Abstract
The flavour of fermented beverages such as beer, cider, saké and wine owe much to the primary fermentation yeast used in their production, Saccharomyces cerevisiae. Where once the role of yeast in fermented beverage flavour was thought to be limited to a small number of volatile esters and higher alcohols, the discovery that wine yeast release highly potent sulfur compounds from non-volatile precursors found in grapes has driven researchers to look more closely at how choice of yeast can influence wine style. This review explores recent progress towards understanding the range of ‘flavour phenotypes’ that wine yeast exhibit, and how this knowledge has been used to develop novel flavour-active yeasts. In addition, emerging opportunities to augment these phenotypes by engineering yeast to produce so-called grape varietal compounds, such as monoterpenoids, will be discussed.
Keywords: Aroma, Flavour, Fermented beverages, Wine, Yeast
Introduction
While purchase of bottled wine is strongly influenced by extrinsic factors such as price (Mueller et al. 2010) and grape variety (King et al. 2012), the intrinsic flavour properties of a wine have a direct impact on how much it is ‘liked’ by consumers (Lattey et al. 2010). The ability to modulate wine style through changed winemaking practice is, therefore, an attractive target, that is dependent upon understanding flavour compound composition and how this influences sensory perception (Francis and Newton 2005). Nykanen (1986) reviewed progress in the field of wine and distillate flavour compound formation over a 25-year period (1960s–1980s), highlighting that while once it was thought alcoholic beverage flavours were composed of a small number of compounds, by 1985 more than 1,300 volatile compounds had been implicated. Many volatiles in wine are grape-derived, or form during processing and maturation — indeed the proportion of wine volatiles modulated by yeast was recently found to be relatively small (Robinson et al. 2011). Nonetheless, Nykanen (1986) contended that “the body of flavour is formed during fermentation by yeast”, and that “formation of the most dominant compounds occurring in beverages depend more on the yeast selected than the raw materials used in fermentation”. Ensuing research over the past 25 years has served to reinforce these observations.
The flavour compounds underlying the so-called 'yeast bouquet'; ethyl esters, acetate esters, fusel alcohols, carbonyls, and volatile fatty acids, are secondary metabolites synthesized by a wide range of microbial species. Depending upon winemaking practices, multiple yeast species from the grapes and winery equipment can be involved in alcoholic fermentation, and potentially contribute to wine flavour (Romano et al. 2003). Wine fermentation is a highly selective environment, however, and as ethanol concentrations rise, the species diversity of the ecosystem is diminished, giving way to predominance of the wine yeast, Saccharomyces cerevisiae (Heard and Fleet 1985). Consequently, most advancement in field over the past 25 years has been made in understanding formation of the core ‘yeast bouquet’ flavour compounds by S. cerevisiae, with production of esters (Saerens et al. 2010; Sumby et al. 2010) and fusel alcohols and acids (Hazelwood et al. 2008) recently reviewed.
Over the same 25-year period, S. cerevisiae emerged as the eukaryotic cell model system of choice, greatly enhancing the understanding of industrial yeast strains (Chambers and Pretorius 2010). Population genomics revealed a close relationship between man and yeast (Liti et al. 2009), and as useful industrial traits have been selected for over time, some consider that S. cerevisiae has been domesticated for brewing, baking, and winemaking (Legras et al. 2007; Verstrepen et al. 2006). Some industrially important phenotypes, such as ability to rapidly produce carbon dioxide (baking strains), ability to degrade maltose (brewing strains), ability to complete fermentation in high sugar grape musts (winemaking strains), are relatively straightforward to score and select for. Wine ‘flavour’ as a phenotype is much more ambiguous, but has nonetheless been a strong driver for wine yeast selection since the concept of single-yeast inoculation was introduced to the wine industry in 1890 (Pretorius 2000). Wines made through single-yeast inoculation differ in sensory properties to those made by spontaneous fermentations, an observation reinforced by differences in chemical composition (Varela et al. 2009). Hyma et al. (2011) recently found that domesticated S. cerevisiae strains made wines that were sensorially distinct from wines made by inoculation with single ‘wild’ S. cerevisiae strains, implying that the ‘flavour’ phenotype has indeed been a target for wine yeast domestication. Even amongst commercial, or domesticated, wine strains of S. cerevisiae, different wine flavour profiles generated solely through choice of yeast inoculum (including single, or multi-strain co-inoculation) can be detected by trained panels and wine professionals (King et al. 2008; Swiegers et al. 2009), and most importantly, by wine consumers (King et al. 2010).
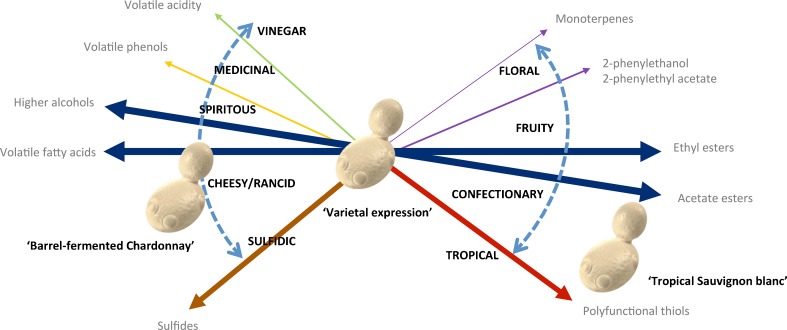
What are the yeast ‘flavour phenotypes’ that have been selected for? In broad terms, wine yeast strains can be categorized on one dimension as 'fruity'–'floral', 'neutral', or 'cheesy'–‘rancid’–spirituous’, depending on their relative capacity to produce esters, higher alcohols, and volatile fatty acids (Fig. 1). Generally, there is a high level of correlation between individual compounds within these broader classes; however, there are exceptions. Wine strains of Saccharomyces bayanus produce relatively high concentrations of 2-phenylethanol and 2-phenylethyl acetate compared to other higher alcohols and acetate esters, which may enhance ‘rose’ and ‘floral’ characters (Masneuf-Pomarede et al. 2010). An additional dimension can be used to separate wine yeast according to their production of sulfur containing compounds, which are associated with ‘tropical’ or ‘sulfidic’ flavours in wine (Fig. 1). Further 'flavour-fault' phenotypes include 'medicinal' phenolic off-flavour, and excessive production of volatile acidity —imparting a flavour associated with vinegar. Finally, some floral nuances can be imparted by yeast able to release glycosidically bound monoterpenes (Ubeda and Briones 2000; Ugliano et al. 2006), or, as a result of mutations in the ergosterol pathway, able to de novo synthesize these 'varietal' compounds at low levels (Chambon et al. 1990, 1991).
Fig. 1.
‘Flavour phenotypes’ that wine yeast have been selected for. Spectrum of flavour phenotypes that wine yeast exhibit (bold, dashed arrows), with flavour compound groups that drive them indicated by solid arrows weighted according to magnitude of impact. Examples of ‘flavour phenotypes’ that may be desirable for different winemaking objectives shown by positioning of yeast
Some of the genetic and environmental factors that affect these phenotypes are well understood — the PAD1 gene from S. cerevisiae was designated Pof through genetic studies that linked it to the phenolic off-flavour phenotype encountered in brewing (Goodey and Tubb 1982; Meaden and Taylor 1991), and Pof-wine yeast are available today. On the other hand, prior to 2005 there was no knowledge of S. cerevisiae genes involved in formation of polyfunctional thiols (Howell et al. 2005). Expanding the knowledge of enzymes involved in flavour compound formation, and the genetic networks that regulate them, is crucial to advancing the capability to develop flavour-active yeasts with the best mix of flavour phenotypes.
This review will explore recent advances in our understanding of yeast influence on formation of flavour compounds, then focus on the emerging opportunity to engineer wine yeast to enhance formation of so-called grape 'varietal' flavour compounds, such as the monoterpenoids and high-impact sulfur-containing polyfunctional thiols.
Recent advances in knowledge of flavour compound formation by yeast
Ethyl esters and acetate esters
There are two classes of flavour-active esters in fermented beverages. First, the acetate esters, where the acyl group is derived from acetate (in the form of acetyl-CoA), and the alcohol group is ethanol or a complex alcohol derived from amino acid metabolism. The most significant acetate esters are ethyl acetate (‘fruity’, ‘solvent-like’ aromas), isoamyl acetate (‘banana’ aroma), and 2-phenylethyl acetate (‘honey’, ‘roses’, ‘flowery’ aromas). The second group comprises the medium-chain fatty acid (MCFA) ethyl esters, where the alcohol group is ethanol, and the acyl group is derived from activated medium-chain fatty acids. Examples are ethyl hexanoate (‘apple-like’ aroma), and ethyl octanoate (‘apple’ aroma).
The rate of ester formation during fermentation is dependent on two primary factors: (1) the concentration of the co-substrates, the acyl-CoA and the alcohol; and (2) the activity of enzymes involved in their synthesis and hydrolysis (acyltransferases and esterases) (Saerens et al. 2006, 2008; Verstrepen et al. 2003). To date, five distinct proteins — Atf1p, Atf2p, Eht1p, Eeb1p and Iah1p — have been identified and characterized in S. cerevisiae as having ester synthesis or hydrolysis activity, with the alcohol acetyltransferase Atf1p having the greatest activity and being the most studied (Lilly et al. 2000, 2006a; Saerens et al. 2010; reviewed by Sumby et al. 2010; Verstrepen et al. 2003).
Overexpression of ATF1 during wine fermentation results in a significant increase (between 10- and 200-fold) in acetate ester production (Lilly et al. 2000, 2006a; Verstrepen et al. 2003), whereas ATF2 appears to play a minor role in ester formation (Lilly et al. 2006a; Verstrepen et al. 2003). Excessively high production of ethyl acetate by yeast overexpressing ATF1 did not improve the fermentation bouquet and aroma of the young wines, however, it was observed that hydrolysis during bottle aging caused a significant decrease in the levels of acetate esters, particularly ethyl acetate (Lilly et al. 2000). Therefore, higher initial levels of esters could lead to wines with a more fruity character (Lilly et al. 2000). On the other hand, the deletion of both ATF1 and ATF2 completely abolishes the formation of isoamyl acetate (Verstrepen et al. 2003). However, the double deletion strain still produced about 50 % as much ethyl acetate the wild-type strain, suggestive of the existence of unknown acetate ester synthases in the yeast genome.
Acetate ester formation by yeast is balanced by the IAH1-encoded esterase. Diploid brewer’s yeast strains deficient in IAH1 accumulate much higher amounts of isoamyl acetate than do the parent strains (Fukuda et al. 1998). Conversely, overexpression of IAH1 results in a significant decrease in the concentration of many esters, including isoamyl acetate, hexyl acetate, ethyl acetate, and 2-phenylethyl acetate, compared to control strains (Lilly et al. 2006a). IAH1 crucially regulates the accumulation of isoamyl acetate and other esters during fermentation and thus determines the flavour quality of wine (Lilly et al. 2006a). Recently, Iah1p has been crystallized (Ma et al. 2011). The hydrolytic activity of Iah1p was shown to be maximal for acetate esters, and was lower with hexanoate esters. Interestingly, a C-terminally truncated version of Iah1p was able to hydrolyze decanoate esters.
A further, largely unexplored, level at which acetate ester formation is modulated is the availability of acetyl-CoA. Increased levels of both CoA and acetyl-CoA were accompanied by a 6-fold increase in production of isoamyl acetate, for an Escherichia coli strain expressing the yeast ATF2 gene and overexpressing its own pantothenate kinase (panK) gene — which regulates CoA biosynthesis (Vadali et al. 2004). A decrease in formation of isoamyl acetate and ethyl acetate by S. cerevisiae was noted when carnitine acetyltransferase, an enzyme that regulates the transfer of activated acetyl groups to the mitochondria and regulates acetyl-CoA/CoA pools within the cells, was overexpressed (Cordente et al. 2007).
EHT1 and EEB1 encode proteins with MCFA ethyl ester synthase and esterase activities. Specifically, EEB1 encodes an ethanol acyltransferase responsible for the synthesis of the majority of MCFA ethyl esters during fermentation (Saerens et al. 2006), while EHT1 encodes for an ethanol hexanoyl transferase, which plays a minor role in MCFA ethyl ester biosynthesis. Both EHT1 and EEB1 also possess short-chain esterase activity (Saerens et al. 2006). Another protein encoded by YMR210w, and similar to both Eeb1p and Eht1p, has been identified as a putative acyltransferase (Saerens et al. 2006). Whereas expression levels seem to be the limiting factor for ATF1 gene regulation and acetate ester production, this does not seem to be the case for EEB1 and EHT1 — overexpression of these genes in an industrial strain had a minor effect on MCFA ethyl ester content, presumably due to their competing synthesis and hydrolysis activities (Lilly et al. 2006a; Saerens et al. 2006). Precursor availability appears to be the limiting factor in ethyl ester biosynthesis (Saerens et al. 2008) since the addition of hexanoic or octanoic acid to the fermentation medium causes a strong increase in the formation of the corresponding ethyl ester (Saerens et al. 2006). In this regard, octanoic (but not hexanoic) acid induces expression of EEB1 and EHT1 (Saerens et al. 2008). Recently, a transcriptomic analysis has revealed that EEB1 was the most strongly induced gene (8.4-fold) after addition of decanoic acid (Legras et al. 2010), which suggests that ethyl ester synthesis plays a complementary role in the detoxification of MCFA. A slight (35 %) but significant induction of YMR210w was also reported in these conditions.
The possible role of YMR210w in ethyl ester synthesis remains unclear. While overexpression of this open reading frame (ORF) does not significantly affect the concentration of ethyl esters at the end of fermentation (Rossouw et al. 2008; Saerens et al. 2006), its native expression levels correlate positively with ethyl acetate, ethyl octanoate and isoamyl acetate (Rossouw et al. 2008). Furthermore, while deletion of YMR210w does not affect the production of MCFA ethyl esters, deletion of this gene in an Δeeb1 or Δeeb1Δeht1 background further decreases formation of both ethyl octanoate and ethyl decanoate.
Higher alcohols and volatile fatty acids
Alcoholic fermentation is also accompanied by the formation of aliphatic and aromatic alcohols known as higher alcohols or fusel alcohols. While fusel alcohols at high concentrations impart off-flavours, low concentrations of these compounds and their esters make a crucial contribution to the flavour and aroma of wine (Lambrechts and Pretorius 2000; Nykanen et al. 1977). In particular, 2-phenylethanol is considered to be one of the most important aromatic alcohols contributing to wine flavour. The higher alcohols are predominantly formed by yeast during fermentation from α-keto acids, involving degradation of an amino acid via the so-called Ehrlich pathway (Ehrlich 1904; reviewed by Hazelwood et al. 2008; Styger et al. 2011b), but can also be synthesised from glucose via pyruvate (Chen 1978; Dickinson et al. 1997; Eden et al. 2001). The Ehrlich pathway involves three steps: (1) an initial transamination that results in the formation of an α-keto acid; (2) decarboxylation of the α-keto acid to form a ‘fusel aldehyde’; and (3) its reduction to generate the ‘fusel alcohol’.
