Abstract
Background:
Radix Paeoniae Alba, Radix Paeoniae Rubra, and Cortex Moutan are important Chinese herbs. Their bioactivities and efficacies are similar. However, they have different superior benefits clinically; so, a comprehensive investigation of the chemical difference is necessary and is of great importance for more reasonable quality assessment and proper clinical application of these three herbal medicines.
Objective:
To establish a high-performance liquid chromatography (HPLC) fingerprint method for the quality control of Radix Paeonia Alba, Radix Paeonia Rubra, and Cortex Moutan, and to compare their main constituents.
Materials and Methods:
The separations of Radix Paeoniae Alba, Radix Paeoniae Rubra, and Cortex Moutan was carried out, respectively, through a gradient elution using a monolithic column and a mobile phase consisting of water (containing 0.1% formic acid) and acetonitrile (containing 0.1% formic acid) at a flow rate of 5.0 ml/min. The detection wavelength was set at 230 nm. The data calculation was performed with CHROMAP v1.51 and Statistical Package for the Social Sciences (SPSS) 18.0 software for principal component analysis.
Results:
A rapid separation method based on high-performance liquid chromatography with diode-array detection (HPLC-DAD) with monolithic columns and a fingerprint analysis method was established. Fifteen Radix Paeonia Alba, 45 Radix Paeonia Rubra, and 21 Cortex Moutan samples were analyzed and 11 chromatographic peaks were identified. Differences of chromatographic peaks among these three herbal medicines in chemical compositions were revealed.
Conclusion:
The separation and analysis method are fast and simple, which can be used for chemical fingerprint comparison of Radix Paeonia Alba, Radix Paeonia Rubra, and Cortex Moutan. The results for the evaluation of the three medicines could provide experimental evidence for chemical affinity.
Keywords: Cortex Moutan, fingerprint, high performance liquid chromatography coupled, monolithic column, principal component analysis, Radix Paeonia Alba, Radix Paeonia Rubra
INTRODUCTION
Radix Paeoniae Alba (Bai-shao in Chinese), Radix Paeoniae Rubra (Chi-shao in Chinese), and Cortex Moutan (Mu- dan-pi in Chinese) are important Chinese herbs used in many herbal preparations.[1] These three herbal medicines all come from Paeonia: Bai-shao is the steamed and dried root of cultivated P. lactiflora; Chi-shao is the dried root of wild P. lactiflora or P. veitchii; and Mu-dan-pi is the root bark of P. suffruticosa. Some unofficial species from Paeonia are also misused as Chi-shao or Mu-dan-pi in some areas of China. They all mainly contain monoterpenoid glucosides, tannins, phenolic acids, triterpenes, and other substances.[2] However, Chi-shao and Mu-dan-pi contain more tannins due to the retained velamen, and Mu-dan-pi possesses a higher level of paeonol compared with the other two herbal medicines. Because of the similarity in original plants and characteristic chemical compositions, their bioactivities and efficacies are similar as well.[3] They all play a significant role in treating blood diseases. Moreover, they are uniformly bitter in taste and cold in property and enter the liver meridians. Clinically, they have different superior benefits, namely: Bai-shao is superior in nourishing the blood; Chi-shao is superior in invigorating the blood circulation; and Mu-dan-pi is superior in cooling the blood.[1,4] Thus, a comprehensive investigation of the chemical difference is necessary and is of great importance for more reasonable quality assessment and proper clinical application of these three herbal medicines.
Fingerprinting spectral analysis represented by high-performance liquid chromatography (HPLC) has been used as a major tool for the quality assurance of crude drugs and compound preparations.[5,6] Several chromatographic fingerprint analytical methods have been developed for the qualitative or quantitative evaluation of Bai-shao[7,8] and Mu-dan-pi,[9,10] respectively, or for the comparison of Bai-shao and Chi-shao.[11–15] However, traditional Chinese medicine (TCM) views multi-compound, multi-ingredient preparations as its features representing the activity of the herbal drugs. Selection of individual or a few analytical compounds for evaluating either efficacy or quality is contrary to the principles of TCM.[5] To date, no simultaneous chemical fingerprint analysis of these three herbals has been conducted. In this research, a simple and quick HPLC method for simultaneous comparison of Radix Paeonia Alba, Radix Paeonia Rubra, and Cortex Moutan using HPLC with monolithic columns and their chemical pattern recognition was established.
