Abstract
The canonical TGF-β/Smad signaling pathway was delineated in the mid 90’s and enriched over the past decade with many findings about its specificity, regulation, networking, and malfunctions in disease. However, a growing understanding of the chromatin status of a critical class of TGF-β target genes –the genes controlling differentiation of embryonic stem cells– recently prompted a reexamination of this pathway and its critical role in the regulation of stem cell differentiation. The new findings reveal master regulators of the pluripotent state set the stage for Smad-mediated activation of master regulators of the next differentiation stage. Furthermore, a novel branch of the TGF-β/Smad pathway has been identified in which a chromatin-reading Smad complex makes the master differentiation genes accessible to canonical Smad complexes for transcriptional activation. These findings provide exciting new insights into the global role of TGF-β signaling in the regulators of stem cell fate.
Keywords: stem cells, chromatin, epigenetics, transcription, differentiation, TGF-β
The canonical model and its regulation
The transforming growth factor β (TGF-β family plays central roles in the development and maintenance of metazoan organisms (Flavell et al., 2010; Heldin et al., 2009; Massagué, 2000; Schier, 2003; Wu and Hill, 2009). This large family of paracrine factors regulates functions that guide the exit of embryonic stem cells from the pluripotent state, and the subsequent differentiation of committed progenitors to more restricted cell fates for the establishment of body axes, mature tissues, and whole organs. In most cell types, TGF-β signaling additionally controls the expression of a plethora of homeostatic genes whose activity determines cell proliferation, extracellular matrix production, paracrine factor secretion, cell-cell contacts, immune function, and tissue repair. Pathway feedback and crosstalk responses are also built into the transcriptional program of TGF-β in most cell types.
Regulation of gene expression is central to all these effects. The TGF-β signal transduction pathway is largely a pathway for transcriptional control. TGF-β factors initiate signaling by binding to a multi-component receptor complex that includes two pairs (type I and type II) of receptor serine/threonine kinases. The type II receptor subunits in this complex phosphorylate and activate the type I receptors, which then phosphorylate and activate Smad transcription factors to propagate the signal. The TGF-β, nodal, activin and myostatin members of the family bind to receptors that phoshorylate Smad2 and Smad3, and in some cases Smad1. Bone morphogenetic protein (BMP) receptor phosphorylate Smads 1, 5 and 8. A common fate of these receptor-regulated Smads (RSmads) is two form complexes with Smad4, which binds tightly to the receptor-phosphorylated C-terminal tail of RSmads. Ancillary Smad-independent pathways are activated by TGF-β in a context dependent manner through interactions with the receptors that remain under investigation (Massagué, 2008)
In the basal state Smads cycle constantly between the cytoplasm and the nucleus, but once activated by receptor action, the resulting RSmad-Smad4 complexes accumulate in the nucleus and bind to specific promoter elements throughout the genome. The N-terminal globular domain of Smad proteins contains a DNA-binding finger that preferentially recognizes the sequence CAGAC, known as Smad Binding Element (SBE). To achieve high affinity and specificity for target sequences, Smad4-RSmad complexes recruit additional DNA binding proteins into the complex. The first such cofactor to be identified was the forkhead-box family member FoxH1 (previously known as FAST1). The FoxH1-Smad2/3-Smad4 complex binds to a composite site known as the “activin response element” (ARE) on target differentiation genes in embryo cells (Silvestri et al., 2008). Other factors belonging to different families of DNA binding proteins have since been identified that, in a similar manner, cooperate with Smad complexes in binding to specific promoter sequences. Each of the resulting complexes targets a particular subset of TGF-β responsive genes, called a “synexpression group”, for coordinated regulation of cellular activities (Massagué and Gomis, 2006).
On the DNA, Smad complexes recruit histone acetyltransferases such as p300, CBP, P/CAF, and GCN5 (Itoh et al., 2000; Kahata et al., 2004; Massagué et al., 2005), Mediator, which is a multi-subunit co-activator complex that bridges with the C-terminal domain of RNA polymerase (Pol) II (Kato et al., 2002), and the SWI/SNF chromatin remodeling complex that repositions nucleosomes to facilitate transcription (Ross et al., 2006; Xi et al., 2008). These interactions stimulate the transcriptional output of the target genes. Alternatively, depending on the promoter context, the Smad complex may recruit histone deacetylases such as HDAC4 for transcriptional inhibition (Kang et al., 2005). The end result of these findings is a model for signal-driven regulation of gene expression that reconciles the biochemical simplicity of the TGF-β receptor/Smad signal transduction process with the broad diversity and context-dependence of its transcriptional targets and cellular responses (Massagué et al., 2005).