Four S. cerevisiae genes have been implicated in the transamination step of the Ehrlich pathway: the mitochondrial and cytosolic branched-chain amino acid (BCAA) aminotransferases (BAT1 and BAT2, respectively) and the aromatic amino acid aminotransferases I and II (ARO8 and ARO9, respectively) (Eden et al. 1996; Iraqui et al. 1998; Kispal et al. 1996). Researchers have looked at the effect of modulating yeast BCAA activity on the production of higher alcohols (Eden et al. 2001; Lilly et al. 2006b). In wines and distillates, the overexpression of BAT1 increased the concentration of isoamyl alcohol, its acetate ester, as well as isobutanol; while overexpression of BAT2 resulted in a substantial increase in the formation of isobutanol and isobutyric acid (Lilly et al. 2006b). Sensory analysis confirmed that the overexpression of BAT1 and BAT2 had an impact on aroma profiles of wines and distillates (Lilly et al. 2006b). The perturbation of the BAT genes not only affects the concentrations of metabolites directly linked to these genes, but also other aroma metabolites not directly related to higher alcohols, highlighting the complexities of the interconnections within such complex metabolic networks (Lilly et al. 2006b; Styger et al. 2011a). BAT2 seems to have a more prominent role than BAT1 in the Ehrlich pathway. In support of this hypothesis, it has been recently shown that BAT2 function is determinant for BCAA catabolism, while BAT1 is involved in the biosynthesis of these amino acids (Colon et al. 2011).
To date, five proteins have been implicated in α-keto decarboxylation: the pyruvate decarboxylases Pdc1p, Pdcp5, and Pdc6p; the phenylpyruvate decarboxylase Aro10p; and the probable carboxylase Thi3p (Styger et al. 2011a), which plays a role as a regulatory protein of the enzymes involved in thiamine biosynthesis (Mojzita and Hohmann 2006; Nosaka et al. 2005).
The final step of the Ehrlich pathway involves either the reduction or oxidation of the fusel aldehydes to form fusel alcohols or fusel acids, respectively. Formation of the fusel alcohols can be catalyzed by several oxidoreductases: the alcohol dehydrogenases (Adh1p to Adh7p) (Dickinson et al. 2003; Kondo et al. 2012; Larroy et al. 2002), the formaldehyde dehydrogenase Sfa1p (Dickinson et al. 2003), the 3-methylbutanal reductase Gre2p (Hauser et al. 2007), and the NADPH-dependent aldo-keto reductase Ypr1p (Ford and Ellis 2002), and at least one of the putative aryl-alcohol dehydrogenases (AAD6) (Styger et al. 2011a). The balance between oxidation and reduction of the fusel aldehydes depends on the global redox status of the yeast cell. In glucose-grown batch cultures of S. cerevisiae, where growth is predominantly fermentative, the formation of fusel alcohols is favoured over that of the acids (Dickinson et al. 1997, 2003), while the opposite is true in aerobic-limited chemostat cultures grown in the presence of various amino acids (Vuralhan et al. 2003).
Recently, Styger et al. (2011a) conducted a targeted screen of genes encoding dehydrogenase, decarboxylase and reductase enzymes potentially involved in flavour compound formation via the Ehrlich pathway. The ten genes with greatest impact on higher alcohol formation were further characterized, including some not previously linked with this pathway: two highly promiscuous carboxylases (PAD1 and SPE1) and two dehydrogenases (OYE2 and HOM2). Discovery of novel flavour-active genes such as these provide excellent targets for biotechnological improvement of aroma production by industrial strains of S. cerevisiae.
Strategies to modify ester, higher alcohol and volatile fatty acid flavour profiles of yeast
Recently, several strategies for genetic engineering of S. cerevisiae to increase productivity of isobutanol from glucose through the endogenous Ehrlich pathway have been reported (Chen et al. 2011; Kondo et al. 2012). Kondo enhanced the Ehrlich pathway activity by overexpressing several combinations of alcohol dehydrogenases and keto-acid decarboxylases. Overexpression of the medium-chain alcohol dehydrogenases ADH6 and ADH7 displayed higher isobutanol productivities, as did the overexpression of the probable decarboxylase THI3. The production of isobutanol was further improved by altering carbon flux towards valine biosynthesis and deleting the pyruvate decarboxylase PDC1. Chen et al. (2011) used a different strategy based upon the overexpression of several genes involved in valine biosynthesis, along with BAT2, achieving similar results.
Another strategy to increase the formation of higher alcohols, specifically 2-phenylethanol (‘flowery’, ‘rose’ aroma), is the expression in yeast of flower and fruit enzymes involved in the production of this aromatic volatile. Farhi et al. (2010) demonstrated that yeast can be harnessed in the field of floral volatiles by expressing the rose phenylacetaldehyde synthase, which was shown to complement the deletion of the native phenylpyruvate decarboxylase ARO10, and to enhance the production of both the alcohol and phenylacetaldehyde compared to the wild-type strain.
To date, there has been almost no application of genetically modified (GM) technology in commercial winemaking (Chambers and Pretorius 2010; Pretorius et al. 2012); therefore, non-GM strategies to develop flavour-active yeast are required. The isolation of yeast mutants, induced or spontaneous, that are resistant to different drugs and amino acids analogues, has proven an effective strategy for modulation of ester production by yeast (Fukuda et al. 1990a, b; Hirooka et al. 2005; Ichikawa et al. 1991). It has been reported that mutant saké yeast resistant to cerulenin, an inhibitor of fatty acid synthesis, overproduced ethyl hexanoate, one of the most important components of saké flavour (Ichikawa et al. 1991). Cerulenin resistance is conferred by a particular dominant mutation in the fatty synthase (FAS2) gene (Fas2pG1250S) (Inokoshi et al. 1994). A self-cloning saké strain bearing this mutation, and no extraneous DNA sequences, has become the first GM microorganism to be approved for use in Japan (Aritomi et al. 2004).
Saké yeast mutants resistant to the l-leucine analog 5,5′,5″-trifluoro-dl-leucine (TFL) (Ashida et al. 1987; Oba et al. 2005) show higher levels of isoamyl acetate production. Resistance to TFL has been linked to mutations in LEU4 gene (Casalone et al. 1997; Oba et al. 2005), which releases leucine feedback inhibition and causes hyper-production of isoamyl alcohol, and thus, an accumulation of the corresponding acetate ester. Saké yeast resistant to o- and p-fluoro-dl-phenylalanine produce higher levels of 2-phenylethanol and 2-phenylethyl acetate (Fukuda et al. 1990a, b). Hirooka et al. (2005) isolated a spontaneous saké mutant resistant to 1-farnesylpyridinium, an analog of the isoprenoid farnesol (Hirooka et al. 2005), with improved production of isoamyl acetate. This mutant has an increased alcohol acetyltransferase activity, and it is currently used for industrial saké brewing (Hirooka et al. 2010).
While much research in this area has been devoted to industrial yeast for saké brewing, cerulenin and TFL resistant S. cerevisiae yeast strains used in the production of cachaça, the Brazilian sugarcane spirit, have been isolated that produce higher levels of both isoamyl acetate and ethyl hexanoate (de Souza et al. 2012; Vicente et al. 2006). Similar strategies have not, to date, been applied in development of flavour-active wine yeast. Ichikawa et al. (1991) noted that ethyl hexanoate overproduction by their FAS2 mutant saké yeast was accompanied by an increase in formation of hexanoic acid — while this may not be detrimental to saké quality, the net effect of similar mutations in wine yeast on flavour profile and balance awaits evaluation.
Monoterpenoids
Terpenoids (isoprenoids) comprise a large and diverse family of naturally occurring compounds, which are involved in the fragrance and aroma of flowers and fruits, plant defense and primary plant metabolism. All terpenoids are synthesized from the universal five carbon precursors, isopentenyl diphosphate (IPP) and dimethylallyl diphosphate (DMAPP). The significance of volatile C10 monoterpenes to the flavour and varietal character of some cultivars of Vitis vinifera is well reviewed (Mateo and Jimenez 2000; Strauss et al. 1986; Versini et al. 1999).
Monoterpenes are present as free as well as glycosylated flavourless conjugates amongst the secondary metabolites of certain grape varieties of V. vinifera. Hence, when these compounds are detected in wine they are considered to originate from grape and not from fermentation. In general, more bound glycosides are found than free terpenoids, and the ratios of bound to free terpenoids can also vary amongst different grape cultivars (Williams et al. 1984). Both bound and free terpenoids can be modified to various degrees during alcoholic and malolactic fermentation (Swiegers et al. 2005). During winemaking, bound terpenoids can be released by glycosidase enzymes produced by grapes, yeast and bacteria, increasing the volatile terpenoid composition of wines and enhancing wine aroma and flavour (van Rensburg and Pretorius 2000). Enzymatic hydrolysis of glycosides occurs in two steps: first, depending on the diglycoside conjugate, either an α-l-arabinofuranosidase, an α-l-rhamnosidase or a β-d-apiosidase release the corresponding monoterpenyl glucosides. Second, monoterpenyl glucosides are then hydrolysed by the action of a β-glucosidase releasing the monoterpene alcohol (Flipphi et al. 1993; LeClinche et al. 1997; Ramachandran et al. 2012; Zietsman et al. 2011). β-Glucosidases do not have endoglucanase activity and therefore can only act on monoterpenyl glucosides (Gunata et al. 1988).
Several S. cerevisiae strains have been shown to secrete enzymes characterised principally by β-glucosidase activity (Fernandez et al. 1999; Ubeda and Briones 2000; Ugliano et al. 2006). However, their activity towards monoterpenyl glycosides is very low (Hernandez et al. 2003). A well-recognised strategy to improve the hydrolysis of glycosylated bound conjugates is the addition of exogenous enzyme preparations from other microorganisms during or after fermentation (Armada et al. 2010; Genoves et al. 2003; van Rensburg and Pretorius 2000; Vasserot et al. 1993). Commercial preparations contain a mix of pectinases, glucanases and xylanases obtained principally from Aspergillus spp. Addition of exogenous enzyme preparations can increase production costs; moreover, the lack of specificity of these enzymes might induce secondary reactions detrimental to wine flavour (Riou et al. 1998).
Another strategy to enhance formation of monoterpenes during winemaking is to engineer S. cerevisiae wine yeast by introducing enzymes able to hydrolyse glycosylated precursors (Manzanares et al. 2003; Pretorius and Bauer 2002; Schuller and Casal 2005). There are several reports in literature assessing the effect of the exogenous expression of these enzymes on the chemical composition and aroma profile of wines fermented with engineered strains. Expression of the β-(1,4)-endoglucanase encoded by the egl1 gene from Trichoderma longibrachiatum changed volatile composition and enhanced perception of fruity aroma (Perez-Gonzalez et al. 1993). Engineered strains expressing the Aspergillus nidulans xlnA gene encoding for a β-(1,4)-endoxylanase showed significative higher concentrations of several esters, higher alcohols and terpenes, particularly, ethyl acetate, 3-methyl butanol, 2-phenylethanol and linalool in Chenin Blanc wines (Ganga et al. 1999). Manzanares et al. (2003) engineered two wine strains: one expressed the α-rhamnosidase gene (rhaA) from Aspergillus aculeatus and the second expressed the β-glucosidase gene from Candida molischiana. Wines co-fermented with both strains showed an increase in the concentration of linalool, α-terpeniol, nerol and geraniol in Muscat wine. Expression of β-glucosidases from Saccharomycopsis fibuligera showed not only higher levels of terpenols but also increased concentrations of esters (van Rensburg et al. 2005). Gil et al. (2005) overexpressed the S. cerevisiae exoglucanase encoded by the EXG1 gene. Wines fermented with engineered strains exhibited the higher concentrations of volatile compounds, including several alcohols and terpenols. Co-expression of the xyn2 gene from Trichoderma reesei which encodes a xylanase and the end1 gene from Butyrivibrio fibriosolvens encoding an endo-β-(1,4)-glucanase showed significant improvement in the aromatic profile of wines fermented by engineered strains (Louw et al. 2006). Zietsman et al. (2011) constructed a wine yeast co-expressing an α-l-arabinofuranosidase from Aspergillus awamori and a β-glucosidase from S. fibuligera. Gewürztraminer wine fermented with the engineered strain showed significative higher concentrations of linalool, citronellol, nerol and α-terpineol and lower concentration of geraniol after fermentation, and resulted in wines exhibiting higher floral and fruity characters than non-engineered wine.
Although genetic engineering approaches can considerably change the volatile composition and enhance the varietal aroma profile of wine they are not used in the commercial production of wine. Therefore, attention has been focussed on the characterisation and development of non-genetically modified wine strains able to increase the release of monoterpenes (Fernandez-Gonzalez et al. 2003; Gamero et al. 2011; Hernandez-Orte et al. 2008).
All the strategies described above, however, are less useful for musts derived from non-aromatic grape varieties having low contents of free and bound monoterpenes. An alternative would be to engineer wine yeast for the de novo biosynthesis of monoterpenes through the existing mevalonate pathway, which results in the formation of IPP and DMAPP.
Unlike plants, S. cerevisiae cannot produce monoterpenes efficiently, and only a few natural S. cerevisiae strains have been shown to produce small amounts of monoterpenes (Carrau et al. 2005; Zea et al. 1995). This is because S. cerevisiae lacks enzymes with monoterpene synthase activity (MTS), which catalyze the conversion of the universal precursor, geranyl diphosphate (GPP) to monoterpenes. In addition, yeast do not carry a specific GPP synthase, and this metabolite only occurs as an intermediate of farnesyl diphosphate (FPP) synthesis, which is the precursor of several classes of essential metabolites such as ergosterol, ubiquinone, dolichols, or heme A (Grabinska and Palamarczyk 2002). In yeast, GPP and FPP synthase activities are shared by one enzyme: farnesyl diphosphate synthase (FPPS). FPPS catalyzes two sequential condensation reactions of the IPP with its isomer DMAPP into GPP, and then GPP with another IPP molecule into FPP. It was thought that tight binding of GPP to the FPPS catalytic site might lead to minimal release of GPP for biosynthesis of monoterpenoids. However, it has been established that S. cerevisiae has enough free GPP to be used by exogenous MTS to produce monoterpenes under laboratory and vinification conditions (Herrero et al. 2008; Oswald et al. 2007).