MATERIALS AND METHODS
Plant materials
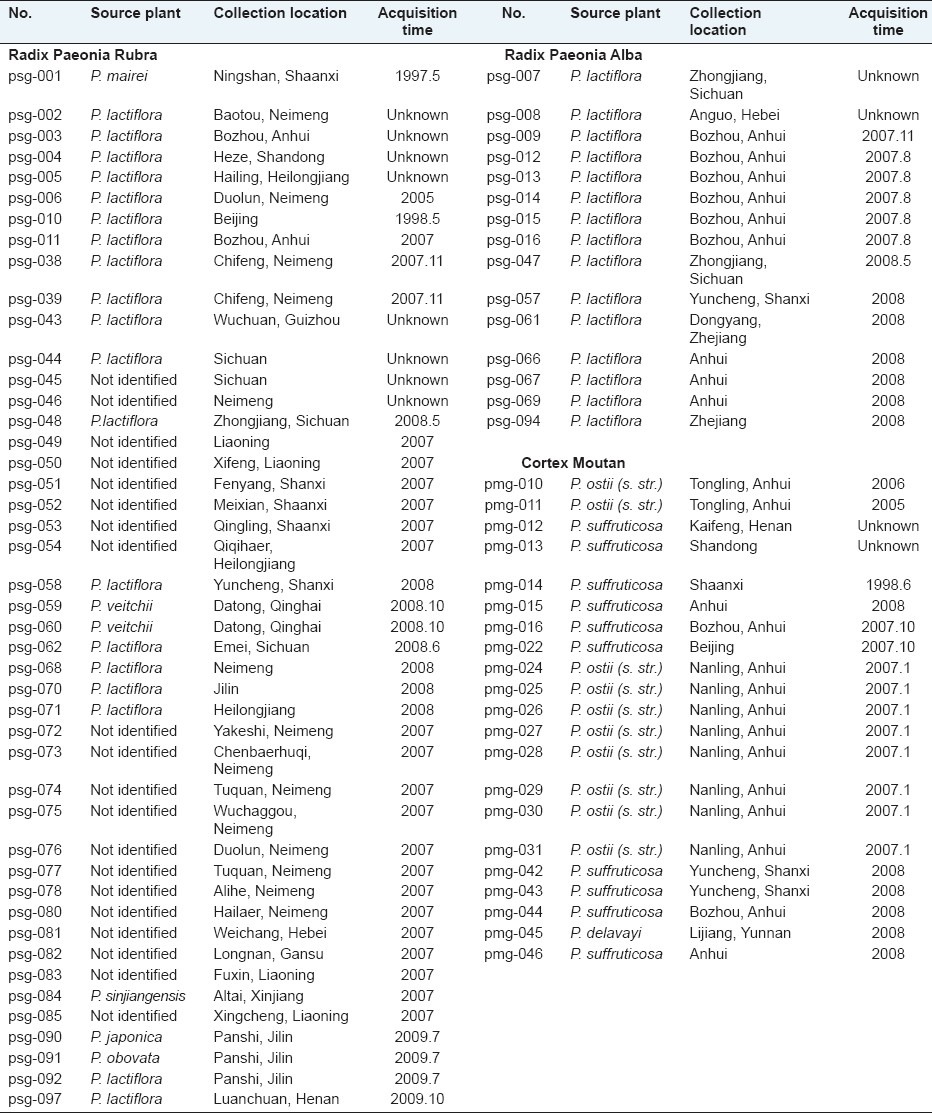
A total of 15 Radix Paeonia Alba, 45 Radix Paeonia Rubra, and 21 Cortex Moutan samples were collected either from production areas or markets in China [Table 1]. Most samples were authenticated by Professor Pei-gen Xiao, Yong Peng, and Dr. Chun-nian He. The voucher specimens were deposited in Professor Xiao's laboratory at the Institute of Medicinal Plant Development, Chinese Academy of Medical Sciences, Beijing, P. R. China.
Table 1.