Many modulators of this process have been identified that control the receptor access and activity of Smad proteins (Furuhashi et al., 2001; Hocevar et al., 2001; Lin et al., 2004; Tang et al., 2003; Tsukazaki et al., 1998; Yamakawa et al., 2002), their movements in and out of the nucleus (Xu and Massagué, 2004; Xu et al., 2007; Yao et al., 2008), their cooperation with other signal-activated transcription factors (Alarcón et al., 2009; Massagué et al., 2005; Varelas et al., 2010), their interactions with transcriptional cofactors (Massagué et al., 2005), the recycling of activated Smads by dephosphorylation (Bruce and Sapkota, 2012) and poly(ADP)-ribosylation (Lönn et al., 2010), or their removal by polyubiquinylation and proteasome-dependent degradation (Alarcón et al., 2009; Fuentealba et al., 2007; Gao et al., 2009; Guo et al., 2008; Sapkota et al., 2007).
Chromatin status of TGF-β target genes
This long-standing model of the TGF-β/Smad pathway is adequate to explain how TGF-β factors regulate genes that control cellular homeostasis (Figure 1A). External signals like those conveyed by the Smad pathway control cell behavior by modulating up or down the basal activity of such genes. These genes are in an active state within the euchromatin, and their Smad binding sites are likely well exposed to incoming Smad4-RSmad complexes. The net effect of TGF-β- or BMP-activated Smads is to increase or decrease Pol II action and the transcriptional output of these genes, typically within a 5-fold range.
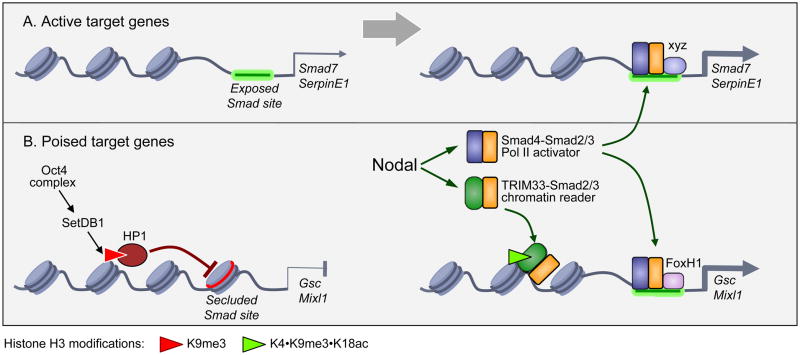
Figure 1. Model of nodal regulation of gene expression in embryonic stem cells.
A. Active target genes. Nodal target genes that control cell homeostasis or Smad pathway feedback (e.g. SerpinE1 and Smad7, respectively) are in a transcriptionally active state with the Smad binding sites exposed. Signal-driven Smad4-Smad2/3 complexes, in associated with DNA binding cofactors (“xyz”), readily bind to these sites for stimulation of transcription.
B. Poised target genes. Nodal target genes that control ESC differentiation (e.g. Gsc and Mixl1) have Smad binding sites secluded by repressive chromatin. The Oct4-Sox2-nanog complex, which is an enforcer of pluripotency in ESCs, recruits SetDB1 to differentiation genes. SetDB1 catalyzes Lys9 trimethylation on histone H3 (H3K9me3 motif), which binds HP1 for chromatin compaction and gene repression. When H3K9me3 is accompanied with unmodified K4 and acetylated K18 (H3K4-K9me3-K18ac motif), it provides a platform for activation of Gsc and Mixl1 by nodal signals. The TRIM33-Smad2/3 complex binds to H3K4-K9me3-K18ac, displacing HP1 to relax the chromatin and provide Smad4-Smad2/3 with access to its binding sites. FoxH1 is a Smad cofactor for specific recognition of these sites. Thus, Gsc and Mixl1 in ESCs are in a “poised” state that is silent but ready for nodal-driven activation through the joint action of a TRIM33-Smad2/3 chromatin-binding complex and a Smad4-Smad2/3 Pol II activating complex.
This general model also applies to TGF-β gene responses that control cell fate. However, recent evidence coming from the field of transcriptional regulation in stem cell differentiation raised questions about the ability of this model to adequately explain the regulation of differentiation genes by TGF-β signals. In contrast to the accessible state of homeostasis modulator genes, the genes that encode master regulators of stem cell differentiation are guarded by chromatin structures that bar access to transcriptional activators (Figure 1B). This level of protection makes sense because the activation of such genes irrevocably commits stem cells to differentiated lineages and end the pluripotent state. However, while master differentiation genes are kept in a repressed state, they are also ready for activation by appropriate signals. These genes may harbor RNA polymerase II (Pol II) at the transcription start site, paused but ready to proceed with transcription elongation under the right command (Young, 2011). Therefore, master differentiation genes and their chromatin are said to be in a “poised state”, that is, a state that is repressed yet set for activation.
The biochemical basis for the poised state of master differentiation genes in embryonic stem cells (ESCs) includes certain chromatin modifications. Oct4, Sox2 and Nanog, which are core enforcers of the pluripotent state, are also responsible for some of these modifications (Young, 2011). These proteins form a DNA binding complex that in turn stimulates chromatin-modifying factors including polycomb group (PcG) proteins, SetDB1, and others. PcG complexes mediate methylation of histone 3 at Lys27, which is a repressive mark, whereas SetDB1 catalyzes trimethylation of Lys9 on histone H3 (H3K9me3) (Figure 1B). H3K9me3 is a hallmark of heterochromatin (Mikkelsen et al., 2007) and serves as a docking site for heterochromatin protein 1 (HP1). HP1 bound to H3K9me3 is thought to form dimers that strap nucleosomes together for compaction of the chromatin, leading to gene repression (Ruthenburg et al., 2007). However, H3K9me3 and HP1 are also present in “heterochromatinic” regions of the euchromatin (Singh, 2011). SetDB1 under the command of Oct4, Sox2 and Nanog may create such regions around master differentiation genes.