The ERG20-encoded FPPS enzyme is essential for S. cerevisiae. Yeast mutants secreting the monoterpene alcohols linalool and geraniol have been characterized previously (Chambon et al. 1990, 1991), which carry a specific mutation in ERG20 (Erg20pK197E) (Blanchard and Karst 1993). This mutation leads to an increase of the available GPP for monoterpene synthesis (Blanchard and Karst 1993; Fischer et al. 2011). Therefore, interrupting the sterol pathway by mutation in ERG20 can alter monoterpene content.
In recent years, many genes have been characterized that encode plant MTS, for example; the linalool synthase gene from Clarkia breweri (Dudareva et al. 1996), the geraniol synthase from Ocimum basilicum (Iijima et al. 2004), and α-terpineol synthase from V. vinifera (Martin and Bohlmann 2004). Since all monoterpenes are produced from the ubiquitous C10 intermediate GPP, it is possible to engineer yeast for the de novo production of specific monoterpene(s). The introduction of MTS in yeast leads to a redirection of the flux of the isoprenoid precursors DMAPP and IPP towards GPP, competing with FPP formation, which is required to produce sterols (Herrero et al. 2008). Yeast has been harnessed in several recent studies to act as a cell factory for production of different terpenes (Farhi et al. 2011; Fischer et al. 2011; Herrero et al. 2008; Oswald et al. 2007; Rico et al. 2010; Tokuhiro et al. 2009), as reviewed by Siddiqui et al. (2012).
An emerging opportunity to engineer wine aroma has arisen through recent work characterizing novel MTS encoding genes from V. vinifera (Martin et al. 2010). Recent analysis of the grapevine genome allowed the prediction of 69 putatively functional terpene synthase (VvTPS) encoding genes, which represent five of the seven plant TPS subfamilies. In addition, 39 of these VvTPS enzymes were functionally characterized, the largest number of TPS characterized for any species, and found to produce different profiles of terpenoids. As other grapevine genome sequences become available, the number of available MTS genes will grow, further expanding the potential for engineering of S. cerevisiae to produce terpene profiles to achieve desired sensory profiles in finished wines.
Volatile sulfur compounds
The propensity of S. cerevisiae yeast to produce negative volatile sulfur compounds (VSCs), particularly ‘rotten-egg’-aroma imparting hydrogen sulfide (H2S), has been well studied (reviewed by Swiegers and Pretorius 2007). VSCs in wine can be considered a ‘double-edged sword’, as some sulfur-containing flavour compounds contribute positively to wine (Swiegers and Pretorius 2005). Prominent examples include furfurylthiol (‘roast coffee’ aroma) (Tominaga et al. 2000); and the ‘fruity’ polyfunctional thiols 3-mercaptohexan-1-ol (3MH), 4-mercapto-4-methyl-pentan-2-one (4MMP), and 3-mercaptohexyl acetate (3MHA), that impart ‘passionfruit’, ‘grapefruit’, ‘gooseberry’, ‘guava’, and ‘box hedge’ aromas (Dubourdieu et al. 2006; Swiegers et al. 2006; Swiegers and Pretorius 2005). Other important VSCs found in wine include methanethiol (‘cooked cabbage’ aroma); dimethylsulfide, dimethyldisulfide, and dimethyltrisulfide (‘cabbage’, ‘cauliflower’, and ‘garlic’ aromas); and methylthioesters (‘cooked cauliflower’, ‘cheesy’ and ‘chives’ aromas).
The production of H2S is a significant problem for the global wine industry since it imparts an undesirable ‘sulfurous’, ‘rotten egg’-like off-flavour (Rauhut 1993), even at low concentrations (1 μg/l) (Siebert et al. 2009). The production of H2S during wine fermentation is a frequently encountered problem in winemaking, and, if it is not treated, the resulting wine will be tainted leading to a loss in quality and the possibility of being rejected by consumers. It is well established that S. cerevisiae is responsible for H2S off-flavour in wine and that the production is strain dependent (Acree et al. 1972; Giudici and Kunkee 1994; Kumar et al. 2010; Mendes-Ferreira et al. 2002; Nowak et al. 2004), even though not all wine yeast produce H2S — about 1 % of naturally occurring wine strains are unable to produce this off-flavour (Zambonelli et al. 1984). Other factors affecting the production of H2S include environmental and nutritional factors such as the availability of sulfur compounds (sulfur dioxide, organic sulfur compounds, and elemental sulfur in the vineyard for plant protection); nitrogen limitation, and vitamin deficiency (Giudici and Kunkee 1994; Rauhut 1993; Rauhut and Kurbel 1994; Spiropoulos et al. 2000; Ugliano et al. 2009; Wang et al. 2003; Winter et al. 2011a). H2S can be formed metabolically by wine yeast from inorganic sulfur compounds, sulfate, and sulfite, or organic compounds, cysteine, and glutathione (Henschke and Jiranek 1993; Rauhut 1993; Spiropoulos et al. 2000). The majority of H2S produced during winemaking occurs as a result of the biosynthesis of the sulfur containing amino acids methionine and cysteine, which occur in low concentrations in grape juice, through the sulfate reduction sequence (SRS). These amino acids are essential for the growth of S. cerevisiae, and if they are not present, or depleted, in the growth medium, then sulfur must be assimilated from inorganic sources (Henschke and Jiranek 1993). The most common sulfur source in S. cerevisiae is extracellular sulfate, which naturally exists in high amounts in grape juice (Vos and Gray 1979).
In the first step of the SRS pathway, sulfate is transported into the cell by two specific permeases before a two-step activation with the aid of two molecules of ATP. The first reduction step produces sulfite, which is, in turn, reduced by sulfite reductase to sulfide. At this point, the sulfide produced is combined with a nitrogenous precursor, O-acetyl serine or O-acetyl homoserine, to ultimately form cysteine and methionine. If there is a deficiency of assimilable nitrogen in the grape must, O-acetyl serine or O-acetyl homoserine becomes limiting, and sulfide builds up and is converted to the volatile gas H2S, which then diffuses from the yeast cell into the wine (Giudici and Kunkee 1994; Henschke and Jiranek 1993).
Several genetic engineering strategies have been used for limiting H2S production, which generally consisted in the overexpression or inactivation of some of the genes involved in the SRS pathway. Constitutive expression of the MET25 gene (alias MET17), which encodes a bifunctional O-acetylserine/O-acetylhomoserine sulfhydrylase, lowered H2S by 2-fold in a brewing yeast (Omura et al. 1995). In a similar study, the overexpression of the same gene in a strain of S. cerevisiae greatly reduced H2S formation in a wine ferment, but this was not the case for another strain (Spiropoulos and Bisson 2000). Overexpression of the CYS4 gene, encoding cystathionine β-synthetase, was also shown to reduce H2S production (Linderholm et al. 2006; Tezuka et al. 1992). Altering sulfite reductase activity has been considered a better approach for limiting H2S formation in yeast, since reducing the production of sulfide is a better approach than trying to consume it in a later metabolic step. The yeast NADPH-dependent sulfite reductase is a heterotetramer protein, consisting of two α- and two β-subunits (α2β2). The α-subunit is encoded by the MET10 gene, whereas the β-subunit is encoded by the MET5 gene. The inactivation of MET10 in a brewer’s yeast resulted in increased sulfite accumulation during beer production and increased flavour stability, and no sign of H2S production (Hansen and Kielland-Brandt 1996). In wine yeast, there have been some efforts to develop commercial yeast with impaired hydrogen sulfide production (Cordente et al. 2009; Linderholm et al. 2010), in which the partial inactivation of either of the two catalytic subunits of the sulfite reductase enzyme led to the desired phenotype. Recently, the MET10 G176A allele, present in one of the low-H2S strains described by Cordente et al. (2009), was found to have a strong dominant effect, which allowed the use of this strain in the breeding of new interspecific hybrids with a low-H2S production phenotype and other desired industrial traits (Bizaj et al. 2012).
The polyfunctional thiols 4MMP, 3MH and 3MHA are extremely potent having perception thresholds in the parts per trillion range (Dubourdieu et al. 2006; Tominaga et al. 1998a, b). These compounds are of particular importance for the varietal character of Sauvignon Blanc wines (reviewed by Coetzee and du Toit 2012), and are found to be highly desired in some styles of Sauvignon Blanc by consumers (King et al. 2012). It has been shown that 4MMP and 3MH exist in grapes in their non-volatile precursor form, conjugated to cysteine or glutathione (Fedrizzi et al. 2009; Peyrot Des Gachons et al. 2002; Roland et al. 2010a; Tominaga et al. 1998a). The wine yeast take up these precursors and cleave them to release the corresponding free thiol during fermentation (Darriet et al. 1995; Grant-Preece et al. 2010; Winter et al. 2011b), although only a small fraction of available precursors are converted to the respective polyfunctional thiols (Dubourdieu et al. 2006; Subileau et al. 2008b; Winter et al. 2011b). No cysteine or glutathione precursor of 3MHA has been identified, and this compound is formed during fermentation and through esterification of 3MH by the alcohol acetyltransferase ATF1. The overexpression of ATF1 in both commercial and laboratory strains results in a significant increase in the amount of 3MHA formed, while the overexpression of the esterase IAH1 had the opposite effect (Swiegers et al. 2006).
Yeast strains vary in their abilities to release polyfunctional thiols, and therefore selection of yeast strain is highly important to modulate their concentration in wine (Dubourdieu et al. 2006; Howell et al. 2005; Swiegers et al. 2006, 2009). Polyfunctional thiol production also depends on other factors, such as fermentation temperature (Masneuf-Pomarede et al. 2006; Swiegers et al. 2006), addition of nutrients to active dry yeast rehydration media (Winter et al. 2011a), pre-fermentation operations such as skin contact (Peyrot Des Gachons et al. 2002), as well as oxygen, phenol, and sulfur dioxide content (Blanchard et al. 2004).
The genetic determinants for release of 3MH and 4MMP from their cysteinylated precursors have been studied in a targeted manner over recent years (Holt et al. 2011; Howell et al. 2005; Roncoroni et al. 2011; Subileau et al. 2008a; Thibon et al. 2008). Uptake of the precursors is assumed to be mediated by amino acid transporters on the plasma membrane. However, the deletion of the general amino acid transporter, GAP1, has a limited effect on 3MH release from the cysteine precursor Cys-3MH in synthetic media (Subileau et al. 2008a), which indicates that other transporters might be involved in its uptake during fermentation. Once inside the cell, the cysteinylated precursor is cleaved by a yeast enzyme with carbon–sulfur β-lyase activity (Swiegers et al. 2007; Tominaga et al. 1995). A gene encoding a yeast β-lyase enzyme, IRC7, was found to be the key determinant of 4MMP release (Roncoroni et al. 2011; Thibon et al. 2008), while also contributing to the release of 3MH. Interestingly, most strains of S. cerevisiae (Liti et al. 2009; Roncoroni et al. 2011), have a deletion in the C terminus of the protein that render IRC7 inactive. This variation might account for the strain variation observed in 4MMP release (Howell et al. 2005; Swiegers et al. 2009).
3MH release, on the other hand, appears to be mediated by more than one gene (Roncoroni et al. 2011; Thibon et al. 2008). It was recently demonstrated that the cystathionine β-lyase STR3, integrated into a commercial wine yeast under the control of a constitutive promoter, increased release of 3MH by 30 % (Holt et al. 2011). The activity of this enzyme against Cys-3MH in vitro was consistent with 3MH release being a side (non-physiological) activity, reinforcing the concept that for highly potent compounds such as 3MH, small contributions by multiple non-specific carbon–sulfur lyase enzymes may be important during winemaking.
In contrast to the growing knowledge of cysteine conjugate release, there have been no detailed studies of polyfunctional thiol release from glutathionated precursors. Based upon equivalent conversion rates, and relative abundance of precursors, it was estimated that up to 20 % of 4MMP was derived from the glutathionated precursor (Roland et al. 2010b). Glutathionylated 3MH can also be released by yeast (Grant-Preece et al. 2010; Roland et al. 2010a), but at lower efficiency than the cysteinylated precursor (Kobayashi et al. 2010; Winter et al. 2011b), the latter estimated to contribute to 3–7 % of the total 3MH found in wine (Subileau et al. 2008b). Nonetheless, this lower conversion of glutathionylated 3MH might be compensated by its high abundance, which in some juices has been reported to be up to 35 times higher than that of Cys-3MH (Capone et al. 2010).
It has been proposed that glutathionated thiol precursors enter the yeast cell via the high affinity glutathione transporter, OPT1, since its deletion resulted in a 2-fold decrease in the formation of 3MH in grape must (Subileau et al. 2008b). Once inside the cell, the mechanism by which the glutathionated thiol precursors are degraded has not been fully elucidated, but is likely to involve a multi-step pathway with the production of the cysteinylated form as an intermediate (Grant-Preece et al. 2010). In support of this, a known carbon–sulfur β-lyase could not directly cleave 3MH from its glutathionylated precursor (Winter et al. 2011b). Such a pathway would be analogous to catabolism of glutathione and of xenobiotic glutathione conjugates, involving sequential degradation of the tripeptide to individual amino acids (Ubiyvovk et al. 2006; Wuenschmann et al. 2010), largely in the vacuole.
Enhanced knowledge of genes involved in polyfunctional thiol precursor uptake and cleavage will provide several new targets for engineering of yeast to enhance varietal flavours. It is also important to note that, as these precursors contain amino acids, transcriptional networks involved in regulation of amino acid metabolism (nitrogen catabolite repression [NCR]) in turn affect polyfunctional thiol release. The abolition/relief of NCR by deleting the transcriptional regulator URE2, results in an increase in the release of both 3MH and 4MMP (Subileau et al. 2008a; Thibon et al. 2008), which was dependent on the presence of an active copy of IRC7 and associated with an up-regulation of the IRC7 transcript (Thibon et al. 2008).
In addition, several observational studies have highlighted natural yeast variation in capacity to release and esterify polyfunctional thiols can be harnessed to modulate wine flavour, for example see Swiegers et al. (2009). Further optimization of polyfunctional thiol release, and formation of the acetate ester of 3MH, has been achieved through co-inoculation of yeast strains and species (Anfang et al. 2009; King et al. 2008; King et al. 2010). It has also been noted that Saccharomyces interspecies hybrid yeast produce relatively high concentrations of polyfunctional thiols (Swiegers et al. 2009). The latter observation may prove particularly useful for development of flavour active wine yeast that produce higher concentrations of positive flavour compounds, whilst producing low levels of H2S (Bizaj et al. 2012).