Samples of radix paeonia alba, radix paeonia rubra and cortex moutan for HPLC analyses
Instrumentation
An Agilent 1200 HPLC system (Agilent Technologies, Palo Alto, CA, USA) equipped with an online vacuum degasser, a quaternary pump, an autosampler, a thermostated column compartment, and a diode array detector (DAD) was used. HPLC separation was performed by a Chromolith performance RP-18e (100×4.6mm, Merck, Germany) using the mobile phase A (purified water containing 0.1% formic acid) and B (acetonitrile containing 0.1% formic acid). The linear gradient elution was optimized as follows: 3→30% B (0→10 min). The flow rate was 5.0 ml/min. The column temperature was set at 30°C. The injection volume was 10 μl and the UV detection wavelength was set at 230 nm. The data were collected and analyzed using Chemstation software (B.02.01).
Chemicals
HPLC grade methanol was purchased from Fisher Scientific (NJ, USA). The purified water was prepared using Millipore purification system (Millipore, Milford, MA, USA) and filtered with 0.45 μm membranes. The 11 chemical reference substances were purified from Cortex Moutan (Paeonia suffruticosa, Andrews) in Professor Xiao's laboratory. They were p-hydroxybenzoic acid (1), methyl gallate (2), catechin (3), oxypaeonifiorin (4), albiflorin (5), paeonifiorin (6), benzoic acid (7), 1,2,3,4,6-penta-O-galloyl-β-D-glucose (8), quercetin (9), paeonol (10), and kaempferol (11). Their structures were identified by ultraviolet (UV), infra-red (IR), mass spectrometry (MS), and nuclear magnetic resonance (NMR). On the basis of UV, NMR and HPLC analyses, the purities of all the reference compounds were more than 95%.
Plant samples preparation
Each dried sample was ground to fine powders (60 mesh) using a pulverizer. An aliquot of 0.5 g of each sample was accurately weighed into an appropriately sized volumetric flask and macerated with 50 ml of 75% ethanol for 30 min. The sample was placed in an ultrasonic water bath for 30 min and conditioned to room temperature. The extracts were filtered through 0.45 μm filter and 10 μl filtrate was injected for each analysis.
Validation
The reliability of the HPLC method for analysis was validated through its stability, precision, and recovery.
Stability
The same sample (pmg-025, P. ostii) was stored in a HPLC vial at 25°C and analyzed at 0, 1, 3, 6, 12, and 24 h. Five chromatographic peaks were chosen to be inspected, of which, paeoniflorin was considered as a reference peak [Figure 1]. The relative standard deviation (RSD) of the relative retention time and of the relative peak area was less than 2%, respectively. Thus, the test solution was stable within 24 h.
Figure 1.
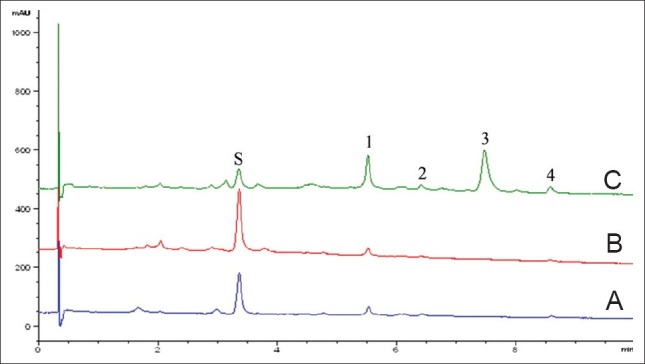
Liquid chromatograms of reference samples (S: paeonifiorin; A: Radix Paeonia Alba, pmg-012; B: Radix Paeonia Rubra, pmg-006; C: Cortex Moutan, pmg-025)
Precision
The precision of the method was evaluated by applying the method developed above to analyze the same sample (pmg-025, P. ostii) on six consecutive injections. The result showed that the RSD of relative peak area was less than 3%, indicating the method could be considered reliable.
Recovery
Six samples from the same source (pmg-025, P. ostii) were extracted and analyzed using the method developed above. The RSD was calculated as the method described above. The result showed that RSD of relative peak area was less than 3%, which showed that the method has excellent repeatability.