Secluded Smad sites
The nature of the poised state implies that activation of ESC differentiation genes by the TGF-β/SMAD pathway has restrictions that may not apply to the regulation of readily accessible homeostasis genes. This knowledge posed a conundrum: if master regulator genes are secluded from access by promoter-binding transcription factors like Smads, how do Smad gain access to these genes?
An early encounter with this situation occurs when the TGF-β family member nodal activates master regulators of mesendodermal differentiation during mouse embryogenesis. Nodal is highly expressed in the node, which is the organizer of gastrulation and differentiation of the three germ layers –ectoderm, mesoderm and endoderm– in vertebrates. In the absence of nodal, embryogenesis is halted right after gastrulation. In the mouse, nodal is also essential for the organization of left-right axial structures, neural patterning, and other developmental events.
Nodal induces mesendodermal differentiation in ESCs by inducing the expression of goosecoid (Gsc), Mix-like homeodomain protein 1 (Mixl1), and other master differentiation genes (Blum et al., 1992; Chen et al., 1997; Hart et al., 2002). Gsc is a member of the bicoid subfamily of paired homeobox transcription factors, and Mixl1 of the Mix/Bix subfamily. Gsc and Mixl1 are expressed in the primitive streak to mediate gastrulation, axial mesendoderm morphogenesis, and endoderm formation. Nodal activates Gsc and Mixl1 expression through nodal/activin receptors, including the type I receptors ALK4 and ALK7, and the type II receptors ActR-II and ActR-IIB (Reissmann et al., 2001; Yeo and Whitman, 2001). The receptors phosphorylate Smads 2 and 3, leading to the formation of a Smad2/3-Smad4-FoxH1 complex (Figure 1B). This complex binds to AREs in the proximal promoters of Gsc and Mixl1 for transcription activation (Chen et al., 1997; Labbe et al., 1998). In ESCs however Gsc and MixL1 are in the poised, repressed state. Therefore, something must happen first in order for Smad2/3-Smad4-FoxH1 to gain access to these AREs.
TRIM33 enters the scene
A recent investigation of how Smad complexes gain access to the AREs in Gsc and Mixl1 turned the focus to TRIM33 (Tripartite Motif protein 33) (Hatakeyama, 2011; Ozato et al., 2008). The TRIM family of proteins includes more than seventy members that are defined by a motif of contiguous RING-finger, B-box, and coiled-coil domains in the N-terminal region of the proteins. The RING domain of many TRIM proteins has E3 ubiquitin ligase activity that can mediate ubiquitylation of the TRIM protein itself and of other proteins. The coiled-coil domain mediates protein TRIM protein homo- and hetero-oligomerization. TRIM family members are involved in a broad range of biological processes, in many cases through the control of gene expression. A prominent member of this family, PML (TRIM19), is involved in the t(15;17) translocation of acute promyelocytic leukemia.
TRIM33 (also known as transcription intermediate factor 1β, TIF1β), TRIM24 (TIF1α), TRIM28 (TIF1β, KAP1), and TRIM66 (TIF1β), form a subfamily that is characterized by the presence of a PHD domain and a Bromodomain (Bromo) in the C-terminal region of the proteins. TRIM24 binds estrogen receptors to activate estrogen-dependent genes associated with cellular proliferation and tumor development (Tsai et al., 2010), and retinoic acid receptor-α (RARα) to attenuate RARα-mediated transcription (Le Douarin et al., 1996). TRIM28 is implicated in gene silencing as a component of the NCoR1 nuclear receptor repressor complex and the NuRD nucleosome-remodeling complex (Hatakeyama, 2011; Ozato et al., 2008). TRIM66 expression is restricted to the testis (Khetchoumian et al., 2004).
The entry of TRIM33 into the TGF-β scene was smooth at first but soon turned confusing. Several studies suggested that TRIM33 is a negative regulator of Smad4 and a general inhibitor of TGF-β family signaling (Agricola et al., 2011; Dupont et al., 2009; Dupont et al., 2005; Morsut et al., 2010). Knockdown of TRIM33 in Xenopus embryos enhanced mesendoderm induction (Dupont et al., 2005). TRIM33 knockdown in mouse resulted in excessive nodal signaling (Morsut et al., 2010). In cell-based assays using promoter constructs driving reporter genes, TRIM33 scored as an inhibitor of TGF-β dependent transcription (Agricola et al., 2011; Dupont et al., 2005; He et al., 2006). These observations and the presence of a RING-finger domain in TRIM33 fit with the notion that TRIM33 might be an inhibitor of Smad4 in TGF-β and BMP pathways. Indeed, in vitro and in overexpressing cells TRIM33 can mediate poly-ubiquitylation and degradation of Smad4 (Dupont et al., 2005), although this was later revised to TRIM33 mediating inhibitory mono-ubiquitylation of Smad4 in a histone-dependent manner (Agricola et al., 2011; Dupont et al., 2009).