Future perspectives — new approaches to unravel the yeast flavour phenotype
Targeted development of yeast strains that enhance varietal wine flavours, or contribute to wine complexity, is an endeavour still in its infancy. Strain development has been mainly based on classical strain selection and modification methods, such as variant selection as a result of spontaneous mutations, mutagenesis, and hybridization (see Table 1 for examples). The advantage of these methods is that they do not give rise to products that are included in the statutory definition of genetically modified organisms (GMOs). On the other hand, these methods are not specific enough to modify wine yeast in a well-controlled manner, and they might improve some of the properties of the yeast strain, while compromising other desired traits. The use of recombinant DNA technology and genetic engineering offers the possibility to change specific properties of a yeast strain (reviewed by Carrascosa et al. 2011), but the resulting strain is a GMO. When compared with efforts to engineer other traits into S. cerevisiae, it is clear that only a handful of modifications have been made to yeast to specifically alter production of flavour-active metabolites (Table 1).
Table 1.
Examples of flavour-active yeast strain development for production of saké, beer and wine
| Target flavour compound(s) | Strain | Description of method | Selection/screening | Phenotype | Target gene/enzymatic activity | Reference(s) |
|---|---|---|---|---|---|---|
| Hydrogen sulfide (H2S) | Commercial wine yeast (Maurivin PDM) | Chemical mutagenesis | Screening of H2S production in a plate assay | Low H2S | MET10, MET5 | (Cordente et al. 2009) (Patent PCT/AU08/01485) |
| Commercial and native wine yeasts | Allele swapping (MET10-932) from naturally occurring low-H2S strain | Low H2S | MET10 | (Linderholm et al. 2010) (Patent WO 2008/115759) | ||
| Commercial wine1 and brewing2 yeast | Overexpression of O-acetyl homoserine-O-acetyl serine sulfhydrylase (MET17) | Low H2S | MET17 | (Spiropoulos and Bisson 20001; (Omura et al. 19952) | ||
| Brewing1 and native wine2 yeast | Transformation of cystathionine β-synthase (CYS4) allelic variants | Low H2S | CYS4 | (Tezuka et al. 19921; Linderholm et al. 20062) | ||
| Commercial brewer’s yeast | Inactivation of sulfite reductase | Low H2S, high SO2 | MET10 | (Hansen and Kielland-Brandt 1996) | ||
| Commercial wine yeast (AWRI 1640, AWRI 1116, AWRI 1539) | Interspecific hybridization between a low-H2S S. cerevisiae strain and high-flavour S. cerevisiae x S. kudriavzevii hybrids) | Complementary selectable markers and screening of H2S production in a plate assay | Low H2S, high flavour | MET10 | (Bizaj et al. 2012) | |
| Polyfunctional thiols | Commercial wine yeast (VIN13) | Overexpression of E. coli tryptophanase | Increased 3MH, 4MMP | Cysteine S-conjugate β-lyase | (Swiegers et al. 2007) | |
| Commercial wine yeast (VIN13) | Overexpression of cystathionine β-lyase STR3 | Increased 3MH | STR3 | (Holt et al. 2011) | ||
| Commercial wine yeast (Zymaflore F15) | Overexpression of β-lyase IRC7 | Increased 3MH. 4MMP, 3MHA | IRC7 | (Roncoroni et al. 2011) | ||
| Commercial wine yeast (VIN13) | Overexpression of alcohol acetyltransferase ATF1 | Increased 3MHA | ATF1 | (Swiegers et al. 2006) | ||
| VL3-1D (derived from commercial wine yeast VL3c) | Deletion of nitrogen catabolite repression transcriptional regulator URE2 | Increased 3MH, 4MMP, 3MHA | URE2 | (Thibon et al. 2008) (Patent WO 2008/068635) | ||
| Monoterpenes (de novo biosynthesis) | Haploid laboratory strain | UV mutagenesis | Resistance to nystatin and screening with radioactive mevalonate | Increased geraniol, linalool | ERG20, ERG9 | (Chambon et al. 1990; Chambon et al. 1991) |
| Haploid laboratory strains | Overexpression of geraniol synthase from Ocimum basilicum, and of farnesyl pyrophosphate synthetase ERG20 mutated allele | Increased geraniol, linalool | Monoterpene synthase, ERG20 | (Fischer et al. 2011; Oswald et al. 2007) | ||
| Wine strain T73-4 | Overexpression of linalool synthase from Clarkia breweri, and deletion regulatory region of HMG-CoA reductase (HMG1)* | Increased linalool | Monoterpene synthase, HMG1 | (Herrero et al. 2008; Rico et al. 2010)* | ||
| Higher alcohols | Diploid saké yeast (K30) | Spontaneous mutations | Resistance to leucine analog 5,5,5-trifluoro-dl-leucine | Increased isoamyl alcohol and its acetate | LEU4 | (Oba et al. 2005) |
| Haploid saké yeast (G1101, G1103) | Chemical mutagenesis | Resistance to leucine analog 5,5,5-trifluoro-dl-leucine | Increased isoamyl alcohol and its acetate | LEU4 | (Ashida et al. 1987) | |
| Diploid saké yeast (Kyokai 9) | Chemical mutagenesis | Resistance to phenylalanine analogs (p- and o-fluoro-dl-phenylalanine) | Increased phenylethanol and its acetate | TYR1 (p-analog) ARO4 (o-analog) | (Fukuda et al. 1990a; Fukuda et al. 1990b) | |
| Haploid laboratory strain (BY4741) | Overexpression of rose phenylacetaldehyde synthase | Increased phenylethanol | Phenylpyruvate decarboxylase | (Farhi et al. 2010) | ||
| Commercial wine strain (VIN13) | Overexpression of branched-chain amino acid transaminases BAT1 and BAT2 | Increased isobutanol, isoamyl alcohol | BAT1, BAT2 | (Lilly et al. 2006b) | ||
| Esters | Haploid saké yeast G1103 (derived from Kyokai 7) | Chemical mutagenesis | Resistance to fatty acid synthase (FAS) 2 inhibitor cerulenin | Increased ethyl hexonate and hexanoic acid | FAS2 | (Ichikawa et al. 1991) |
| Diploid saké yeast (Kyokai 7) | Allele swapping (FAS2 G1250S) from cerulenin-resistant yeast (self-cloning) | Increased ethyl hexonate and hexanoic acid | FAS2 | (Aritomi et al. 2004) | ||
| Diploid saké yeast (Kyokai 901) | Chemical mutagenesis | Screening of flavour profile of bank of mutants | Increased isoamyl acetate or ethyl hexanoate | (Arikawa et al. 2000) | ||
| Saké yeast 2NF | Spontaneous mutations | Resistance to farnesol analog 1-farnesylpyridinium1 or to copper2 | Increased isoamyl acetate | Increased alcohol acetyltransferase activity | (Hirooka et al. 2005)1; (Hirooka et al. 2010)2 | |
| Commercial wine strains (VIN13, VIN7)1 Commercial lager strain (CMBS33)2 | Overexpression alcohol acetyltransferases ATF1 and ATF2 | Increased acetate esters | ATF1, ATF2 | (Lilly et al. 2000)1; (Lilly et al. 2006a)1; (Verstrepen et al. 2003)2 | ||
| Diploid saké yeast (Kyokai 7) | Overexpression alcohol acetyltransferase ATF1(self-cloning) | Increased isoamyl acetate | ATF1 | (Hirosawa et al. 2004) |
This is partly due to the diversity of ‘flavour’ phenotypes and highly specialized analytical techniques required to objectively measure chemical targets linked to them. Consequently, few large-scale systematic studies have been performed to identify gene targets for modification. Indeed, screening of entire yeast deletion libraries for mutations affecting flavour compound formation has, thus far, only been applied to H2S production (Linderholm et al. 2008). Higher alcohol formation was probed using a targeted subset of deletion strains by Styger et al. (2011a), due to the limitations imposed by chemical analysis. Further development in the area of high throughput metabolite analyses, coupled with fermentation miniaturization (Liccioli et al. 2011) will be required before broader studies are likely to be performed. Similarly, while a proven approach to map the genetic variation corresponding to phenotypic variation in S. cerevisiae, including wine yeast phenotypes (Ambroset et al. 2011), quantitative trait loci (QTL) mapping has seen limited application in understanding complex ‘flavour phenotypes’ involving multiple flavour compounds. QTL guided breeding has been successfully applied to minimize acetic acid and H2S production, and the release of volatile phenols from odourless precursors (Marullo et al. 2006, 2007), highlighting the potential of this approach.
With the development of low-cost, high-throughput DNA sequencing technologies, the genomes of several wine yeasts have become available (Borneman et al. 2008, 2011, 2012; Novo et al. 2009). Comparative genomic studies of yeast strains have already shown that not only there is a substantial nucleotide variation within the S. cerevisiae species, but also the presence of several regions of DNA that are specific and are predicted to encode proteins that are unique to certain industrial strains. An example are the presence of novel aryl-alcohol dehydrogenase (AAD) proteins in the wine strain AWRI796, when compared with other wine strains, which may have a direct impact on the production of higher alcohols and other flavour compounds during fermentation (Borneman et al. 2011). It is likely, though, that ‘flavour phenotype’ variation amongst wine yeast strains will be determined by differences in transcriptional network regulation. It has been proposed that some of the primary evolutionary targets of strain diversification are transcription factors and their binding sites (Dermitzakis and Clark 2002). Data show that although S. cerevisiae and Saccharomyces mikatae have similar genome sequences, they are significantly different in their transcription factor binding profiles (Borneman et al. 2007a, b). Recent studies have provided some insight into transcriptional networks involved in flavour compound formation (Rossouw et al. 2008, 2009) by wine yeast, and it was shown that the metabolic phenotype of a strain can be shifted by changing expression levels of individual (key) transcription factors (Rossouw et al. 2012).
Increased availability of genome sequences in combination with QTL studies will also reveal allelic variants of genes known to be involved in flavour compound formation, that may explain variation in ‘flavour phenotypes’ amongst wine yeast strains. Hydrogen sulfide formation by wine yeast was recently linked to allelic variants of MET5 and MET10 (Cordente et al. 2009; Linderholm et al. 2010), while formation of the polyfunctional thiol 4MMP relies upon an apparently rare allele of IRC7 (Roncoroni et al. 2011). Investigation of the impact known allelic flavour gene variants (Linderholm et al. 2006; Linderholm et al. 2010) have upon ‘flavour phenotypes’, while extending the search for flavour-active alleles beyond S. cerevisiae, has the potential to greatly expand the toolkit of synthetic biologists and provide options for multi-yeast starter cultures. Recent examples where flavour-impact of non-S. cerevisiae wine yeast have been evaluated include S. bayanus (Masneuf-Pomarede et al. 2010), Torulaspora delbruekii (Renault et al. 2009) and Pichia kluyverii (Anfang et al. 2009). As the genomes of non-Saccharomyces wine yeasts are sequenced and made available, the understanding of the global wine metabolic network will provide wine yeast strain developers with a broader range of options to confer desirable ‘flavour phenotypes’.
Indeed, looking over the horizon at emerging technologies and how they might impact future strain development strategies, it may soon be possible to bring all ‘flavour-active’ genes, or interesting alleles from diverse species, together in a single ‘re-programmed’ yeast strain. Recently, a chemically synthesized chromosome of the bacterium Mycoplasma mycoides — all 1.08 mega basepairs of its DNA — was successfully transplanted into a closely related bacterial cell, Mycoplasma capricolum (Gibson et al. 2010). This marked a world-first: a ‘synthetic’ genome, created in silico, giving life to another living organism with no ancestor. The emerging field of synthetic biology is revolutionizing biotechnology, providing the means to systematically reprogram the genetic makeup of biological systems using ‘off-the-shelf’ functional genetic modules. Within such a context, wine yeast ‘flavour phenotypes’ could be effectively uncoupled, making it possible to develop yeast strains that produce wines with flavour profiles that are difficult to achieve currently — or indeed provide the means to rapidly develop new wine styles. Pending societal acceptance of wines made using GM organisms, the potential for future advances will be limited only by knowledge of flavour compounds and their formation.
Acknowledgments
Research at the Australian Wine Research Institute (AWRI) is financially supported by Australia’s grapegrowers and winemakers through their investment body, the Grape and Wine Research and Development Corporation, with matching funds from the Australian Government. Systems biology research at the AWRI uses resources provided as part of the National Collaborative Research Infrastructure Strategy (NCRIS), an initiative of the Australian Government, in addition to funds from the South Australian State Government. Our collaborating partners within this NCRIS-funded initiative — which is overseen by Bioplatforms Australia — are Genomics Australia, Proteomics Australia, Metabolomics Australia and Bioinformatics Australia. Some AWRI yeast strain development research projects have been funded by yeast supplier companies; Anchor Yeast, AB Mauri, Laffort and Lallemand. The AWRI is part of the Wine Innovation Cluster in Adelaide. This paper draws upon results from several AWRI research projects and publications; therefore, the authors gratefully acknowledge the contributions from several past and present colleagues, particularly, Paul Chambers, Paul Henschke, Anthony Borneman, Simon Schmidt, Hentie Swiegers, Jenny Bellon and Robyn Kievit.
Open Access
This article is distributed under the terms of the Creative Commons Attribution License which permits any use, distribution, and reproduction in any medium, provided the original author(s) and the source are credited.