Sample analysis and HPLC chromatogram data processing methods
The newly established analytical method was subsequently applied to analyzing 81 samples including 15 Radix Paeonia Alba, 45 Radix Paeonia Rubra, and 21 Cortex Moutan samples collected from different areas in China. Representative chromatograms are shown in Figure 1.
The fingerprint common patterns were generated by inputting the original data suits of all samples acquired from the HPLC workstation to the fingerprint solution software (CHROMAP v1.51, Chromap Institute of Herbal Medicine Research, Zhuhai, China). The following data processing simulated all the HPLC profiles one by one and the results were exhibited on the computer's screen. The major peaks were manually aligned by clicking the peak apexes in order to ensure correct recognition and the common pattern among the species was obtained by averaging all of the computer-simulated profiles. The similarity can be calculated and expressed by the correlative coefficient.
Preparation of chemical reference substance solutions
Q.S. milligrams of each of 11 chemical reference substances were dissolved in 10 ml of methanol for analysis, respectively. Liquid chromatograms of 11 chemical reference substances were obtained [Figure 2].
Figure 2.
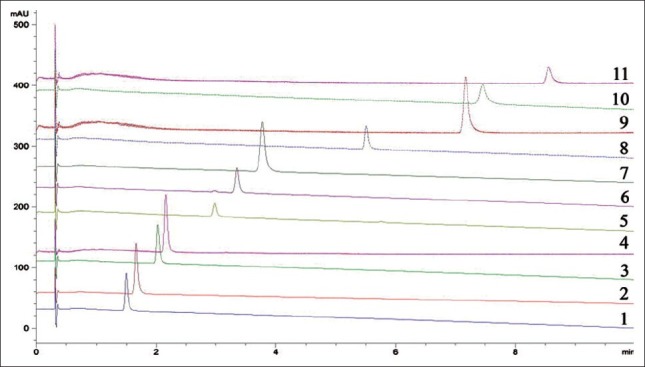
Liquid chromatograms of 11 chemical reference substances chemical reference substances - retention time (min): 1 - 1.497, 2 - 1.665, 3 - 2.023, 4 - 2.16, 5 - 2.980, 6 - 3.348, 7 - 3.774, 8 - 5.506, 9 - 7.164, 10 - 7.450 and 11 - 8.561
RESULTS AND DISCUSSION
The common pattern of the three herbal medicines
The common patterns of the HPLC profiles of the 15 Radix Paeonia Alba showed that 34 peaks were detected and 10 out of them were common peaks. Fifty six peaks arose in the common patterns of HPLC profiles of the 46 Radix Paeonia Rubra, of which, 4 peaks were common peaks; 46 peaks arose in the common patterns of HPLC profiles of the 21 Cortex Moutan, of which, 13 peaks were common peaks; and a total of 63 peaks were detected from the three herbal medicines, of which, 4 peaks were common peaks. Some common peaks were identified according to retention time and UV absorption spectra of reference standard compounds [Figure 3].
Figure 3.
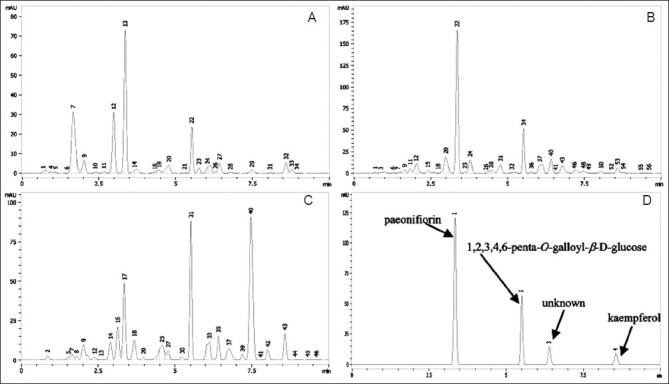
Common Pattern chromatogram of the three herbal medicines A: Radix Paeonia Alba (common peaks: 9-3, 12-5, 13-6, 22-8, 32-11, 20, 24, 25, 27, 33-unknown); B: Radix Paeonia Rubra (common peaks: 12-6, 34-8, 40-unknown, 53-11); C: Cortex Moutan (common peaks: 9-3, 17-6, 31-8, 39-9, 40-10, 43-11, 14, 18, 25, 32, 33, 35, 37-unknown,); D: common peaks of the three herbal medicines
