRNA.
- Premature termination codon (PTC)
a translational stop codon found in transcripts upstream of the authentic stop codon; PTCs can be generated by mutations in DNA, errors in transcription or splicing, or alternative splicing.
- Serine/arginine-rich (SR) proteins
constitutive and alternative splicing factors containing RNA-binding motifs and a RS-rich domain.
- Small-interfering peptides (siPEPs)
short peptides which interfere with cellular processes.
References
- 1.Black D.L. Mechanisms of alternative pre-messenger RNA splicing. Annu. Rev. Biochem. 2003;72:291–336. doi: 10.1146/annurev.biochem.72.121801.161720. [DOI] [PubMed] [Google Scholar]
- 2.Graveley B.R. Alternative splicing: increasing diversity in the proteomic world. Trends Genet. 2001;17:100–107. doi: 10.1016/s0168-9525(00)02176-4. [DOI] [PubMed] [Google Scholar]
- 3.Lareau L.F. The evolving roles of alternative splicing. Curr. Opin. Struct. Biol. 2004;14:273–282. doi: 10.1016/j.sbi.2004.05.002. [DOI] [PubMed] [Google Scholar]
- 4.Stamm S. Function of alternative splicing. Gene. 2005;344:1–20. doi: 10.1016/j.gene.2004.10.022. [DOI] [PubMed] [Google Scholar]
- 5.Pan Q. Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing. Nat. Genet. 2008;40:1413–1415. doi: 10.1038/ng.259. [DOI] [PubMed] [Google Scholar]
- 6.Barash Y. Deciphering the splicing code. Nature. 2010;465:53–59. doi: 10.1038/nature09000. [DOI] [PubMed] [Google Scholar]
- 7.Wang B.B., Brendel V. Genome-wide comparative analysis of alternative splicing in plants. Proc. Natl. Acad. Sci. U.S.A. 2006;103:7175–7180. doi: 10.1073/pnas.0602039103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang B.B. Cross-species EST alignments reveal novel and conserved alternative splicing events in legumes. BMC Plant Biol. 2008;8:17. doi: 10.1186/1471-2229-8-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Filichkin S.A. Genome-wide mapping of alternative splicing in Arabidopsis thaliana. Genome Res. 2010;20:45–58. doi: 10.1101/gr.093302.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Barbazuk W.B. Genome-wide analyses of alternative splicing in plants: opportunities and challenges. Genome Res. 2008;18:1381–1392. doi: 10.1101/gr.053678.106. [DOI] [PubMed] [Google Scholar]
- 11.Marquez Y. Transcriptome survey reveals increased complexity of the alternative splicing landscape in Arabidopsis. Genome Res. 2012;22:1184–1195. doi: 10.1101/gr.134106.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ali G.S., Reddy A.S. Regulation of alternative splicing of pre-mRNAs by stresses. Curr. Top. Microbiol. Immunol. 2008;326:257–275. doi: 10.1007/978-3-540-76776-3_14. [DOI] [PubMed] [Google Scholar]
- 13.Chen F.C. Plant gene and alternatively spliced variant annotator. A plant genome annotation pipeline for rice gene and alternatively spliced variant identification with cross-species expressed sequence tag conservation from seven plant species. Plant Physiol. 2007;143:1086–1095. doi: 10.1104/pp.106.092460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Halterman D.A. Powdery mildew-induced Mla mRNAs are alternatively spliced and contain multiple upstream open reading frames. Plant Physiol. 2003;131:558–567. doi: 10.1104/pp.014407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lorkovic Z.J. Role of plant RNA-binding proteins in development, stress response and genome organization. Trends Plant Sci. 2009;14:229–236. doi: 10.1016/j.tplants.2009.01.007. [DOI] [PubMed] [Google Scholar]
- 16.Quesada V. Autoregulation of FCA pre-mRNA processing controls Arabidopsis flowering time. EMBO J. 2003;22:3142–3152. doi: 10.1093/emboj/cdg305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhu J. Interplay between cold-responsive gene regulation, metabolism and RNA processing during plant cold acclimation. Curr. Opin. Plant Biol. 2007;10:290–295. doi: 10.1016/j.pbi.2007.04.010. [DOI] [PubMed] [Google Scholar]