References
- Acree TE, Sonoff EP, Splittst DF. Effect of yeast strain and type of sulfur compound on hydrogen sulfide production. Am J Enol Vitic. 1972;23:6–9. [Google Scholar]
- Ambroset C, Petit M, Brion C, Sanchez I, Delobel P, Guerin C, Chiapello H, Nicolas P, Bigey F, Dequin S, Blondin B (2011) Deciphering the molecular basis of wine yeast fermentation traits using a combined genetic and genomic approach. G3 1:263–81 [DOI] [PMC free article] [PubMed]
- Anfang N, Brajkovich M, Goddard M. Co-fermentation with Pichia kluyveri increases varietal thiol concentrations in Sauvignon Blanc. Aust J Grape Wine Res. 2009;15:1–8. doi: 10.1111/j.1755-0238.2008.00031.x. [DOI] [Google Scholar]
- Arikawa Y, Yamada M, Shimosaka M, Okazaki M, Fukuzawa M. Isolation of saké yeast mutants producing a high level of ethyl caproate and/or isoamyl acetate. J Biosci Bioeng. 2000;90:675–677. doi: 10.1263/jbb.90.675. [DOI] [PubMed] [Google Scholar]
- Aritomi K, Hirosawa I, Hoshida H, Shiigi M, Nishizawa Y, Kashiwagi S, Akada R. Self-cloning yeast strains containing novel FAS2 mutations produce a higher amount of ethyl caproate in Japanese saké. Biosci Biotech Bioch. 2004;68:206–214. doi: 10.1271/bbb.68.206. [DOI] [PubMed] [Google Scholar]
- Armada L, Fernandez E, Falque E. Influence of several enzymatic treatments on aromatic composition of white wines. LWT-Food Sci Technol. 2010;43:1517–1525. doi: 10.1016/j.lwt.2010.06.009. [DOI] [Google Scholar]
- Ashida S, Ichikawa E, Suginami K, Imayasu S. Isolation and application of mutants producing sufficient isoamyl acetate, a sake flavor component. Agric Biol Chem. 1987;51:2061–2065. doi: 10.1271/bbb1961.51.2061. [DOI] [Google Scholar]
- Bizaj E, Cordente AG, Bellon JR, Raspor P, Curtin CD, Pretorius IS. A breeding strategy to harness flavor diversity of Saccharomyces interspecific hybrids and minimize hydrogen sulfide production. FEMS Yeast Res. 2012;12:456–65. doi: 10.1111/j.1567-1364.2012.00797.x. [DOI] [PubMed] [Google Scholar]
- Blanchard L, Karst F. Characterization of a lysine-to-glutamic acid mutation in a conservative sequence of farnesyl diphosphate synthase from Saccharomyces cerevisiae. Gene. 1993;125:185–189. doi: 10.1016/0378-1119(93)90326-X. [DOI] [PubMed] [Google Scholar]
- Blanchard L, Darriet P, Dubourdieu D. Reactivity of 3-mercaptohexanol in red wine: Impact of oxygen, phenolic fractions, and sulfur dioxide. Am J Enol Vitic. 2004;55:115–120. [Google Scholar]
- Borneman AR, Gianoulis TA, Zhang ZD, Yu H, Rozowsky J, Seringhaus MR, Wang LY, Gerstein M, Snyder M. Divergence of transcription factor binding sites across related yeast species. Science. 2007;317:815–819. doi: 10.1126/science.1140748. [DOI] [PubMed] [Google Scholar]
- Borneman AR, Zhang ZD, Rozowsky J, Seringhaus MR, Gerstein M, Snyder M. Transcription factor binding site identification in yeast: a comparison of high-density oligonucleotide and PCR-based microarray platforms. Funct Integr Genomic. 2007;7:335–345. doi: 10.1007/s10142-007-0054-7. [DOI] [PubMed] [Google Scholar]
- Borneman AR, Forgan AH, Pretorius IS, Chambers PJ. Comparative genome analysis of a Saccharomyces cerevisiae wine strain. FEMS Yeast Res. 2008;8:1185–1195. doi: 10.1111/j.1567-1364.2008.00434.x. [DOI] [PubMed] [Google Scholar]
- Borneman AR, Desany BA, Riches D, Affourtit JP, Forgan AH, Pretorius IS, Egholm M, Chambers PJ (2011) Whole-genome comparison reveals novel genetic elements that characterize the genome of industrial strains of Saccharomyces cerevisiae. Plos Genet 7 [DOI] [PMC free article] [PubMed]
- Borneman AR, Desany BA, Riches D, Affourtit JP, Forgan AH, Pretorius IS, Egholm M, Chambers PJ. The genome sequence of the wine yeast VIN7 reveals an allotriploid hybrid genome with Saccharomyces cerevisiae and Saccharomyces kudriavzevii origins. FEMS Yeast Res. 2012;12:88–96. doi: 10.1111/j.1567-1364.2011.00773.x. [DOI] [PubMed] [Google Scholar]
- Capone DL, Sefton MA, Hayasaka Y, Jeffery DW. Analysis of precursors to wine odorant 3-mercaptohexan-1-ol using HPLC-MS/MS: resolution and quantitation of diastereomers of 3-S-cysteinylhexan-1-ol and 3-S-glutathionylhexan-1-ol. J Agr Food Chem. 2010;58:1390–1395. doi: 10.1021/jf903720w. [DOI] [PubMed] [Google Scholar]
- Carrascosa A, Muñoz R, Gonzalez R (2011) Molecular wine microbiology. Academic Press, London, UK
- Carrau FM, Medina K, Boido E, Farina L, Gaggero C, Dellacassa E, Versini G, Henschke PA. De novo synthesis of monoterpenes by Saccharomyces cerevisiae wine yeasts. FEMS Microbiol Lett. 2005;243:107–115. doi: 10.1016/j.femsle.2004.11.050. [DOI] [PubMed] [Google Scholar]
- Casalone E, Fia G, Barberio C, Cavalieri D, Turbanti L, Polsinelli M. Genetic and biochemical characterization of Saccharomyces cerevisiae mutants resistant to trifluoroleucine. Res Microbiol. 1997;148:613–623. doi: 10.1016/S0923-2508(97)88085-0. [DOI] [PubMed] [Google Scholar]
- Chambers PJ, Pretorius IS. Fermenting knowledge: the history of winemaking, science and yeast research. EMBO Rep. 2010;11:914–920. doi: 10.1038/embor.2010.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambon C, Ladeveze V, Oulmouden A, Servouse M, Karst F. Isolation and properties of yeast mutants affected in farnesyl diphosphate synthetase. Curr Genet. 1990;18:41–46. doi: 10.1007/BF00321113. [DOI] [PubMed] [Google Scholar]
- Chambon C, Ladeveze V, Servouse M, Blanchard L, Javelot C, Vladescu B, Karst F. Sterol pathway in yeast. Identification and properties of mutant strains defective in mevalonate diphosphate decarboxylase and farnesyl diphosphate synthetase. Lipids. 1991;26:633–636. doi: 10.1007/BF02536428. [DOI] [PubMed] [Google Scholar]
- Chen E. The relative contribution of Ehrlich and biosynthetic pathways to the formation of fusel alcohols. J Am Soc Brew Chem. 1978;36:39–43. [Google Scholar]
- Chen X, Nielsen KF, Borodina I, Kielland-Brandt MC, Karhumaa K. Increased isobutanol production in Saccharomyces cerevisiae by overexpression of genes in valine metabolism. Biotechnol Biofuels. 2011;4:21. doi: 10.1186/1754-6834-4-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coetzee C, du Toit WJ. A comprehensive review on Sauvignon blanc aroma with a focus on certain positive volatile thiols. Food Res Int. 2012;45:287–298. doi: 10.1016/j.foodres.2011.09.017. [DOI] [Google Scholar]
- Colon M, Hernandez F, Lopez K, Quezada H, Gonzalez J, Lopez G, Aranda C, Gonzalez A (2011) Saccharomyces cerevisiae Bat1 and Bat2 aminotransferases have functionally diverged from the ancestral-like Kluyveromyces lactis orthologous enzyme. Plos One 6 [DOI] [PMC free article] [PubMed]
- Cordente AG, Swiegers JH, Hegardt FG, Pretorius IS. Modulating aroma compounds during wine fermentation by manipulating carnitine acetyltransferases in Saccharomyces cerevisiae. FEMS Microbiol Lett. 2007;267:159–166. doi: 10.1111/j.1574-6968.2006.00548.x. [DOI] [PubMed] [Google Scholar]
- Cordente AG, Heinrich A, Pretorius IS, Swiegers JH. Isolation of sulfite reductase variants of a commercial wine yeast with significantly reduced hydrogen sulfide production. FEMS Yeast Res. 2009;9:446–459. doi: 10.1111/j.1567-1364.2009.00489.x. [DOI] [PubMed] [Google Scholar]
- Darriet P, Tominaga T, Lavigne V, Boidron J-N, Dubourdieu D. Identification of a powerful aromatic component of Vitis vinifera L. var. Sauvignon wines: 4-mercapto-4-methylpentan-2-one. Flavour Fragr J. 1995;10:385–392. doi: 10.1002/ffj.2730100610. [DOI] [Google Scholar]
- de Souza APG, Vicente MA, Klein RC, Fietto LG, Coutrim MX, Afonso RJCF, Araujo LD, Silva PHA, Bouillet LEM, Castro IM, Brandao RL. Strategies to select yeast starters cultures for production of flavor compounds in cachaca fermentations. Antoine Van Leeuw. 2012;101:379–392. doi: 10.1007/s10482-011-9643-5. [DOI] [PubMed] [Google Scholar]
- Dermitzakis ET, Clark AG. Evolution of transcription factor binding sites in mammalian gene regulatory regions: conservation and turnover. Mol Biol Evol. 2002;19:1114–1121. doi: 10.1093/oxfordjournals.molbev.a004169. [DOI] [PubMed] [Google Scholar]
- Dickinson JR, Lanterman MM, Danner DJ, Pearson BM, Sanz P, Harrison SJ, Hewlins MJE. A 13C nuclear magnetic resonance investigation of the metabolism of leucine to isoamyl alcohol in Saccharomyces cerevisiae. J Biol Chem. 1997;272:26871–26878. doi: 10.1074/jbc.272.43.26871. [DOI] [PubMed] [Google Scholar]
- Dickinson JR, Eshantha L, Salgado J, Hewlins MJE. The catabolism of amino acids to long chain and complex alcohols in Saccharomyces cerevisiae. J Biol Chem. 2003;278:8028–8034. doi: 10.1074/jbc.M211914200. [DOI] [PubMed] [Google Scholar]
- Dubourdieu D, Torninaga T, Masneuf I, Des Gachons CP, Murat ML. The role of yeasts in grape flavor development during fermentation: the example of Sauvignon blanc. Am J Enol Vitic. 2006;57:81–88. [Google Scholar]
- Dudareva N, Cseke L, Blanc VM, Pichersky E. Evolution of floral scent in Clarkia: novel patterns of S-linalool synthase gene expression in the C. breweri flower. Plant Cell. 1996;8:1137–1148. doi: 10.1105/tpc.8.7.1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eden A, Simchen G, Benvenisty N. Two yeast homologs of ECA39, a target for c-Myc regulation, code for cytosolic and mitochondrial branched-chain amino acid aminotransferases. J Biol Chem. 1996;271:20242–20245. doi: 10.1074/jbc.271.34.20242. [DOI] [PubMed] [Google Scholar]
- Eden A, Van Nedervelde L, Drukker M, Benvenisty N, Debourg A. Involvement of branched-chain amino acid aminotransferases in the production of fusel alcohols during fermentation in yeast. Appl Microbiol Biotechnol. 2001;55:296–300. doi: 10.1007/s002530000506. [DOI] [PubMed] [Google Scholar]
- Ehrlich F. Uber das natürliche isomere des leucins. Ber Dtsch Chem Ges. 1904;37:1809–1840. doi: 10.1002/cber.19040370295. [DOI] [Google Scholar]
- Farhi M, Lavie O, Masci T, Hendel-Rahmanim K, Weiss D, Abeliovich H, Vainstein A. Identification of rose phenylacetaldehyde synthase by functional complementation in yeast. Plant Mol Biol. 2010;72:235–245. doi: 10.1007/s11103-009-9564-0. [DOI] [PubMed] [Google Scholar]
- Farhi M, Marhevka E, Masci T, Marcos E, Eyal Y, Ovadis M, Abeliovich H, Vainstein A. Harnessing yeast subcellular compartments for the production of plant terpenoids. Metab Eng. 2011;13:474–481. doi: 10.1016/j.ymben.2011.05.001. [DOI] [PubMed] [Google Scholar]
- Fedrizzi B, Pardon KH, Sefton MA, Elsey GM, Jeffery DW. First identification of 4-S-glutathionyl-4-methylpentan-2-one, a potential precursor of 4-mercapto-4-methylpentan-2-one, in Sauvignon blanc juice. J Agr Food Chem. 2009;57:991–995. doi: 10.1021/jf802799w. [DOI] [PubMed] [Google Scholar]
- Fernandez E, Cortes SM, Castro M, Gil M, Gil ML (1999). Distribution of free and glycosidically bound mono-terpenes and norisoprenoides in the skin and pulp of Albariño grapes during 1998 maduration. 6◦ Symposium International d’Oenologie, Paris, TEC & DOC:161–164
- Fernandez-Gonzalez M, Di Stefano R, Briones A. Hydrolysis and transformation of terpene glycosides from muscat must by different yeast species. Food Microbiol. 2003;20:35–41. doi: 10.1016/S0740-0020(02)00105-3. [DOI] [Google Scholar]
- Fischer MJC, Meyer S, Claudel P, Bergdoll M, Karst F. Metabolic engineering of monoterpene synthesis in yeast. Biotechnol Bioeng. 2011;108:1883–1892. doi: 10.1002/bit.23129. [DOI] [PubMed] [Google Scholar]
- Flipphi MJA, Vanheuvel M, Vanderveen P, Visser J, Degraaff LH. Cloning and characterization of the abfB gene coding for the major α-l-arabinofuranosidase of Aspergillus niger. Curr Genet. 1993;24:525–532. doi: 10.1007/BF00351717. [DOI] [PubMed] [Google Scholar]
- Ford G, Ellis EM. Characterization of Ypr1p from Saccharomyces cerevisiae as a 2-methylbutyraldehyde reductase. Yeast. 2002;19:1087–1096. doi: 10.1002/yea.899. [DOI] [PubMed] [Google Scholar]
- Francis IL, Newton JL. Determining wine aroma from compositional data. Aust J Grape Wine Res. 2005;11:114–126. doi: 10.1111/j.1755-0238.2005.tb00283.x. [DOI] [Google Scholar]
- Fukuda K, Watanabe M, Asano K. Altered regulation of aromatic amino acid biosynthesis in p-phenethyl-alcohol overproducing mutants of sake yeast Saccharomyces cerevisiae. Agr Biol Chem. 1990;54:3151–3156. doi: 10.1271/bbb1961.54.3151. [DOI] [Google Scholar]
- Fukuda K, Watanabe M, Asano K, Ueda H, Ohta S. Breeding of brewing yeast producing a large amount of β-phenylethyl alcohol and β-phenylethyl acetate. Agr Biol Chem. 1990;54:269–271. doi: 10.1271/bbb1961.54.269. [DOI] [Google Scholar]