Similarity analysis of the three herbal medicines
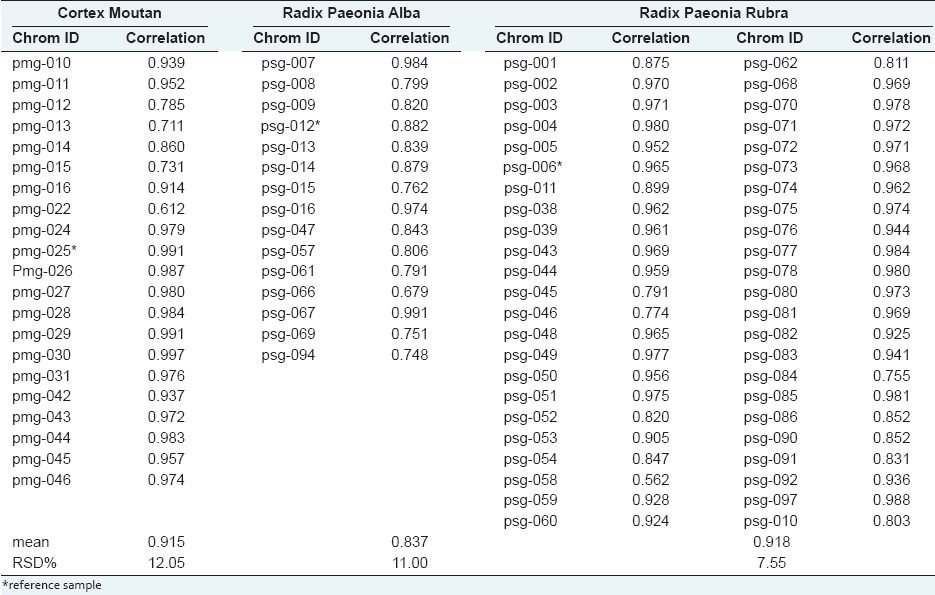
Similarity analysis showed that the similarity coefficient of Radix Paeonia Alba ranged from 0.679 to 0.991 with that of Radix Paeonia Rubra ranging from 0.562 to 0.988 and Cortex Moutan ranging from 0.612 to 0.997. The RSD of the similarity coefficient of the three herbal medicines were more than 5.0%, respectively [Table 2].
Table 2.
The correlation coefficient between each chromatogram of the three herbal medicines and the simulative mean chromatogram
Principal component analysis of 81 samples of Paeonia
Principal component analysis (PCA), an unsupervised pattern recognition method, was performed for the analysis of Radix Paeonia Alba, Radix Paeonia Rubra, and Cortex Moutan. PCA uses an N-dimensional vector approach to separate samples on the basis of the cumulative correlation of all metabolite data and then identifies the vector (eigenvector) that yields the greatest separation among samples without requiring prior knowledge of the data sets.[16] Taking the HPLC profile of pmg-025 (P. ostii) as reference, the data matrix obtained from the 81 samples above by CHROMAP v1.51 was input into Statistical Package for the Social Sciences (SPSS) 18.0 software for the principal component analysis and the analysis result was visualized through Spotfire 5.0 software.
A three-component PCA model cumulatively accounted for 82.6% of variation. The PCA scores plot in Figure 4 could be readily divided into three relative clusters: Radix Paeonia Alba (I), Radix Paeonia Rubra, and Cortex Moutan (III) indicating that the content and distribution of secondary metabolites highly varied in the different Paeonia herbs. Radix Paeonia Alba was clearly separated from the other herbs by principal component 1 (PC1), while Cortex Moutan was easily discriminated from the others by principal component 3 (PC3). The corresponding PCA loadings plot were utilized to identify the differential metabolic compositions for the discrimination of groups [Figure 5]. For example, the preferential distribution of marker in group A of the loadings plot accounted primarily for the difference of Radix Paeonia Alba. Analogously, the distribution of the loadings plot in group C indicated the variation of cortex moutan. A comparative study on HPLC fingerprinting among these three herbal medicines showed:
Figure 4.