- 18.Kalyna M. Alternative splicing and nonsense-mediated decay modulate expression of important regulatory genes in Arabidopsis. Nucleic Acids Res. 2011;40:2454–2469. doi: 10.1093/nar/gkr932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lorkovic Z.J. Pre-mRNA splicing in higher plants. Trends Plant Sci. 2000;5:160–167. doi: 10.1016/s1360-1385(00)01595-8. [DOI] [PubMed] [Google Scholar]
- 20.Schwartz S. Chromatin organization marks exon–intron structure. Nat. Struct. Mol. Biol. 2009;16:990–995. doi: 10.1038/nsmb.1659. [DOI] [PubMed] [Google Scholar]
- 21.Tilgner H. Nucleosome positioning as a determinant of exon recognition. Nat. Struct. Mol. Biol. 2009;16:996–1001. doi: 10.1038/nsmb.1658. [DOI] [PubMed] [Google Scholar]
- 22.Matlin A.J. Understanding alternative splicing: towards a cellular code. Nat. Rev. Mol. Cell Biol. 2005;6:386–398. doi: 10.1038/nrm1645. [DOI] [PubMed] [Google Scholar]
- 23.Richardson D.N. Comparative analysis of serine/arginine-rich proteins across 27 eukaryotes: insights into sub-family classification and extent of alternative splicing. PLoS ONE. 2011;6:e24542. doi: 10.1371/journal.pone.0024542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kalyna M., Barta A. A plethora of plant serine/arginine-rich proteins: redundancy or evolution of novel gene functions? Biochem. Soc. Trans. 2004;32:561–564. doi: 10.1042/BST0320561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Martinez-Contreras R. hnRNP proteins and splicing control. Adv. Exp. Med. Biol. 2007;623:123–147. doi: 10.1007/978-0-387-77374-2_8. [DOI] [PubMed] [Google Scholar]
- 26.Feng Y. Phosphorylation switches the general splicing repressor SRp38 to a sequence-specific activator. Nat. Struct. Mol. Biol. 2008;15:1040–1048. doi: 10.1038/nsmb.1485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xue Y. Genome-wide analysis of PTB–RNA interactions reveals a strategy used by the general splicing repressor to modulate exon inclusion or skipping. Mol. Cell. 2009;36:996–1006. doi: 10.1016/j.molcel.2009.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Barta A. Plant SR proteins and their functions. Curr. Top. Microbiol. Immunol. 2008;326:83–102. doi: 10.1007/978-3-540-76776-3_5. [DOI] [PubMed] [Google Scholar]
- 29.Lazar G., Goodman H.M. The Arabidopsis splicing factor SR1 is regulated by alternative splicing. Plant Mol. Biol. 2000;42:571–581. doi: 10.1023/a:1006394207479. [DOI] [PubMed] [Google Scholar]
- 30.Palusa S.G. Alternative splicing of pre-mRNAs of Arabidopsis serine/arginine-rich proteins: regulation by hormones and stresses. Plant J. 2007;49:1091–1107. doi: 10.1111/j.1365-313X.2006.03020.x. [DOI] [PubMed] [Google Scholar]
- 31.Tanabe N. Differential expression of alternatively spliced mRNAs of Arabidopsis SR protein homologs, atSR30 and atSR45a, in response to environmental stress. Plant Cell Physiol. 2007;48:1036–1049. doi: 10.1093/pcp/pcm069. [DOI] [PubMed] [Google Scholar]
- 32.Duque P. A role for SR proteins in plant stress responses. Plant Signal. Behav. 2011;6:49–54. doi: 10.4161/psb.6.1.14063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stamm S. Signals and their transduction pathways regulating alternative splicing: a new dimension of the human genome. Hum. Mol. Genet. 2002;11:2409–2416. doi: 10.1093/hmg/11.20.2409. [DOI] [PubMed] [Google Scholar]
- 34.Stamm S. Regulation of alternative splicing by reversible protein phosphorylation. J. Biol. Chem. 2008;283:1223–1227. doi: 10.1074/jbc.R700034200. [DOI] [PubMed] [Google Scholar]