- Fukuda K, Yamamoto N, Kiyokawa Y, Yanagiuchi T, Wakai Y, Kitamoto K, Inoue Y, Kimura A. Brewing properties of sake yeast whose EST2 gene encoding isoamyl acetate-hydrolyzing esterase was disrupted. J Ferrment Bioeng. 1998;85:101–106. doi: 10.1016/S0922-338X(97)80362-5. [DOI] [Google Scholar]
- Gamero A, Hernandez-Orte P, Querol A, Ferreira V. Effect of aromatic precursor addition to wine fermentations carried out with different Saccharomyces species and their hybrids. Int J Food Microbiol. 2011;147:33–44. doi: 10.1016/j.ijfoodmicro.2011.02.035. [DOI] [PubMed] [Google Scholar]
- Ganga MA, Pinaga F, Valles S, Ramon D, Querol A. Aroma improving in microvinification processes by the use of a recombinant wine yeast strain expressing the Aspergillus nidulans xlnA gene. Int J Food Microbiol. 1999;47:171–178. doi: 10.1016/S0168-1605(98)00202-5. [DOI] [PubMed] [Google Scholar]
- Genoves S, Gil JV, Manzanares P, Aleixandre JL, Valles S. Candida molischiana β-glucosidase production by Saccharomyces cerevisiae and its application in winemaking. J Food Sci. 2003;68:2096–2100. doi: 10.1111/j.1365-2621.2003.tb07025.x. [DOI] [Google Scholar]
- Gibson DG, Glass JI, Lartigue C, Noskov VN, Chuang RY, Algire MA, Benders GA, Montague MG, Ma L, Moodie MM, Merryman C, Vashee S, Krishnakumar R, Assad-Garcia N, Andrews-Pfannkoch C, Denisova EA, Young L, Qi ZQ, Segall-Shapiro TH, Calvey CH, Parmar PP, Hutchison CA, Smith HO, Venter JC. Creation of a bacterial cell controlled by a chemically synthesized genome. Science. 2010;329:52–56. doi: 10.1126/science.1190719. [DOI] [PubMed] [Google Scholar]
- Gil JV, Manzanares P, Genoves S, Valles S, Gonzalez-Candelas L. Over-production of the major exoglucanase of Saccharomyces cereviside leads to an increase in the aroma of wine. Int J Food Microbiol. 2005;103:57–68. doi: 10.1016/j.ijfoodmicro.2004.11.026. [DOI] [PubMed] [Google Scholar]
- Giudici P, Kunkee RE. The effect of nitrogen deficiency and sulfur-containing amino acids on the reduction of sulfate to hydrogen sulfide by wine yeasts. Am J Enol Vitic. 1994;45:107–112. [Google Scholar]
- Goodey AR, Tubb RS. Genetic and biochemical analysis of the ability of Saccharomyces cerevisiae to decarboxylate cinnamic acids. J Gen Microbiol. 1982;128:2615–2620. [Google Scholar]
- Grabinska K, Palamarczyk G. Dolichol biosynthesis in the yeast Saccharomyces cerevisiae: an insight into the regulatory role of farnesyl diphosphate synthase. FEMS Yeast Res. 2002;2:259–265. doi: 10.1016/S1567-1356(02)00110-1. [DOI] [PubMed] [Google Scholar]
- Grant-Preece PA, Pardon KH, Capone DL, Cordente AG, Sefton MA, Jeffery DW, Elsey GM. Synthesis of wine thiol conjugates and labeled analogues: fermentation of the glutathione conjugate of 3-mercaptohexan-1-ol yields the corresponding cysteine conjugate and free thiol. J Agr Food Chem. 2010;58:1383–1389. doi: 10.1021/jf9037198. [DOI] [PubMed] [Google Scholar]
- Gunata Z, Bitteur S, Brillouet JM, Bayonove C, Cordonnier R. Sequential enzymic-hydrolysis of potentially aromatic glycosides from grape. Carbohydr Res. 1988;184:139–149. doi: 10.1016/0008-6215(88)80012-0. [DOI] [Google Scholar]
- Hansen J, Kielland-Brandt MC. Inactivation of MET10 in brewer's yeast specifically increases SO2 formation during beer production. Nat Biotechnol. 1996;14:1587–1591. doi: 10.1038/nbt1196-1587. [DOI] [PubMed] [Google Scholar]
- Hauser M, Horn P, Tournu H, Hauser NC, Hoheisel JD, Brown AJP, Dickinson JR. A transcriptome analysis of isoamyl alcohol-induced filamentation in yeast reveals a novel role for Gre2p as isovaleraldehyde reductase. FEMS Yeast Res. 2007;7:84–92. doi: 10.1111/j.1567-1364.2006.00151.x. [DOI] [PubMed] [Google Scholar]
- Hazelwood LA, Daran JM, van Maris AJA, Pronk JT, Dickinson JR. The Ehrlich pathway for fusel alcohol production: a century of research on Saccharomyces cerevisiae metabolism. Appl Environ Microb. 2008;74:2259–2266. doi: 10.1128/AEM.02625-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heard GM, Fleet GH. Growth of natural yeast flora during the fermentation of inoculated wines. Appl Environ Microb. 1985;50:727–728. doi: 10.1128/aem.50.3.727-728.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henschke P, Jiranek V (1993) Yeast growth during fermentation. In: Fleet G (ed) Wine microbiology and biotechnology. Harwood Academic Publishers, Chur, Switzerland, pp 27–54
- Hernandez LF, Espinosa JC, Fernandez-Gonzalez M, Briones A. β-Glucosidase activity in a Saccharomyces cerevisiae wine strain. Int J Food Microbiol. 2003;80:171–176. doi: 10.1016/S0168-1605(02)00149-6. [DOI] [PubMed] [Google Scholar]
- Hernandez-Orte P, Cersosimo M, Loscos N, Cacho J, Garcia-Moruno E, Ferreira V. The development of varietal aroma from non-floral grapes by yeasts of different genera. Food Chem. 2008;107:1064–1077. doi: 10.1016/j.foodchem.2007.09.032. [DOI] [Google Scholar]
- Herrero O, Ramon D, Orejas M. Engineering the Saccharomyces cerevisiae isoprenoid pathway for de novo production of aromatic monoterpenes in wine. Metab Eng. 2008;10:78–86. doi: 10.1016/j.ymben.2007.11.001. [DOI] [PubMed] [Google Scholar]
- Hirooka K, Yamamoto Y, Tsutsui N, Tanaka T. Improved production of isoamyl acetate by a sake yeast mutant resistant to an isoprenoid analog and its dependence on alcohol acetyltransferase activity, but not on isoamyl alcohol production. J Biosci Bioeng. 2005;99:125–129. doi: 10.1263/jbb.99.125. [DOI] [PubMed] [Google Scholar]
- Hirooka K, Ogita A, Fujita K-I, Yamamoto Y, Tanaka T. Isolation of a copper-resistant saké yeast mutant with improved flavour compound production. J Inst Brewing. 2010;116:261–264. doi: 10.1002/j.2050-0416.2010.tb00429.x. [DOI] [Google Scholar]
- Hirosawa I, Aritomi K, Hoshida H, Kashiwagi S, Nishizawa Y, Akada R. Construction of a self-cloning sake yeast that overexpresses alcohol acetyltransferase gene by a two-step gene replacement protocol. Appl Microbiol Biotechnol. 2004;65:68–73. doi: 10.1007/s00253-004-1563-0. [DOI] [PubMed] [Google Scholar]
- Holt S, Cordente AG, Williams SJ, Capone DL, Jitjaroen W, Menz IR, Curtin C, Anderson PA. Engineering Saccharomyces cerevisiae to release 3-mercaptohexan-1-ol during fermentation through overexpression of an S. cerevisiae gene, STR3, for improvement of wine aroma. Appl Environ Microbiol. 2011;77:3626–3632. doi: 10.1128/AEM.03009-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howell KS, Klein M, Swiegers JH, Hayasaka Y, Elsey GM, Fleet GH, Høj PB, Pretorius IS, de Barros Lopes MA. Genetic determinants of volatile-thiol release by Saccharomyces cerevisiae during wine fermentation. Appl Environ Microbiol. 2005;71:5420–5426. doi: 10.1128/AEM.71.9.5420-5426.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyma KE, Saerens SM, Verstrepen KJ, Fay JC. Divergence in wine characteristics produced by wild and domesticated strains of Saccharmoyces cerevisiae. FEMS Yeast Res. 2011;11:540–551. doi: 10.1111/j.1567-1364.2011.00746.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichikawa E, Hosokawa N, Hata Y, Abe Y, Suginami K, Imayasu S. Breeding of a saké yeast with improved ethyl caproate productivity. Agr Biol Chem. 1991;55:2153–2154. doi: 10.1271/bbb1961.55.2153. [DOI] [Google Scholar]
- Iijima Y, Gang DR, Fridman E, Lewinsohn E, Pichersky E. Characterization of geraniol synthase from the peltate glands of sweet basil. Plant Physiol. 2004;134:370–379. doi: 10.1104/pp.103.032946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inokoshi J, Tomoda H, Hashimoto H, Watanabe A, Takeshima H, Omura S. Cerulenin-resistant mutants of Saccharomyces cerevisiae with an altered fatty acid synthase gene. Mol Gen Genet. 1994;244:90–96. doi: 10.1007/BF00280191. [DOI] [PubMed] [Google Scholar]
- Iraqui I, Vissers S, Cartiaux M, Urrestarazu A. Characterisation of Saccharomyces cerevisiae ARO8 and ARO9 genes encoding aromatic aminotransferases I and II reveals a new aminotransferase subfamily. Mol Gen Genet. 1998;257:238–248. doi: 10.1007/s004380050644. [DOI] [PubMed] [Google Scholar]
- King ES, Swiegers JH, Travis B, Francis IL, Bastian SEP, Pretorius IS. Coinoculated fermentations using Saccharomyces yeasts affect the volatile composition and sensory properties of Vitis vinifera L. cv. Sauvignon blanc wines. J Agr Food Chem. 2008;56:10829–10837. doi: 10.1021/jf801695h. [DOI] [PubMed] [Google Scholar]
- King ES, Kievit RL, Curtin C, Swiegers JH, Pretorius IS, Bastian SEP, Francis IL. The effect of multiple yeasts co-inoculations on Sauvignon Blanc wine aroma composition, sensory properties and consumer preference. Food Chem. 2010;122:618–626. doi: 10.1016/j.foodchem.2010.03.021. [DOI] [Google Scholar]
- King ES, Johnson TE, Bastian SEP, Osidacz P, Francis IL. Consumer liking of white wines: segmentation using self-reported wine liking and wine knowledge. Int J Wine Bus Res. 2012;24:33–46. doi: 10.1108/17511061211213774. [DOI] [Google Scholar]
- Kispal G, Steiner H, Court DA, Rolinski B, Lill R. Mitochondrial and cytosolic branched-chain amino acid transaminases from yeast, homologs of the myc oncogene-regulated Eca39 protein. J Biol Chem. 1996;271:24458–24464. doi: 10.1074/jbc.271.51.32605. [DOI] [PubMed] [Google Scholar]
- Kobayashi H, Takase H, Kaneko K, Tanzawa F, Takata R, Suzuki S, Konno T. Analysis of S-3-(hexan-1-ol)-glutathione and S-3-(hexan-1-ol)-l-cysteine in Vitis vinifera L. cv. Koshu for aromatic wines. Am J Enol Vitic. 2010;61:176–185. [Google Scholar]
- Kondo T, Tezuka H, Ishii J, Matsuda F, Ogino C, Kondo A. Genetic engineering to enhance the Ehrlich pathway and alter carbon flux for increased isobutanol production from glucose by Saccharomyces cerevisiae. J Biotechnol. 2012;159:32–37. doi: 10.1016/j.jbiotec.2012.01.022. [DOI] [PubMed] [Google Scholar]
- Kumar GR, Ramakrishnan V, Bisson LF. Survey of hydrogen sulfide production in wine strains of Saccharomyces cerevisiae. Am J Enol Vitic. 2010;61:365–371. [Google Scholar]
- Lambrechts MG, Pretorius IS. Yeast and its importance to wine aroma. S Afr J Enol Vitic. 2000;21:97–129. [Google Scholar]
- Larroy C, Pares X, Biosca JA. Characterization of a Saccharomyces cerevisiae NADP(H)-dependent alcohol dehydrogenase (ADHVII), a member of the cinnamyl alcohol dehydrogenase family. Eur J Biochem. 2002;269:5738–5745. doi: 10.1046/j.1432-1033.2002.03296.x. [DOI] [PubMed] [Google Scholar]
- Lattey KA, Bramley BR, Francis IL. Consumer acceptability, sensory properties and expert quality judgements of Australian Cabernet Sauvignon and Shiraz wines. Aust J Grape Wine Res. 2010;16:189–202. doi: 10.1111/j.1755-0238.2009.00069.x. [DOI] [Google Scholar]
- LeClinche F, Pinaga F, Ramon D, Valles S. α-l-Arabinofuranosidases from Aspergillus terreus with potential application in enology: induction, purification, and characterization. J Agr Food Chem. 1997;45:2379–2383. doi: 10.1021/jf970046k. [DOI] [Google Scholar]
- Legras J, Merdinoglu D, Cornuet J, Karst F. Bread, beer and wine: Saccharomyces cerevisiae diversity reflects human history. Mol Ecol. 2007;16:2091–2102. doi: 10.1111/j.1365-294X.2007.03266.x. [DOI] [PubMed] [Google Scholar]
- Legras JL, Erny C, Le Jeune C, Lollier M, Adolphe Y, Demuyter C, Delobel P, Blondin B, Karst F. Activation of two different resistance mechanisms in Saccharomyces cerevisiae upon exposure to octanoic and decanoic acids. Appl Environ Microb. 2010;76:7526–7535. doi: 10.1128/AEM.01280-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liccioli T, Tran TMT, Cozzolino D, Jiranek V, Chambers PJ, Schmidt SA. Microvinification—how small can we go? Appl Microbiol Biot. 2011;89:1621–1628. doi: 10.1007/s00253-010-2992-6. [DOI] [PubMed] [Google Scholar]
- Lilly M, Lambrechts MG, Pretorius IS. Effect of increased yeast alcohol acetyltransferase activity on flavor profiles of wine and distillates. Appl Environ Microbiol. 2000;66:744–753. doi: 10.1128/AEM.66.2.744-753.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lilly M, Bauer FF, Lambrechts MG, Swiegers JH, Cozzolino D, Pretorius IS. The effect of increased yeast alcohol acetyltransferase and esterase activity on the flavour profiles of wine and distillates. Yeast. 2006;23:641–659. doi: 10.1002/yea.1382. [DOI] [PubMed] [Google Scholar]
- Lilly M, Bauer FF, Styger G, Lambrechts MG, Pretorius IS. The effect of increased branched-chain amino acid transaminase activity in yeast on the production of higher alcohols and on the flavour profiles of wine and distillates. FEMS Yeast Res. 2006;6:726–743. doi: 10.1111/j.1567-1364.2006.00057.x. [DOI] [PubMed] [Google Scholar]