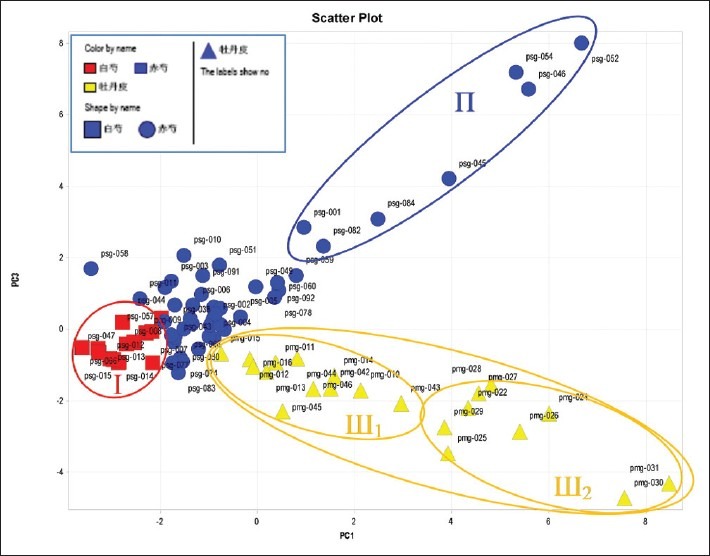
Scores plot of PCA of Paeonia samples on the first three PCs. Accumulated mean square. PC1 = 46.5%; PC2 = 71.6%; PC3 = 82.6%
Figure 5.
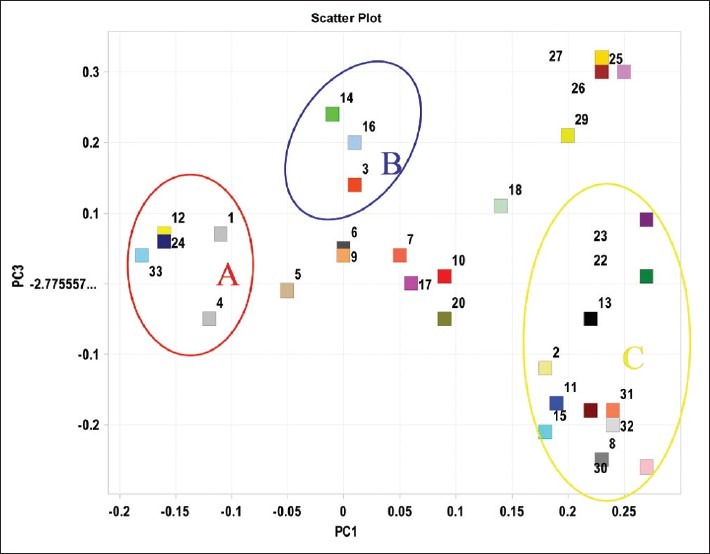
Loadings plot of PCA for the three herbal medicines’ components. Identified by reference standard compounds as: 4-1, 5-2, 7-3, 8-4, 12-5, 14-6, 16-7, 23-8, 29-9, 30-10 and 32-11
(1) Radix Paeonia Alba samples were clustered together closely (I), which showed that its main chemical compositions were stable and could be clearly separated from Radix Paeonia Rubra. According to the load diagram, the contents of chemical compositions of area A in Radix Paeonia Alba samples were higher than those in Radix Paeonia Rubra. It has been identified that area A contains hydroxy acid and albiflorin.
(2) Radix Paeonia Rubra samples appeared to be very scattered, especially psg-001, 045, 046, 052, 054, 084, and 082 (Π area). Psg-084 was P.sinjiangensis from the Altai Mountains in Xinjiang, while psg-001 was P.mairei from Ningshan city, Shaanxi province. Moreover, the other samples were wild peony from an unidentified source, suggesting that several samples may be different from the P. lactiflora or P. veitchii. Furthermore, the samples psg-059 and 060 from P. veitchii had a certain distance from the main Radix Paeonia Rubra sample, so it was assumed that P. veitchii had different chemical compositions from red paeony. This has also been supported by literature.[12,13] P.obovata (psg-091) and P.japonica (psg-090) were similar to most of the samples; in addition, two cultivated P. lactiflora (psg-011 and 058) used as Chishao were much closer to Radix Paeonia Alba, indicating it was inappropriate to apply cultivated P. lactiflora as Radix Paeonia Rubra. The contents of the chemical compositions of area B in the Radix Paeonia Rubra samples were higher than the others samples. It has been identified that area B contains paeoniflorin and benzoic acid.