- 35.de la Fuente van Bentem S. Phosphoproteomics reveals extensive in vivo phosphorylation of Arabidopsis proteins involved in RNA metabolism. Nucleic Acids Res. 2006;34:3267–3278. doi: 10.1093/nar/gkl429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.de la Fuente van Bentem S. The subcellular localization of plant protein phosphatase 5 isoforms is determined by alternative splicing. Plant Physiol. 2003;133:702–712. doi: 10.1104/pp.103.026617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Savaldi-Goldstein S. Alternative splicing modulation by a LAMMER kinase impinges on developmental and transcriptome expression. Plant Cell. 2003;15:926–938. doi: 10.1105/tpc.011056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stauffer E. Polypyrimidine tract-binding protein homologues from Arabidopsis underlie regulatory circuits based on alternative splicing and downstream control. Plant J. 2010;64:243–255. doi: 10.1111/j.1365-313X.2010.04321.x. [DOI] [PubMed] [Google Scholar]
- 39.Schoning J.C. Auto-regulation of the circadian slave oscillator component AtGRP7 and regulation of its targets is impaired by a single RNA recognition motif point mutation. Plant J. 2007;52:1119–1130. doi: 10.1111/j.1365-313X.2007.03302.x. [DOI] [PubMed] [Google Scholar]
- 40.Schuttpelz M. Changes in conformational dynamics of mRNA upon AtGRP7 binding studied by fluorescence correlation spectroscopy. J. Am. Chem. Soc. 2008;130:9507–9513. doi: 10.1021/ja801994z. [DOI] [PubMed] [Google Scholar]
- 41.Schoning J.C. Reciprocal regulation of glycine-rich RNA-binding proteins via an interlocked feedback loop coupling alternative splicing to nonsense-mediated decay in Arabidopsis. Nucleic Acids Res. 2008;36:6977–6987. doi: 10.1093/nar/gkn847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Raczynska K.D. Involvement of the nuclear cap-binding protein complex in alternative splicing in Arabidopsis thaliana. Nucleic Acids Res. 2010;38:265–278. doi: 10.1093/nar/gkp869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang X.N., Mount S.M. Two alternatively spliced isoforms of the Arabidopsis SR45 protein have distinct roles during normal plant development. Plant Physiol. 2009;150:1450–1458. doi: 10.1104/pp.109.138180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Carrillo Oesterreich F. Global analysis of nascent RNA reveals transcriptional pausing in terminal exons. Mol. Cell. 2010;40:571–581. doi: 10.1016/j.molcel.2010.11.004. [DOI] [PubMed] [Google Scholar]
- 45.Luco R.F. Epigenetics in alternative pre-mRNA splicing. Cell. 2011;144:16–26. doi: 10.1016/j.cell.2010.11.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Luco R.F., Misteli T. More than a splicing code: integrating the role of RNA, chromatin and non-coding RNA in alternative splicing regulation. Curr. Opin. Genet. Dev. 2011;21:366–372. doi: 10.1016/j.gde.2011.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.de Almeida S.F., Carmo-Fonseca M. The CTD role in cotranscriptional RNA processing and surveillance. FEBS Lett. 2008;582:1971–1976. doi: 10.1016/j.febslet.2008.04.019. [DOI] [PubMed] [Google Scholar]
- 48.Fong N. A 10 residue motif at the C-terminus of the RNA pol II CTD is required for transcription, splicing and 3′ end processing. EMBO J. 2003;22:4274–4282. doi: 10.1093/emboj/cdg396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Goldstrohm A.C. The transcription elongation factor CA150 interacts with RNA polymerase II and the pre-mRNA splicing factor SF1. Mol. Cell. Biol. 2001;21:7617–7628. doi: 10.1128/MCB.21.22.7617-7628.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Howe K.J. RNA polymerase II conducts a symphony of pre-mRNA processing activities. Biochim. Biophys. Acta. 2002;1577:308–324. doi: 10.1016/s0167-4781(02)00460-8. [DOI] [PubMed] [Google Scholar]
- 51.McCracken S. The C-terminal domain of RNA polymerase II couples mRNA processing to transcription. Nature. 1997;385:357–361. doi: 10.1038/385357a0. [DOI] [PubMed] [Google Scholar]