TRIM33 deficient mouse embryos lack mesoderm, and this was hard to explain in the face of seemingly high levels of nodal signaling (Morsut et al., 2010). The notion that TRIM33 is an inhibitor of Smad4 function had been challenged by the finding that TRIM33 binds to Smad3, and it does so with higher affinity than for Smad4 (He et al., 2006). Upon TGF-β stimulation, the pool of phosphorylated Smad3 is distributed between separate TRIM33-Smad2/3 and Smad4-Smad2/3 complexes. Knockdown of TRIM33 had little or no effect on classical TGF-β target genes controlling cell homeostasis. In human hematopoietic progenitor cells TRIM33 was dispensable for TGF-β mediated cell cycle arrest (a homeostatic response that is driven by Smad2/3-Smad4). Notably, TRIM33 was required in these cells for TGF-β stimulation of erythroid differentiation. Based on these observations, it was proposed that the TGF-β-activated Smad pathway has two arms, a canonical Smad4-Smad2/3 arm that mediates homeostatic gene responses and a TRIM33-Smad2/3 arm that, in hematopoietic progenitors at least, stimulates differentiation (He et al., 2006). This resonated with evidence that in zebrafish TRIM33 is required for erythropoiesis (Ransom et al., 2004) and it acts as a transcriptional elongation of erythroid differentiation genes (Bai et al., 2010). Furthermore, the conditional knockout of TRIM33 in premalignant pancreatic progenitors phenocopies the effect of Smad4 conditional knockout, suggesting that TRIM33 and Smad4 converge on pancreatic tumor suppression (Bardeesy et al., 2006; Vincent et al., 2009). However, without hard evidence for a direct involvement of TRIM33 in TGF-β-dependent gene regulation, the controversy about the role of TRIM33 in Smad signaling went on.
TRIM33 as a direct TGF-β signal transducer
Hard evidence for a role of TRIM33 as a signal transducer and direct mediator of transcription in the TGF-β pathway was recently provided by studies in ESCs. A combination of genetic, biochemical and structural evidence demonstrated an essential role for TRIM33 in nodal activation of Gsc and Mixl1 during ESC differentiation (Xi et al., 2011). TRIM33 is present in the majority of nuclei in all regions of the gastrulating embryo, including the node and primitive streak (Xi et al., 2011). In mouse ESCs derived from the inner cell mass, ligand activation of nodal/activin receptors rapidly induces the formation of biochemically separate TRIM33-Smad2/3 and Smad4-Smad2/3 protein complexes (Figures 1B and 2). Under conditions that allow differentiation, ESCs form structures known as embryoid bodies (EBs) that in response to external factors differentiate into all three germ layer fates (Murry and Keller, 2008). Notably, TRIM33-deficient mouse and human ESCs form EBs that undergo ectoderm differentiation but fail to undergo mesendodermal differentiation in response to autocrine nodal (Xi et al., 2011). Stimulation of nodal/activin receptors by added ligand in EBs acutely activates the expression of mesendodermal differentiation genes such as Gsc, Mixl1 and others, together with stimulation of homeostasis genes such as SerpinE1, Skil and Smad7 (Figure 2). Whereas Smad2/3 and Smad4 are required for the vast majority of these gene responses, TRIM33 was required for the differentiation gene responses but not the homeostatic responses. Genome-wide transcriptomic analysis showed that a large proportion of nodal gene responses in EBs require both Smad4 and TRIM33. In contrast, almost all TGF-β responses in human keratinocyte and breast carcinoma cell lines are homeostatic and require Smad2/3 and Smad4 but not TRIM33.
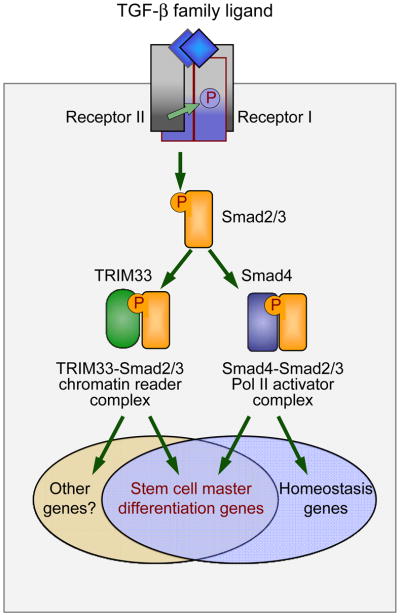
Figure 2. Complementary branches of the TGF-β/Smad pathway in embryonic stem cells.