- Linderholm A, Olineka T, Hong Y, Bisson LF. Allele diversity among genes of the sulfate reduction pathway in wine strains of Saccharomyces cerevisiae. Am J Enol Vitic. 2006;57:431–440. [Google Scholar]
- Linderholm AL, Findleton CL, Kumar G, Hong Y, Bisson LF. Identification of genes affecting hydrogen sulfide formation in Saccharomyces cerevisiae. Appl Environ Microbiol. 2008;74:1418–1427. doi: 10.1128/AEM.01758-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linderholm A, Dietzel K, Hirst M, Bisson LF. Identification of MET10-932 and characterization as an allele reducing hydrogen sulfide formation in wine strains of Saccharomyces cerevisiae. Appl Environ Microbiol. 2010;76:7699–7707. doi: 10.1128/AEM.01666-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liti G, Carter DM, Moses AM, Warringer J, Parts L, James SA, Davey RP, Roberts IN, Burt A, Koufopanou V, Tsai IJ, Bergman CM, Bensasson D, O'Kelly MJT, van Oudenaarden A, Barton DBH, Bailes E, Ba ANN, Jones M, Quail MA, Goodhead I, Sims S, Smith F, Blomberg A, Durbin R, Louis EJ. Population genomics of domestic and wild yeasts. Nature. 2009;458:337–341. doi: 10.1038/nature07743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louw C, La Grange D, Pretorius IS, van Rensburg P. The effect of polysaccharide-degrading wine yeast transformants on the efficiency of wine processing and wine flavour. J Biotechnol. 2006;125:447–461. doi: 10.1016/j.jbiotec.2006.03.029. [DOI] [PubMed] [Google Scholar]
- Ma J, Lu Q, Yuan Y, Ge H, Li K, Zhao W, Gao Y, Niu L, Teng M. Crystal structure of isoamyl acetate-hydrolyzing esterase from Saccharomyces cerevisiae reveals a novel active site architecture and the basis of substrate specificity. Proteins. 2011;79:662–668. doi: 10.1002/prot.22865. [DOI] [PubMed] [Google Scholar]
- Manzanares P, Orejas M, Gil JV, de Graaff LH, Visser J, Ramon D. Construction of a genetically modified wine yeast strain expressing the Aspergillus aculeatus rhaA gene, encoding an α-l-rhamnosidase of enological interest. Appl Environ Microbiol. 2003;69:7558–7562. doi: 10.1128/AEM.69.12.7558-7562.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin DM, Bohlmann J. Identification of Vitis vinifera (−)-alpha-terpineol synthase by in silico screening of full-length cDNA ESTs and functional characterization of recombinant terpene synthase. Phytochemistry. 2004;65:1223–1229. doi: 10.1016/j.phytochem.2004.03.018. [DOI] [PubMed] [Google Scholar]
- Martin DM, Aubourg S, Schouwey MB, Daviet L, Schalk M, Toub O, Lund ST, Bohlmann J. Functional annotation, genome organization and phylogeny of the grapevine (Vitis vinifera) terpene synthase gene family based on genome assembly, FLcDNA cloning, and enzyme assays. BMC Plant Biol. 2010;10:226. doi: 10.1186/1471-2229-10-226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marullo P, Bely M, Masneuf-Pomarede I, Pons M, Aigle M, Dubourdieu D. Breeding strategies for combining fermentative qualities and reducing off-flavor production in a wine yeast model. FEMS Yeast Res. 2006;6:268–279. doi: 10.1111/j.1567-1364.2006.00034.x. [DOI] [PubMed] [Google Scholar]
- Marullo P, Aigle M, Bely M, Masneuf-Pomarede I, Durrens P, Dubourdieu D, Yvert G. Single QTL mapping and nucleotide-level resolution of a physiologic trait in wine Saccharomyces cerevisiae strains. FEMS Yeast Res. 2007;7:941–952. doi: 10.1111/j.1567-1364.2007.00252.x. [DOI] [PubMed] [Google Scholar]
- Masneuf-Pomarede I, Mansour C, Murat ML, Tominaga T, Dubourdieu D. Influence of fermentation temperature on volatile thiols concentrations in Sauvignon blanc wines. Int J Food Microbiol. 2006;108:385–390. doi: 10.1016/j.ijfoodmicro.2006.01.001. [DOI] [PubMed] [Google Scholar]
- Masneuf-Pomarede I, Bely M, Marullo P, Lonvaud-Funel A, Dubourdieu D. Reassessment of phenotypic traits for Saccharomyces bayanus var. uvarum wine yeast strains. Int J Food Microbiol. 2010;139:79–86. doi: 10.1016/j.ijfoodmicro.2010.01.038. [DOI] [PubMed] [Google Scholar]
- Mateo JJ, Jimenez M. Monoterpenes in grape juice and wines. J Chromatogr A. 2000;881:557–567. doi: 10.1016/S0021-9673(99)01342-4. [DOI] [PubMed] [Google Scholar]
- Meaden PG, Taylor NR. Cloning of a yeast gene which causes phenolic off-flavors in beer. J Inst Brew. 1991;97:353–357. [Google Scholar]
- Mendes-Ferreira A, Mendes-Faia A, Leao C. Survey of hydrogen sulphide production by wine yeasts. J Food Prot. 2002;65:1033–1037. doi: 10.4315/0362-028x-65.6.1033. [DOI] [PubMed] [Google Scholar]
- Mojzita D, Hohmann S. Pdc2 coordinates expression of the THI regulon in the yeast Saccharomyces cerevisiae. Mol Genet Genom. 2006;276:147–161. doi: 10.1007/s00438-006-0130-z. [DOI] [PubMed] [Google Scholar]
- Mueller S, Osidacz P, Francis IL, Lockshin L. Combining discrete choice and informed sensory testing in a two-stage process: can it predict wine market share? Food Qual Prefer. 2010;21:741–754. doi: 10.1016/j.foodqual.2010.06.008. [DOI] [Google Scholar]
- Nosaka K, Onozuka M, Konno H, Kawasaki Y, Nishimura H, Sano M, Akaji K. Genetic regulation mediated by thiamin pyrophosphate-binding motif in Saccharomyces cerevisiae. Mol Microbiol. 2005;58:467–479. doi: 10.1111/j.1365-2958.2005.04835.x. [DOI] [PubMed] [Google Scholar]
- Novo M, Bigey F, Beyne E, Galeote V, Gavory F, Mallet S, Cambon B, Legras J-L, Wincker P, Casaregola S, Dequin S. Eukaryote-to-eukaryote gene transfer events revealed by the genome sequence of the wine yeast Saccharomyces cerevisiae EC1118. Proc Natl Acad Sci USA. 2009;106:16333–16338. doi: 10.1073/pnas.0904673106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowak A, Kusewicz D, Kalinowska H, Turkiewicz M, Patelski P. Production of H2S and properties of sulfite reductase from selected strains of wine-producing yeasts. Eur Food Res Technol. 2004;219:84–89. doi: 10.1007/s00217-004-0885-6. [DOI] [Google Scholar]
- Nykanen L. Formation and occurrence of flavor compounds in wine and distilled alcoholic beverages. Am J Enol Vitic. 1986;37:84–96. [Google Scholar]
- Nykanen L, Nykanen I, Suomalainen H. Distribution of esters produced during sugar fermentation between the yeast cell and the medium. J Inst Brew. 1977;83:32–34. [Google Scholar]
- Oba T, Nomiyama S, Hirakawa H, Tashiro K, Kuhara S. Asp578 in LEU4p is one of the key residues for leucine feedback inhibition release in sake yeast. Biosci Biotechnol Bioch. 2005;69:1270–1273. doi: 10.1271/bbb.69.1270. [DOI] [PubMed] [Google Scholar]
- Omura F, Shibano Y, Fukui N, Nakatani K. Reduction of hydrogen sulfide production in brewing yeast by constitutive expression of MET25 gene. J Am Soc Brew Chem. 1995;53:58–62. [Google Scholar]
- Oswald M, Fischer M, Dirninger N, Karst F. Monoterpenoid biosynthesis in Saccharomyces cerevisiae. FEMS Yeast Res. 2007;7:413–421. doi: 10.1111/j.1567-1364.2006.00172.x. [DOI] [PubMed] [Google Scholar]
- Perez-Gonzalez JA, Gonzalez R, Querol A, Sendra J, Ramon D. Construction of a recombinant wine yeast strain expressing β-(1,4)-endoglucanase and its use in microvinification processes. Appl Environ Microbiol. 1993;59:2801–2806. doi: 10.1128/aem.59.9.2801-2806.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peyrot Des Gachons C, Tominaga T, Dubourdieu D. Sulfur aroma precursor present in S-glutathione conjugate form: identification of S-3-(hexan-1-ol)-glutathione in must from Vitis vinifera L. cv. Sauvignon blanc. J Agr Food Chem. 2002;50:4076–4079. doi: 10.1021/jf020002y. [DOI] [PubMed] [Google Scholar]
- Pretorius IS. Tailoring wine yeast for the new millennium: novel approaches to the ancient art of winemaking. Yeast. 2000;16:675–729. doi: 10.1002/1097-0061(20000615)16:8<675::AID-YEA585>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- Pretorius IS, Bauer FF. Meeting the consumer challenge through genetically customized wine yeast strains. Trends Biotechnol. 2002;20:426–432. doi: 10.1016/S0167-7799(02)02049-8. [DOI] [PubMed] [Google Scholar]
- Pretorius IS, Curtin C, Chambers PJ. The winemaker’s bug: from ancient wisdom to opening new vistas with frontier yeast science. Bioeng Bugs. 2012;3:1–10. doi: 10.4161/bbug.19687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramachandran P, Tiwari MK, Singh RK, Haw J-R, Jeya M, Lee J-K. Cloning and characterization of a putative β-glucosidase (NfBGL595) from Neosartorya fischeri. Process Biochem. 2012;47:99–105. doi: 10.1016/j.procbio.2011.10.015. [DOI] [Google Scholar]
- Rauhut D. Yeasts-production of sulfur compounds. In: Fleet G, editor. Wine microbiology and biotechnology. Chur: Harwood Academic Publishers; 1993. pp. 183–223. [Google Scholar]
- Rauhut D, Kurbel H. The production of H2S from elemental sulfur residues during fermentation and its influence on the formation of sulfur metabolites causing off-flavors in wines. Wein-Wiss. 1994;49:27–36. [Google Scholar]
- Renault P, Miot-Sertier C, Marullo P, Hernández-Orte P, Lagarrigue L, Lonvaud-Funel A, Bely M. Genetic characterization and phenotypic variability in Torulaspora delbrueckii species: potential applications in the wine industry. Int J Food Microbiol. 2009;134:201–210. doi: 10.1016/j.ijfoodmicro.2009.06.008. [DOI] [PubMed] [Google Scholar]
- Rico J, Pardo E, Orejas M. Enhanced production of a plant monoterpene by overexpression of the 3-hydroxy-3-methylglutaryl coenzyme A reductase catalytic domain in Saccharomyces cerevisiae. Appl Environ Microbiol. 2010;76:6449–6454. doi: 10.1128/AEM.02987-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riou C, Salmon JM, Vallier MJ, Gunata Z, Barre P. Purification, characterization, and substrate specificity of a novel highly glucose tolerant β-glucosidase from Aspergillus oryzae. Appl Environ Microbiol. 1998;64:3607–3614. doi: 10.1128/aem.64.10.3607-3614.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson AL, Boss PK, Heymann H, Solomon PS, Trengove RD. Influence of yeast strain, canopy management, and site on the volatile composition and sensory attributes of Cabernet Sauvignon wines from Western Australia. J Agr Food Chem. 2011;59:3273–3284. doi: 10.1021/jf104324d. [DOI] [PubMed] [Google Scholar]
- Roland A, Schneider R, Le Guerneve C, Razungles A, Cavelier F. Identification and quantification by LC-MS/MS of a new precursor of 3-mercaptohexan-1-ol (3MH) using stable isotope dilution assay: elements for understanding the 3MH production in wine. Food Chem. 2010;121:847–855. doi: 10.1016/j.foodchem.2009.12.095. [DOI] [Google Scholar]
- Roland A, Schneider R, Razungles A, Le Guerneve C, Cavelier F. Straightforward synthesis of deuterated precursors to demonstrate the biogenesis of aromatic thiols in wine. J Agr Food Chem. 2010;58:10684–10689. doi: 10.1021/jf101996p. [DOI] [PubMed] [Google Scholar]
- Romano P, Fiore C, Paraggio M, Caruso M, Capece A. Function of yeast species and strains in wine flavour. Int J Food Microbiol. 2003;86:169–180. doi: 10.1016/S0168-1605(03)00290-3. [DOI] [PubMed] [Google Scholar]
- Roncoroni M, Santiago M, Hooks DO, Moroney S, Harsch MJ, Lee SA, Richards KD, Nicolau L, Gardner RC. The yeast IRC7 gene encodes a beta-lyase responsible for production of the varietal thiol 4-mercapto-4-methylpentan-2-one in wine. Food Microbiol. 2011;28:926–935. doi: 10.1016/j.fm.2011.01.002. [DOI] [PubMed] [Google Scholar]
- Rossouw D, Naes T, Bauer FF. Linking gene regulation and the exo-metabolome: a comparative transcriptomics approach to identify genes that impact on the production of volatile aroma compounds in yeast. BMC Genomics. 2008;9:530. doi: 10.1186/1471-2164-9-530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossouw D, Olivares-Hernandes R, Nielsen J, Bauer FF. Comparative transcriptomic approach to investigate differences in wine yeast physiology and metabolism during fermentation. Appl Environ Microbiol. 2009;75:6600–6612. doi: 10.1128/AEM.01251-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossouw D, Jacobson D, Bauer FF. Transcriptional regulation and the diversification of metabolism in wine yeast strains. Genetics. 2012;190:251–261. doi: 10.1534/genetics.111.132720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saerens SMG, Verstrepen KJ, Van Laere SDM, Voet ARD, Van Dijck P, Delvaux FR, Thevelein JM. The Saccharomyces cerevisiae EHT1 and EEB1 genes encode novel enzymes with medium-chain fatty acid ethyl ester synthesis and hydrolysis capacity. J Biol Chem. 2006;281:4446–4456. doi: 10.1074/jbc.M512028200. [DOI] [PubMed] [Google Scholar]