- 52.Schwartz S., Ast G. Chromatin density and splicing destiny: on the cross-talk between chromatin structure and splicing. EMBO J. 2010;29:1629–1636. doi: 10.1038/emboj.2010.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Alexander R.D. Splicing-dependent RNA polymerase pausing in yeast. Mol. Cell. 2010;40:582–593. doi: 10.1016/j.molcel.2010.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Spies N. Biased chromatin signatures around polyadenylation sites and exons. Mol. Cell. 2009;36:245–254. doi: 10.1016/j.molcel.2009.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Luco R.F. Regulation of alternative splicing by histone modifications. Science. 2010;327:996–1000. doi: 10.1126/science.1184208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Airoldi C.A. Single amino acid change alters the ability to specify male or female organ identity. Proc. Natl. Acad. Sci. U.S.A. 2010;107:18898–18902. doi: 10.1073/pnas.1009050107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Severing E.I. Predicting the impact of alternative splicing on plant MADS domain protein function. PLoS ONE. 2012;7:e30524. doi: 10.1371/journal.pone.0030524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Brummell D.A. Induction of vacuolar invertase inhibitor mRNA in potato tubers contributes to cold-induced sweetening resistance and includes spliced hybrid mRNA variants. J. Exp. Bot. 2011;62:3519–3534. doi: 10.1093/jxb/err043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kriechbaumer V. Alternative splicing of the auxin biosynthesis gene YUCCA4 determines its subcellular compartmentation. Plant J. 2012;70:292–302. doi: 10.1111/j.1365-313X.2011.04866.x. [DOI] [PubMed] [Google Scholar]
- 60.Rebbapragada I., Lykke-Andersen J. Execution of nonsense-mediated mRNA decay: what defines a substrate? Curr. Opin. Cell Biol. 2009;21:394–402. doi: 10.1016/j.ceb.2009.02.007. [DOI] [PubMed] [Google Scholar]
- 61.Nicholson P. Nonsense-mediated mRNA decay in human cells: mechanistic insights, functions beyond quality control and the double-life of NMD factors. Cell. Mol. Life Sci. 2010;67:677–700. doi: 10.1007/s00018-009-0177-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Arciga-Reyes L. UPF1 is required for nonsense-mediated mRNA decay (NMD) and RNAi in Arabidopsis. Plant J. 2006;47:480–489. doi: 10.1111/j.1365-313X.2006.02802.x. [DOI] [PubMed] [Google Scholar]
- 63.Hori K., Watanabe Y. UPF3 suppresses aberrant spliced mRNA in Arabidopsis. Plant J. 2005;43:530–540. doi: 10.1111/j.1365-313X.2005.02473.x. [DOI] [PubMed] [Google Scholar]
- 64.Kerenyi Z. Inter-kingdom conservation of mechanism of nonsense-mediated mRNA decay. EMBO J. 2008;27:1585–1595. doi: 10.1038/emboj.2008.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Riehs N. Arabidopsis SMG7 protein is required for exit from meiosis. J. Cell Sci. 2008;121:2208–2216. doi: 10.1242/jcs.027862. [DOI] [PubMed] [Google Scholar]
- 66.Wu J. Nonsense-mediated mRNA decay (NMD) silences the accumulation of aberrant trypsin proteinase inhibitor mRNA in Nicotiana attenuata. Plant J. 2007;51:693–706. doi: 10.1111/j.1365-313X.2007.03173.x. [DOI] [PubMed] [Google Scholar]
- 67.Yoine M. Arabidopsis UPF1 RNA helicase for nonsense-mediated mRNA decay is involved in seed size control and is essential for growth. Plant Cell Physiol. 2006;47:572–580. doi: 10.1093/pcp/pcj035. [DOI] [PubMed] [Google Scholar]
- 68.Yoine M. The lba1 mutation of UPF1 RNA helicase involved in nonsense-mediated mRNA decay causes pleiotropic phenotypic changes and altered sugar signalling in Arabidopsis. Plant J. 2006;47:49–62. doi: 10.1111/j.1365-313X.2006.02771.x. [DOI] [PubMed] [Google Scholar]