(3) Scattered Cortex Moutan examples can be divided into two groups (III1 and III2). Other than pmz-022 being collected from Beijing, the III2 group samples were collected from the production areas of genuine herbal medicine (Nanling area, Anhui province) in 2007. The III1 group samples were taken from the drugstore (epidermis of some cortex moutan examples was removed). The PCA analysis showed that the contents of main chemical compositions of cortex moutan were affected by processing methods and storage time, and the latter factor had been confirmed by experiments.[17] According to factor loading plot [Figure 5] derived from the mentation impact factors above, it has been identified that area C contains paeonol, kaempferol, 1,2,3,4,6-penta-O-galloyl-β-D-glucose, oxypaeonifiorin, etc..
CONCLUSION
A total of 15 Radix Paeonia Alba, 45 Radix Paeonia Rubra, and 21 Cortex Moutan samples were analyzed by HPLC-DAD with monolithic columns. A rapid separation and fingerprint analysis method has been established which could compare the similarity and PCAs of these three herbal medicines. The results showed that these three herbal medicines could be readily divided into three relative clusters.
HPLC monolithic packing material is a new approach which is increasingly being explored to accelerate the speed of the analysis. Monolithic columns have different structures when compared with conventional silica-based columns. Higher flow rates (up to 9.9 ml/min) can be used while the resolution of the silica rod column is less affected in comparison to particulate materials. After increasing the flow rate, column back pressure is still low (lower than 400 bar). This makes monolithic columns attractive for high throughput applications without a loss of column efficiency.[18–20] Compared with the normal reversed-phase column taking more than 60 minutes for one fingerprint analysis of Paeonia herbal medicines, in this research, a very fast method employing monolithic columns has been developed for the analysis of all samples in less than 10 minutes and given the high peak capacity (>30). This method can be applied to the determination of chemical fingerprint chromatogram.
In this investigation, this analytical method has demonstrated its potential for the rapid differentiation and identification of complex TCM extracts that contain similar chemical ingredients as well as the potential for the discrimination of subtle variations in the same plant species with different production areas, cultivation methods, processing methods, and collection time. Through analysis on the sources, Radix Paeonia Alba, Radix Paeonia Rubra, and Cortex Moutan can be clearly distinguished and the main features of their chemical compositions can be revealed by PCA: Cortex Moutan contains high contents of paeonol, kaempferol, 1,2,3,4,6-penta-O-galloyl-β-D-glucose and oxypaeonifiorin, etc.; Radix Paeonia Alba contains high contents of p-hydroxybenzoic acid and albiflorin; Radix Paeonia Rubra contains high paeoniflorin and benzoic acid. Therefore, speculations about their different bioactivities can be more accurate due to the knowledge of their different chemical compositions.
ACKNOWLEDGEMENT
This research work was financially supported by grants from the National Basic Research Program of China (Grant No. 2006CB504701) and the Key Program of the National Natural Science Foundation of China (Grant No. 30530860).
Footnotes
Source of Support: Grants from the National Basic Research Program of China (Grant No. 2006CB504701) and the Key Program of the National Natural Science Foundation of China (Grant No. 30530860)
Conflict of Interest: None declared.