- Saerens SMG, Delvaux F, Verstrepen KJ, Van Dijck P, Thevelein JM, Delvaux FR. Parameters affecting ethyl ester production by Saccharomyces cerevisiae during fermentation. Appl Environ Microbiol. 2008;74:454–461. doi: 10.1128/AEM.01616-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saerens SMG, Delvaux FR, Verstrepen KJ, Thevelein JM. Production and biological function of volatile esters in Saccharomyces cerevisiae. Microbiol Biotechnol. 2010;3:165–177. doi: 10.1111/j.1751-7915.2009.00106.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuller D, Casal M. The use of genetically modified Saccharomyces cerevisiae strains in the wine industry. Appl Microbiol Biotechnol. 2005;68:292–304. doi: 10.1007/s00253-005-1994-2. [DOI] [PubMed] [Google Scholar]
- Siddiqui MS, Thodey K, Trenchard I, Smolke CD. Advancing secondary metabolite biosynthesis in yeast with synthetic biology tools. FEMS Yeast Res. 2012;12:144–170. doi: 10.1111/j.1567-1364.2011.00774.x. [DOI] [PubMed] [Google Scholar]
- Siebert T, Bramley B, Solomon M. Hydrogen sulfide: aroma detection threshold study in red and white wine. AWRI Tech Rev. 2009;183:14–16. [Google Scholar]
- Spiropoulos A, Bisson LF. MET17 and hydrogen sulfide formation in Saccharomyces cerevisiae. Appl Environ Microbiol. 2000;66:4421–4426. doi: 10.1128/AEM.66.10.4421-4426.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spiropoulos A, Tanaka J, Flerianos I, Bisson LF. Characterization of hydrogen sulfide formation in commercial and natural wine isolates of Saccharomyces. Am J Enol Vitic. 2000;51:233–248. [Google Scholar]
- Strauss CR, Wilson B, Gooley PR, Williams PJ. Role of monoterpenes in grape and wine flavor. In: Parliment TH, Croteau R, editors. Biogeneration of aromas. Washington, DC: American Chemical Society; 1986. pp. 222–242. [Google Scholar]
- Styger G, Jacobson D, Bauer FF. Identifying genes that impact on aroma profiles produced by Saccharomyces cerevisiae and the production of higher alcohols. Appl Microbiol Biotechnol. 2011;91:713–730. doi: 10.1007/s00253-011-3237-z. [DOI] [PubMed] [Google Scholar]
- Styger G, Prior B, Bauer FF. Wine flavor and aroma. J Ind Microbiol Biotechnol. 2011;38:1145–1159. doi: 10.1007/s10295-011-1018-4. [DOI] [PubMed] [Google Scholar]
- Subileau M, Schneider R, Salmon J-M, Degryse E. Nitrogen catabolite repression modulates the production of aromatic thiols characteristic of Sauvignon blanc at the level of precursor transport. FEMS Yeast Res. 2008;8:771–780. doi: 10.1111/j.1567-1364.2008.00400.x. [DOI] [PubMed] [Google Scholar]
- Subileau M, Schneider R, Salmon J-M, Degryse E. New insights on 3-mercaptohexanol (3MH) biogenesis in Sauvignon blanc wines: Cys-3MH and (E)-Hexen-2-al are not the major precursors. J Agr Food Chem. 2008;56:9230–9235. doi: 10.1021/jf801626f. [DOI] [PubMed] [Google Scholar]
- Sumby KM, Grbin PR, Jiranek V. Microbial modulation of aromatic esters in wine: current knowledge and future prospects. Food Chem. 2010;121:1–16. doi: 10.1016/j.foodchem.2009.12.004. [DOI] [Google Scholar]
- Swiegers JH, Pretorius IS (2005) Yeast modulation of wine flavor. Advances in Applied Microbiology, pp 131–175 [DOI] [PubMed]
- Swiegers JH, Pretorius IS. Modulation of volatile sulfur compounds by wine yeast. Appl Microbiol Biotechnol. 2007;74:954–960. doi: 10.1007/s00253-006-0828-1. [DOI] [PubMed] [Google Scholar]
- Swiegers JH, Bartowsky EJ, Henschke PA, Pretorius IS. Yeast and bacterial modulation of wine aroma and flavour. Austral J Grape Wine Res. 2005;11:139–173. doi: 10.1111/j.1755-0238.2005.tb00285.x. [DOI] [Google Scholar]
- Swiegers J, Willmott R, Hill-Ling A, Capone D, Pardon K, Elsey G, Howell K, de Barros LM, Sefton M, Lilly M, Pretorius IS. Modulation of volatile thiol and ester aromas in wine by modified wine yeast. In: Bredie W, Petersen M, editors. Developments in food science; Flavour science: recent advances and trends. Amsterdam: Elsevier; 2006. pp. 113–116. [Google Scholar]
- Swiegers JH, Capone DL, Pardon KH, Elsey GM, Sefton MA, Francis IL, Pretorius IS. Engineering volatile thiol release in Saccharomyces cerevisiae for improved wine aroma. Yeast. 2007;24:561–574. doi: 10.1002/yea.1493. [DOI] [PubMed] [Google Scholar]
- Swiegers JH, Kievit RL, Siebert T, Lattey KA, Bramley BR, Francis IL, King ES, Pretorius IS. The influence of yeast on the aroma of Sauvignon blanc wine. Food Microbiol. 2009;26:204–211. doi: 10.1016/j.fm.2008.08.004. [DOI] [PubMed] [Google Scholar]
- Tezuka H, Mori T, Okumura Y, Kitabatake K, Tsumura Y. Cloning of a gene suppressing hydrogen sulfide production by Saccharomyces cerevisiae and its expression in a brewing yeast. J Am Soc Brew Chem. 1992;50:130–133. [Google Scholar]
- Thibon C, Marullo P, Claisse O, Cullin C, Dubourdieu D, Tominaga T. Nitrogen catabolic repression controls the release of volatile thiols by Saccharomyces cerevisiae during wine fermentation. FEMS Yeast Res. 2008;8:1076–1086. doi: 10.1111/j.1567-1364.2008.00381.x. [DOI] [PubMed] [Google Scholar]
- Tokuhiro K, Muramatsu M, Ohto C, Kawaguchi T, Obata S, Muramoto N, Hirai M, Takahashi H, Kondo A, Sakuradani E, Shimizu S. Overproduction of geranylgeraniol by metabolically engineered Saccharomyces cerevisiae. Appl Environ Microbiol. 2009;75:5536–5543. doi: 10.1128/AEM.00277-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tominaga T, Masneuf I, Dubourdieu D. A S-cysteine conjugate, precursor of aroma of white Sauvignon. J Int Sci Vigne Vin. 1995;29:227–232. [Google Scholar]
- Tominaga T, des Gachons CP, Dubourdieu D. A new type of flavor precursors in Vitis vinifera L cv Sauvignon blanc: S-cysteine conjugates. J Agr Food Chem. 1998a;46:5215–5219. doi: 10.1021/jf980481u. [DOI] [Google Scholar]
- Tominaga T, Furrer A, Henry R, Dubourdieu D. Identification of new volatile thiols in the aroma of Vitis vinifera L. var. Sauvignon blanc wines. Flavour Frag J. 1998;13:159–162. doi: 10.1002/(SICI)1099-1026(199805/06)13:3<159::AID-FFJ709>3.0.CO;2-7. [DOI] [Google Scholar]
- Tominaga T, Blanchard L, Darriet P, Dubourdieu D. A powerful aromatic volatile thiol, 2-furanmethanethiol, exhibiting roast coffee aroma in wines made from several Vitis vinifera grape varieties. J Agr Food Chem. 2000;48:1799–1802. doi: 10.1021/jf990660r. [DOI] [PubMed] [Google Scholar]
- Ubeda J, Briones A. Characterization of differences in the formation of volatiles during fermentation within synthetic and grape musts by wild Saccharomyces strains. Lebensm Wiss Technol. 2000;33:408–414. doi: 10.1006/fstl.2000.0680. [DOI] [Google Scholar]
- Ubiyvovk VM, Blazhenko OV, Gigot D, Penninckx M, Sibirny AA. Role of gamma-glutamyltranspeptidase in detoxification of xenobiotics in the yeasts Hansenula polymorpha and Saccharomyces cerevisiae. Cell Biol Int. 2006;30:665–671. doi: 10.1016/j.cellbi.2006.04.006. [DOI] [PubMed] [Google Scholar]
- Ugliano M, Bartowsky EJ, McCarthy J, Moio L, Henschke PA. Hydrolysis and transformation of grape glycosidically bound volatile compounds during fermentation with three Saccharomyces yeast strains. J Agr Food Chem. 2006;54:6322–6331. doi: 10.1021/jf0607718. [DOI] [PubMed] [Google Scholar]
- Ugliano M, Fedrizzi B, Siebert T, Travis B, Magno F, Versini G, Henschke PA. Effect of nitrogen supplementation and Saccharomyces species on hydrogen sulfide and other volatile sulfur compounds in Shiraz fermentation and wine. J Agr Food Chem. 2009;57:4948–4955. doi: 10.1021/jf8037693. [DOI] [PubMed] [Google Scholar]
- Vadali RV, Bennett GN, San KY. Applicability of CoA/acetyl-CoA manipulation system to enhance isoamyl acetate production in Escherichia coli. Metab Eng. 2004;6:294–299. doi: 10.1016/j.ymben.2004.02.006. [DOI] [PubMed] [Google Scholar]
- van Rensburg P, Pretorius IS. Enzymes in winemaking: Harnessing natural catalysts for efficient biotransformations: a review. S Afr J Enol Vitic. 2000;21:52–73. [Google Scholar]
- van Rensburg P, Stidwell T, Lambrechts MG, Otero RC, Pretorius IS. Development and assessment of a recombinant Saccharomyces cerevisiae wine yeast producing two aroma enhancing β-glucosidases encoded by the Saccharomycopsis fibuligera BGL1 and BGL2 genes. Ann Microbiol. 2005;55:33–42. [Google Scholar]
- Varela C, Siebert T, Cozzolino D, Rose L, McLean H, Henschke PA. Discovering a chemical basis for differentiating wines made by fermentation with 'wild' indigenous and inoculated yeasts: role of yeast volatile compounds. Aust J Grape Wine Res. 2009;15:238–248. doi: 10.1111/j.1755-0238.2009.00054.x. [DOI] [Google Scholar]
- Vasserot Y, Arnaud A, Galzy P. Evidence for Muscat marc monoterpenol glucosides hydrolysis by free or immobilized yeast β-glucosidase. Bioresour Technol. 1993;43:269–271. doi: 10.1016/0960-8524(93)90040-I. [DOI] [Google Scholar]
- Versini G, Carlin S, Nicolini G, Dellacassa E, Carrau F. Updating of varietal aroma components in wines. Mendoza: VII Latin. Congress Enol. Vitic; 1999. pp. 325–349. [Google Scholar]
- Verstrepen KJ, Van Laere SDM, Vanderhaegen BMP, Derdelinckx G, Dufour JP, Pretorius IS, Winderickx J, Thevelein JM, Delvaux FR. Expression levels of the yeast alcohol acetyltransferase genes ATF1, Lg-ATF1, and ATF2 control the formation of a broad range of volatile esters. Appl Environ Microbiol. 2003;69:5228–5237. doi: 10.1128/AEM.69.9.5228-5237.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verstrepen KJ, Chambers P, Pretorius IS. The development of superior yeast strains for the food and beverage industries: challenges, opportunities and potential benefits. In: Querol A, Fleet GH, editors. The yeast handbook, yeasts in food and beverages. Heidelberg: Springer-Verlag; 2006. pp. 399–444. [Google Scholar]
- Vicente MD, Fietto LG, Castro ID, dos Santos ANG, Coutrim MX, Brandao RL. Isolation of Saccharomyces cerevisiae strains producing higher levels of flavoring compounds for production of "cachaca" the Brazilian sugarcane spirit. Int J Food Microbiol. 2006;108:51–59. doi: 10.1016/j.ijfoodmicro.2005.10.018. [DOI] [PubMed] [Google Scholar]
- Vos PJA, Gray RS. Origin and control of hydrogen sulfide during fermentation of grape must. Am J Enol Vitic. 1979;30:187–197. [Google Scholar]
- Vuralhan Z, Morais MA, Tai SL, Piper MDW, Pronk JT. Identification and characterization of phenylpyruvate decarboxylase genes in Saccharomyces cerevisiae. Appl Environ Microbiol. 2003;69:4534–4541. doi: 10.1128/AEM.69.8.4534-4541.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang XD, Bohlscheid JC, Edwards CG. Fermentative activity and production of volatile compounds by Saccharomyces grown in synthetic grape juice media deficient in assimilable nitrogen and/or pantothenic acid. J Appl Microbiol. 2003;94:349–359. doi: 10.1046/j.1365-2672.2003.01827.x. [DOI] [PubMed] [Google Scholar]
- Williams PJ, Strauss CR, Wilson B, Dimitriadis E. Recent studies into grape terpene glycosides. In: Adda J, editor. Progress in flavour research. Amsterdam: Elsevier; 1984. pp. 349–357. [Google Scholar]
- Winter G, Henschke PA, Higgins VJ, Ugliano M, Curtin CD. Effects of rehydration nutrients on H2S metabolism and formation of volatile sulfur compounds by the wine yeast VL3. AMB Express. 2011;1:36. doi: 10.1186/2191-0855-1-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter G, van der Westhuizen T, Higgins VJ, Curtin C, Ugliano M. Contribution of cysteine and glutathione conjugates to the formation of the volatile thiols 3-mercaptohexan-1-ol (3MH) and 3-mercaptohexyl acetate (3MHA) during fermentation by Saccharomyces cerevisiae. Aust J Grape Wine Res. 2011;17:285–290. doi: 10.1111/j.1755-0238.2011.00127.x. [DOI] [Google Scholar]
- Wuenschmann J, Krajewski M, Letzel T, Huber EM, Ehrmann A, Grill E, Lendzian KJ. Dissection of glutathione conjugate turnover in yeast. Phytochemistry. 2010;71:54–61. doi: 10.1016/j.phytochem.2009.09.034. [DOI] [PubMed] [Google Scholar]
- Zambonelli C, Soli M, Guerra D. A study of H2S non-producing strains of wine yeasts. Arch Microbiol. 1984;34:7–15. [Google Scholar]
- Zea L, Moreno J, Ortega JM, Medina M. Content of free terpenic compounds in cells and musts during vinification with three Saccharomyces cerevisiae races. J Agr Food Chem. 1995;43:1110–1114. doi: 10.1021/jf00052a046. [DOI] [Google Scholar]
- Zietsman AJJ, de Klerk D, van Rensburg P. Coexpression of α-1-arabinofuranosidase and β-glucosidase in Saccharomyces cerevisiae. FEMS Yeast Res. 2011;11:88–103. doi: 10.1111/j.1567-1364.2010.00694.x. [DOI] [PubMed] [Google Scholar]