TGF-βsignaling involves the binding of ligand to two pairs of receptor serine/threonine kinases for the assembly of a receptor complex that phosphorylates RSmad proteins (Smads 2 and 3 in the case of TGF-β and nodal receptors). The receptor-phosphorylated RSmads bind to Smad4, assembling complexes that recognize specific promoter elements to stimulate or inhibit transcription. In a newfound second branch of this pathway, receptor-phosphorylated Smad2/3 bind to TRIM33, assembling complexes that recognize certain repressive marks on the chromatin. The Smad4-Smad2/3 complex is necessary and sufficient for TGF-β regulation of cell homeostasis genes that are in an active state. However, master differentiation genes in ESCs are secluded by repressive chromatin marks. Activation of these genes by TGF-β signals requires TRIM33-Smad2/3 in addition to the Smad4-Smad2/3. Smad4-independent gene responses may also exist.
The PHD-Bromo cassette of TRIM33 is essential for its function as a Smad signal co-transducer in the nodal TGF-β pathway (Xi et al., 2011). In contrast, mutation of the RING finger domain did not inhibit the transcriptional activity of TRIM33. The PHD and Bromo domains typically bind to post-translationally modified histones (Taverna et al., 2007; Yap and Zhou, 2010). Analysis of the histone binding ability of the TRIM33 PHD-Bromo cassette revealed a high affinity for histone H3 containing unmodified K4, K9me3 and K18ac. Notably, the mark of poised master regulators, H3K9me3, is critical for the TRIM33-histone H3 interaction.
The X-ray crystal structure of the PHD-Bromo cassette bound to its cognate histone H3 modifications provided key insights into the mechanism of action of TRIM33. The structure shows that the TRIM33 PHD binds to unmodified K4 and K9me3 whereas Bromo binds the K18ac mark on the same H3 tail (Xi et al., 2011). The structure also explains why methylation of K4 disrupts recognition by TRIM33. The specificity of the TRIM33-histone interaction is noteworthy. Whereas the TRIM24 Bromo recognizes the H3K23ac mark (Tsai et al., 2010), the TRIM33 Bromo recognizes H3K18ac, as determined by the anchoring interaction of the PHD with K9me3 (Xi et al., 2011). The spacer length between the methylation and acetylation marks on histone H3, as well as the sequence context of these marks, are key determinants of molecular recognition by the TRIM33 PHD/Bromo cassette. A recent proposal that TRIM33 binds to K18ac and K23ac acetylation marks on the same histone H3 tail (Agricola et al., 2011), is not tenable based on the crystal structure. Thus, the combination of unmodified K4, K9me3 and K18ac marks on histone H3 tails constitutes a unique recognition code for the binding of TRIM33. The combined binding specificity for modified histone and DNA mediated by TRIM33 and Smad2/3, respectively, ensures further selectivity in target gene recognition.
A dance of complexes
The Gsc and Mixl1 promoters in ESCs contain key features of the poised state, including the H3K9me3 mark of quiescent chromatin, the chromatin compacting factor HP1 bound to this mark, and a basal level of RNA Pol II loaded on the start site (Xi et al., 2011) (Figure 1B). The recent evidence suggests that nodal TGF-β signals activate these master mesendoderm regulator genes by means of two cooperative Smad complexes in four steps. On binding ligand, the nodal receptors phosphorylate Smad2/3 leading to the generation of TRIM33-Smad2/3 and Smad4-Smad2/3 complexes. Next, TRIM33-Smad2/3 binds to nucleosomes containing histone H3 with K4-K9me3-K18ac in the Gsc and Mixl1 promoters. The superior affinity of TRIM33 for H3K9me3-K18ac displaces bound HP1 form these nucleosomes. This requires Smad2/3 and nodal input, suggesting the action of the complex depends on TRIM33 binding to H3K9me3-K18ac and Smad2/3 binding to an adjacent SBE. Third, through as yet unknown mechanisms, but possibly just by evicting HP1, the TRIM3-Smad2/3 complex regionally remodels the chromatin to expose the AREs. Then, and only then, can the Smad4-Smad2/3-FoxH1 complex bind to the AREs for stimulation of Pol II dependent transcription of Gsc and Mixl1(Xi et al., 2011). TRIM33-independent, homeostatic gene responses (Smad7, SerpinE1 and others) go on in parallel (Figure 1). Smad4-Smad2/3 access to these gene promoters may solely depend on the availability of specific DNA-binding Smad partners, following the classical principles of the canonical TGF-β pathway as previously known (Massagué et al., 2005).
The end result of these events is that nodal switches Gsc and Mixl1 from the poised state to the activated state, triggering mesendodermal differentiation. In this context, the observed ability of TRIM33 to mediate Smad4 ubiquitylation (Agricola et al., 2011; Dupont et al., 2005) might provide a negative feedback activity for the inactivation of Smad4 and signal turnover, a point that is open to investigation. As precedent, the Pol II kinases CDK8 and CDK9 phosphorylate Smads1~3 for full activation but, in the process, these phosphorylations prime Smads for ubiquitylation and degradation (Alarcón et al., 2009; Aragón et al., 2011). Smad4 inactivation by poly-(ADP)-ribosyltation provides another mechanism for decommissioning Smad4 in transcriptional complexes (Lönn et al., 2010).