- 69.Kertesz S. Both introns and long 3′-UTRs operate as cis-acting elements to trigger nonsense-mediated decay in plants. Nucleic Acids Res. 2006;34:6147–6157. doi: 10.1093/nar/gkl737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Nyiko T. Plant upstream ORFs can trigger nonsense-mediated mRNA decay in a size-dependent manner. Plant Mol. Biol. 2009;71:367–378. doi: 10.1007/s11103-009-9528-4. [DOI] [PubMed] [Google Scholar]
- 71.Schwartz A.M. Stability of plant mRNAs depends on the length of the 3′-untranslated region. Biochemistry (Mosc) 2006;71:1377–1384. doi: 10.1134/s0006297906120145. [DOI] [PubMed] [Google Scholar]
- 72.Lareau L.F. Unproductive splicing of SR genes associated with highly conserved and ultraconserved DNA elements. Nature. 2007;446:926–929. doi: 10.1038/nature05676. [DOI] [PubMed] [Google Scholar]
- 73.Ni J.Z. Ultraconserved elements are associated with homeostatic control of splicing regulators by alternative splicing and nonsense-mediated decay. Genes Dev. 2007;21:708–718. doi: 10.1101/gad.1525507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Palusa S.G., Reddy A.S. Extensive coupling of alternative splicing of pre-mRNAs of serine/arginine (SR) genes with nonsense-mediated decay. New Phytol. 2010;185:83–89. doi: 10.1111/j.1469-8137.2009.03065.x. [DOI] [PubMed] [Google Scholar]
- 75.Reddy A.S. Alternative splicing of pre-messenger RNAs in plants in the genomic era. Annu. Rev. Plant Biol. 2007;58:267–294. doi: 10.1146/annurev.arplant.58.032806.103754. [DOI] [PubMed] [Google Scholar]
- 76.Doherty C.J., Kay S.A. Circadian control of global gene expression patterns. Annu. Rev. Genet. 2010;44:419–444. doi: 10.1146/annurev-genet-102209-163432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Petrillo E. Alternative splicing adds a new loop to the circadian clock. Commun. Integr. Biol. 2011;4:284–286. doi: 10.4161/cib.4.3.14777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Staiger D., Green R. RNA-based regulation in the plant circadian clock. Trends Plant Sci. 2011;16:517–523. doi: 10.1016/j.tplants.2011.06.002. [DOI] [PubMed] [Google Scholar]
- 79.Hong S. Type II protein arginine methyltransferase 5 (PRMT5) is required for circadian period determination in Arabidopsis thaliana. Proc. Natl. Acad. Sci. U.S.A. 2010;107:21211–21216. doi: 10.1073/pnas.1011987107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Sanchez S.E. A methyl transferase links the circadian clock to the regulation of alternative splicing. Nature. 2010;468:112–116. doi: 10.1038/nature09470. [DOI] [PubMed] [Google Scholar]
- 81.James A.B. Alternative splicing mediates responses of the Arabidopsis circadian clock to temperature changes. Plant Cell. 2012;24:961–981. doi: 10.1105/tpc.111.093948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Ingolia N.T. Ribosome profiling of mouse embryonic stem cells reveals the complexity and dynamics of mammalian proteomes. Cell. 2011;147:789–802. doi: 10.1016/j.cell.2011.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Ner-Gaon H. Intron retention is a major phenomenon in alternative splicing in Arabidopsis. Plant J. 2004;39:877–885. doi: 10.1111/j.1365-313X.2004.02172.x. [DOI] [PubMed] [Google Scholar]
- 84.Seo P.J. Two splice variants of the IDD14 transcription factor competitively form nonfunctional heterodimers which may regulate starch metabolism. Nat. Commun. 2011;2:303. doi: 10.1038/ncomms1303. [DOI] [PubMed] [Google Scholar]
- 85.Seo P.J. Competitive inhibition of transcription factors by small interfering peptides. Trends Plant Sci. 2011;16:541–549. doi: 10.1016/j.tplants.2011.06.001. [DOI] [PubMed] [Google Scholar]
- 86.Staudt A.C., Wenkel S. Regulation of protein function by ‘microProteins’. EMBO Rep. 2011;12:35–42. doi: 10.1038/embor.2010.196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Laitem C. Ets-1 p27: a novel Ets-1 isoform with dominant-negative effects on the transcriptional properties and the subcellular localization of Ets-1 p51. Oncogene. 2009;28:2087–2099. doi: 10.1038/onc.2009.72. [DOI] [PubMed] [Google Scholar]