REFERENCES
- 1.Xiao PG. Modern Chinese Materia Medica. Beijing: Chemical Industry Press; 2002. [Google Scholar]
- 2.He CN, Peng Y, Zhang YC, Xu LJ, Gu J, Xiao PG. Phytochemical and biological studies of Paeoniaceae. Chem Biodivers. 2010;7:805–38. doi: 10.1002/cbdv.200800341. [DOI] [PubMed] [Google Scholar]
- 3.Yu J, Xiao PG. A preliminary study of the chemistry and systematics of paeoniaceae. Acta Phytotaxon Sin. 1987;25:172–9. [Google Scholar]
- 4.Cheng WJ. Treatise on exogenous febrile diseases with notes. Beijing: China Medical Science Press; 2011. [Google Scholar]
- 5.Xie PS, Chen SB, Liang YZ, Wang XH, Tian RT, Upton R. Chromatographic fingerprint analysis: A rational approach for quality assessment of traditional Chinese herbal medicine. J Chromatogr A. 2006;1112:171–80. doi: 10.1016/j.chroma.2005.12.091. [DOI] [PubMed] [Google Scholar]
- 6.Liu YZ, Dan Y, Peng Y, Xiao PG. Reflecting the steps of modernization of Chinese material medica from the papers in Chinese traditional and herbal drugs in 2009. CHM. 2010;2:180–8. [Google Scholar]
- 7.Yang L, Xu SJ, Tian RT, Xie PS, Wang ZT. HPLC fingerprinting of radix paeoniae alba. Acta Pharm Sin. 2007;42:71–4. [PubMed] [Google Scholar]
- 8.He Y, Zhao HD, Tang LY, Wang ZJ, Zhang QW. Quantititive study for chromatographic fingerprints of processed products of Paeonia lactiflora. China J Chin Mater Med. 2007;32:1161–4. [PubMed] [Google Scholar]
- 9.Fan XH, Wang ZZ, Li Q, Ma TC, Bi KS, Jia Y. UPLC characteristic chromatographic profile of Moutan Cortex. China J Chin Mater Med. 2011;36:715–7. [PubMed] [Google Scholar]
- 10.Wu MZ, He Y, Tang LY, Wang ZJ. Clustering analysis of HPLC chromatographic fingerprints of Paeonia suffruticosa. China J Chin Mater Med. 2007;32:1054–6. [PubMed] [Google Scholar]
- 11.Wang Q, Liu RX, Yu HL, Liu P, Liu ZT, Bi KS, et al. Studies on HPLC fingerprint of white peony roots and red peony roots. Chin Pharm J. 2007;42:581–4. [Google Scholar]
- 12.Wang Q, Liu RX, Guo HZ, Ye M, Huo CH, Bi KS, et al. Simultaneous LC determination of major constituents in red and white peony root. Chromatographia. 2005;62:581–8. [Google Scholar]
- 13.Chuang WC, Lin WC, Sheu SJ, Chiou SH, Chang HC, Chen YP. A comparative study on commercial samples of the roots of Paeonia vitchii and P. lactiflora. Planta Med. 1996;62:347–51. doi: 10.1055/s-2006-957899. [DOI] [PubMed] [Google Scholar]
- 14.Li SL, Song JZ, Choi FF, Qiao CF, Zhou Y, Han QB, et al. Chemical profiling of Radix Paeoniae evaluated by ultra-performance liquid chromatography/photo-diode-array/ quadrupole time-of-flight mass spectrometry. J Pharma Biomed Anal. 2009;49:253–66. doi: 10.1016/j.jpba.2008.11.007. [DOI] [PubMed] [Google Scholar]
- 15.Zhou X, Chen X, Ge L, Gong H, Tian S, An D. An D. Rapid determination of paeoniflorin from Paeonia sinjiang K. Y. Pan. by rapid resolution liquid chromatography. Phcog Mag. 2010;6:98–101. doi: 10.4103/0973-1296.62893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang Y, Zhang Y. Process monitoring, fault diagnosis and quality prediction methods based on the multivariate statistical techniques. IETE Tech Rev. 2010;27:406–20. [Google Scholar]
- 17.Han XY, Liu ZA, Wang LS. Studies on the content of effective composition in moutan changes during storage. J Chin Med Mater. 2008;31:823–5. [Google Scholar]
- 18.Guiochon G. Monolithic columns in high-performance liquid chromatography. J Chromatogr A. 2007;1168:101–68. doi: 10.1016/j.chroma.2007.05.090. [DOI] [PubMed] [Google Scholar]
- 19.Kaminski L, el Deeb S, Watzig H. Repeatability of monolithic HPLC columns while using a flow program. J Sep Sci. 2008;31:1745–9. doi: 10.1002/jssc.200700681. [DOI] [PubMed] [Google Scholar]
- 20.Noha NA, Peter Y, Brian JC. Development and validation of a rapid and efficient method for simultaneous determination of methylxanthines and their metabolites in urine using monolithic HPLC columns. J Sep Sci. 2009;32:93–8. doi: 10.1002/jssc.200800655. [DOI] [PubMed] [Google Scholar]