These insights also suggest a biochemical definition of the poised state of master regulator genes: the H3K9me3 mark provides a binding site for HP1 proteins that impose gene repression but, at the same time, H3K9me3 provides an entry point for TRIM33 to displace HP1 and allow signal-driven gene expression. The external TGF-β signals therefore use a repressive chromatin mark as a platform for activation of master regulators of differentiation.
Master programs of TGF-β responsiveness
The work reviewed above has shown that master enforcers of the pluripotent state implement chromatin modification that serve to poise master regulators of the next stage, such as Gsc and Mixl1, for activation by nodal signaling. In a remarkable convergence of complementary findings, recent studies on genome-wide binding patterns of lineage-specific transcription factors have provided a global view of the extent to which Oct4 determines the cell type-specific responsiveness to TGF-β signals (Mullen et al., 2011; Trompouki et al., 2011). Using chromatin immunopreciptation coupled with massive parallel sequence analysis (ChIP-seq), it was shown that nodal-activated Smad3 in ESC co-occupies the genome with Oct4. These observations were extended lineage-specific master enforcers of various committed progenitor cells, including MyoD1 in myoblasts and PU.1 in pro-B cells (Mullen et al., 2011). Furthermore, this paradigm also applies to BMP signaling and Wnt signaling, whose respective mediators, Smad1 and TCF7L2, in hematopoietic progenitors bind to the genome right next to master regulators of this lineage (Trompouki et al., 2011). Moreover, Smads can occupy different cell-specific enhancers at the same gene in different cell lineages as dictated by the stage/lineage-specific master regulator. One example of this is provided by Smad3 binding to Id3, which occurs at an Oct4 targeted enhancer in ESCs and at a PU.1 targeted enhancer in pre-B cells (Mullen et al., 2011). These results argue that cell type-specific master transcription factors determine what genes will be competent for binding signal-activated Smads, thus orchestrating at a global level the lineage-specific effects of TGF-β signaling.
At first glance these results may seem at odds with the extensive genetic and biochemical evidence that Smad binding is determined by DNA-binding cofactors that lend added affinity and selectivity to canonical Smad4-RSmad complexes (Massagué et al., 2005). A straightforward resolution of this apparent conflict is suggested by the different nature of two types of Smad3 binding events at gene promoters. Oct4 imposes chromatin marks such as H3K9me3 that would determine the pattern of TRIM33-Smad3 binding, as demonstrated in the case of Gsc and Mixl1(Xi et al., 2011). Thus, Oct4 (with Sox2 and nanog) licenses specific sites for Smad3 binding as part of a TRIM33-Smad3 complex. This would explain the observed cohabitation of Oct4 and Smad3 in common promoter regions throughout the genome. The Oct4-mediated licensing of specific enhancers for Smad3 binding would be necessary but not sufficient for transcriptional activation of genes such as Gsc and Mixl1. Activation additionally requires binding of the Smad4-Smad2/3-FoxH1 complex. FoxH1 adds affinity and selectivity for binding the Smad4-Smad2/3-FoxH1 complex to the ARE, which is a composite of Smad binding and FoxH1 binding elements. AREs, also called Smad/FoxH1 enhancers (SFEs), are present throughout the genome and are define Smad/FoxH1 gene synexpression groups (Silvestri et al., 2008). The Smad4-Smad2/3-FoxH1 complex would then trigger Pol II transcriptional action. In this model, Oct4 would direct Smad3 (with TRIM33) to regionally relax the chromatin and to license these promoters for subsequent binding of Smad4-Smad2/3-FoxH1 complexes to one set of genes, and other complexes with other DNA-binding partners to access different sets of genes.
Outlook
Progress in this area over the past few years has brought to a new level of understanding how TGF-β signals control intersect with the chromatin of master differentiation genes in stem cells. The collusion of Smad transcriptional complexes with master regulator genes and their products exemplified by the co-occupancy of common sites in target genes and the involvement of a chromatin reader Smad complexes in gene activation are pioneering elements of new knowledge that open doors for a better understanding of how stem cell read TGF-β signals.
From the standpoint of TRIM33-Smad2/3 as a mediator of access to AREs, it will be important to define exactly how this complex regionally remodels the chromatin to expose the AREs. Is this based just on evicting HP1 from H3K9 methylated nucleosomes, or is there more to it? TRIM33 has been shown to mediate transcription elongation of erythroid differentiation genes in hematopoietic progenitors (Bai et al., 2010). The hematopoietic transcription factor SCL and the elongation factors p-TEFb (including cyclin-T1 and CDK9) and FACT participate in this process (Bai et al., 2010). Are access to poised genes and transcription elongation two independent functions of TRIM33, or two sequential functions of the same TRIM33 complex?
Questions also emerge from the role of master enforcers of a particular developmental stage (e.g. Oct4 and Sox2 in the pluripotent stage) as determinants of Smad3 binding to sites throughout the genome. For example, does the bound Smad3 correspond to TRIM33-Smad3 complexes or Smad4-Smad3 complexes? And, how come that thousands of Oct4/Smad3 co-ocupied sites are detected in the genome of ESCs exposed to nodal, yet only about 100 genes respond to nodal?