- 88.Lee G.M. The structural and dynamic basis of Ets-1 DNA binding autoinhibition. J. Biol. Chem. 2005;280:7088–7099. doi: 10.1074/jbc.M410722200. [DOI] [PubMed] [Google Scholar]
- 89.Yang Y. Three-amino acid extension loop homeodomain proteins Meis2 and TGIF differentially regulate transcription. J. Biol. Chem. 2000;275:20734–20741. doi: 10.1074/jbc.M908382199. [DOI] [PubMed] [Google Scholar]
- 90.Hamant O., Pautot V. Plant development: a TALE story. C. R. Biol. 2010;333:371–381. doi: 10.1016/j.crvi.2010.01.015. [DOI] [PubMed] [Google Scholar]
- 91.Magnani E., Hake S. KNOX lost the OX: the Arabidopsis KNATM gene defines a novel class of KNOX transcriptional regulators missing the homeodomain. Plant Cell. 2008;20:875–887. doi: 10.1105/tpc.108.058495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Lu S.X. CIRCADIAN CLOCK ASSOCIATED1 and LATE ELONGATED HYPOCOTYL function synergistically in the circadian clock of Arabidopsis. Plant Physiol. 2009;150:834–843. doi: 10.1104/pp.108.133272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Yakir E. Posttranslational regulation of CIRCADIAN CLOCK ASSOCIATED1 in the circadian oscillator of Arabidopsis. Plant Physiol. 2009;150:844–857. doi: 10.1104/pp.109.137414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.O’Neill J.S. Circadian clock parameter measurement: characterization of clock transcription factors using surface plasmon resonance. J. Biol. Rhythms. 2011;26:91–98. doi: 10.1177/0748730410397465. [DOI] [PubMed] [Google Scholar]
- 95.Gan X. Multiple reference genomes and transcriptomes for Arabidopsis thaliana. Nature. 2011;477:419–423. doi: 10.1038/nature10414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Adams K.L., Wendel J.F. Allele-specific, bidirectional silencing of an alcohol dehydrogenase gene in different organs of interspecific diploid cotton hybrids. Genetics. 2005;171:2139–2142. doi: 10.1534/genetics.105.047357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Zhou R. Extensive changes to alternative splicing patterns following allopolyploidy in natural and resynthesized polyploids. Proc. Natl. Acad. Sci. U.S.A. 2011;108:16122–16127. doi: 10.1073/pnas.1109551108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Kalyna M. Evolutionary conservation and regulation of particular alternative splicing events in plant SR proteins. Nucleic Acids Res. 2006;34:4395–4405. doi: 10.1093/nar/gkl570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Iida K., Go M. Survey of conserved alternative splicing events of mRNAs encoding SR proteins in land plants. Mol. Biol. Evol. 2006;23:1085–1094. doi: 10.1093/molbev/msj118. [DOI] [PubMed] [Google Scholar]
- 100.Blencowe B.J. Alternative splicing: new insights from global analyses. Cell. 2006;126:37–47. doi: 10.1016/j.cell.2006.06.023. [DOI] [PubMed] [Google Scholar]
- 101.Zhu W. Refined annotation of the Arabidopsis genome by complete expressed sequence tag mapping. Plant Physiol. 2003;132:469–484. doi: 10.1104/pp.102.018101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Iida K. Genome-wide analysis of alternative pre-mRNA splicing in Arabidopsis thaliana based on full-length cDNA sequences. Nucleic Acids Res. 2004;32:5096–5103. doi: 10.1093/nar/gkh845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Xiao Y.L. Analysis of the cDNAs of hypothetical genes on Arabidopsis chromosome 2 reveals numerous transcript variants. Plant Physiol. 2005;139:1323–1337. doi: 10.1104/pp.105.063479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Campbell M.A. Comprehensive analysis of alternative splicing in rice and comparative analyses with Arabidopsis. BMC Genomics. 2006;7:327. doi: 10.1186/1471-2164-7-327. [DOI] [PMC free article] [PubMed] [Google Scholar]