The newly delineated mechanism of TGF-β control of stem cell differentiation genes may be relevant beyond the TGF-β pathway. Similar events involving signal-driven chromatin reader complexes that open the way for signal-driven Pol II activating complexes may operate in other developmental pathways whose target genes are also secluded by repressive chromatin. The new insights into the mechanism provide a general framework for further analysis of these critical questions.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Agricola E, Randall RA, Gaarenstroom T, Dupont S, Hill CS. Recruitment of TIF1gamma to Chromatin via Its PHD Finger-Bromodomain Activates Its Ubiquitin Ligase and Transcriptional Repressor Activities. Mol Cell. 2011;43:85–96. doi: 10.1016/j.molcel.2011.05.020. [DOI] [PubMed] [Google Scholar]
- Alarcón C, Zaromytidou AI, Xi Q, Gao S, Yu J, Fujisawa S, Barlas A, Miller AN, Manova-Todorova K, Macias MJ, et al. Nuclear CDKs drive Smad transcriptional activation and turnover in BMP and TGF-beta pathways. Cell. 2009;139:757–769. doi: 10.1016/j.cell.2009.09.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aragón E, Goerner N, Zaromytidou AI, Xi Q, Escobedo A, Massagué J, Macias MJ. A Smad action turnover switch operated by WW domain readers of a phosphoserine code. Genes Dev. 2011;25:1275–1288. doi: 10.1101/gad.2060811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai X, Kim J, Yang Z, Jurynec MJ, Akie TE, Lee J, LeBlanc J, Sessa A, Jiang H, DiBiase A, et al. TIF1gamma controls erythroid cell fate by regulating transcription elongation. Cell. 2010;142:133–143. doi: 10.1016/j.cell.2010.05.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bardeesy N, Cheng KH, Berger JH, Chu GC, Pahler J, Olson P, Hezel AF, Horner J, Lauwers GY, Hanahan D, et al. Smad4 is dispensable for normal pancreas development yet critical in progression and tumor biology of pancreas cancer. Genes Dev. 2006;20:3130–3146. doi: 10.1101/gad.1478706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blum M, Gaunt SJ, Cho KW, Steinbeisser H, Blumberg B, Bittner D, De Robertis EM. Gastrulation in the mouse: the role of the homeobox gene goosecoid. Cell. 1992;69:1097–1106. doi: 10.1016/0092-8674(92)90632-m. [DOI] [PubMed] [Google Scholar]
- Bruce DL, Sapkota GP. Phosphatase in SMAD Regulation. FEBS letter. 2012 doi: 10.1016/j.febslet.2012.02.001. [DOI] [PubMed] [Google Scholar]
- Chen X, Weisberg E, Fridmacher V, Watanabe M, Naco G, Whitman M. Smad4 and FAST-1 in the assembly of activin-responsive factor. Nature. 1997;389:85–89. doi: 10.1038/38008. [DOI] [PubMed] [Google Scholar]
- Dupont S, Mamidi A, Cordenonsi M, Montagner M, Zacchigna L, Adorno M, Martello G, Stinchfield MJ, Soligo S, Morsut L, et al. FAM/USP9x, a deubiquitinating enzyme essential for TGFbeta signaling, controls Smad4 monoubiquitination. Cell. 2009;136:123–135. doi: 10.1016/j.cell.2008.10.051. [DOI] [PubMed] [Google Scholar]
- Dupont S, Zacchigna L, Cordenonsi M, Soligo S, Adorno M, Rugge M, Piccolo S. Germ-layer specification and control of cell growth by Ectodermin, a Smad4 ubiquitin ligase. Cell. 2005;121:87–99. doi: 10.1016/j.cell.2005.01.033. [DOI] [PubMed] [Google Scholar]
- Flavell RA, Sanjabi S, Wrzesinski SH, Licona-Limon P. The polarization of immune cells in the tumour environment by TGFbeta. Nat Rev Immunol. 2010;10:554–567. doi: 10.1038/nri2808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuentealba LC, Eivers E, Ikeda A, Hurtado C, Kuroda H, Pera EM, De Robertis EM. Integrating patterning signals: Wnt/GSK3 regulates the duration of the BMP/Smad1 signal. Cell. 2007;131:980–993. doi: 10.1016/j.cell.2007.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furuhashi M, Yagi K, Yamamoto H, Furukawa Y, Shimada S, Nakamura Y, Kikuchi A, Miyazono K, Kato M. Axin facilitates Smad3 activation in the transforming growth factor beta signaling pathway. Mol Cell Biol. 2001;21:5132–5141. doi: 10.1128/MCB.21.15.5132-5141.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao S, Alarcón C, Sapkota G, Rahman S, Chen PY, Goerner N, Macias MJ, Erdjument-Bromage H, Tempst P, Massagué J. Ubiquitin ligase Nedd4L targets activated Smad2/3 to limit TGF-beta signaling. Mol Cell. 2009;36:457–468. doi: 10.1016/j.molcel.2009.09.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo X, Waddell DS, Wang W, Wang Z, Liberati NT, Yong S, Liu X, Wang XF. Ligand-dependent ubiquitination of Smad3 is regulated by casein kinase 1 gamma 2, an inhibitor of TGF-beta signaling. Oncogene. 2008;27:7235–7247. doi: 10.1038/onc.2008.337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hart AH, Hartley L, Sourris K, Stadler ES, Li R, Stanley EG, Tam PP, Elefanty AG, Robb L. Mixl1 is required for axial mesendoderm morphogenesis and patterning in the murine embryo. Development. 2002;129:3597–3608. doi: 10.1242/dev.129.15.3597. [DOI] [PubMed] [Google Scholar]
- Hatakeyama S. TRIM proteins and cancer. Nat Rev Cancer. 2011;11:792–804. doi: 10.1038/nrc3139. [DOI] [PubMed] [Google Scholar]
- He W, Dorn DC, Erdjument-Bromage H, Tempst P, Moore MA, Massagué J. Hematopoiesis controlled by distinct TIF1gamma and Smad4 branches of the TGFbeta pathway. Cell. 2006;125:929–941. doi: 10.1016/j.cell.2006.03.045. [DOI] [PubMed] [Google Scholar]
- Heldin CH, Landstrom M, Moustakas A. Mechanism of TGF-beta signaling to growth arrest, apoptosis, and epithelial-mesenchymal transition. Curr Opin Cell Biol. 2009;21:166–176. doi: 10.1016/j.ceb.2009.01.021. [DOI] [PubMed] [Google Scholar]
- Hocevar BA, Smine A, Xu XX, Howe PH. The adaptor molecule Disabled-2 links the transforming growth factor beta receptors to the Smad pathway. EMBO J. 2001;20:2789–2801. doi: 10.1093/emboj/20.11.2789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh S, Ericsson J, Nishikawa J, Heldin CH, ten Dijke P. The transcriptional co-activator P/CAF potentiates TGF-beta/Smad signaling. Nucleic Acids Res. 2000;28:4291–4298. doi: 10.1093/nar/28.21.4291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kahata K, Hayashi M, Asaka M, Hellman U, Kitagawa H, Yanagisawa J, Kato S, Imamura T, Miyazono K. Regulation of transforming growth factor-beta and bone morphogenetic protein signalling by transcriptional coactivator GCN5. Genes Cells. 2004;9:143–151. doi: 10.1111/j.1365-2443.2004.00706.x. [DOI] [PubMed] [Google Scholar]
- Kang JS, Alliston T, Delston R, Derynck R. Repression of Runx2 function by TGF-beta through recruitment of class II histone deacetylases by Smad3. EMBO J. 2005;24:2543–2555. doi: 10.1038/sj.emboj.7600729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato Y, Habas R, Katsuyama Y, Naar AM, He X. A component of the ARC/Mediator complex required for TGF beta/Nodal signalling. Nature. 2002;418:641–646. doi: 10.1038/nature00969. [DOI] [PubMed] [Google Scholar]
- Khetchoumian K, Teletin M, Mark M, Lerouge T, Cervino M, Oulad-Abdelghani M, Chambon P, Losson R. TIF1delta, a novel HP1-interacting member of the transcriptional intermediary factor 1 (TIF1) family expressed by elongating spermatids. J Biol Chem. 2004;279:48329–48341. doi: 10.1074/jbc.M404779200. [DOI] [PubMed] [Google Scholar]
- Labbe E, Silvestri C, Hoodless PA, Wrana JL, Attisano L. Smad2 and Smad3 positively and negatively regulate TGF beta-dependent transcription through the forkhead DNA-binding protein FAST2. Mol Cell. 1998;2:109–120. doi: 10.1016/s1097-2765(00)80119-7. [DOI] [PubMed] [Google Scholar]
- Le Douarin B, Nielsen AL, Garnier JM, Ichinose H, Jeanmougin F, Losson R, Chambon P. A possible involvement of TIF1 alpha and TIF1 beta in the epigenetic control of transcription by nuclear receptors. EMBO J. 1996;15:6701–6715. [PMC free article] [PubMed] [Google Scholar]
- Lin HK, Bergmann S, Pandolfi PP. Cytoplasmic PML function in TGF-beta signalling. Nature. 2004;431:205–211. doi: 10.1038/nature02783. [DOI] [PubMed] [Google Scholar]
- Lönn P, van der Heide LP, Dahl M, Hellman U, Heldin CH, Moustakas A. PARP-1 attenuates Smad-mediated transcription. Mol Cell. 2010;40:521–532. doi: 10.1016/j.molcel.2010.10.029. [DOI] [PubMed] [Google Scholar]
- Massagué J. How cells read TGF-beta signals. Nat Rev Mol Cell Biol. 2000;1:169–178. doi: 10.1038/35043051. [DOI] [PubMed] [Google Scholar]
- Massagué J. TGFbeta in Cancer. Cell. 2008;134:215–230. doi: 10.1016/j.cell.2008.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massagué J, Gomis RR. The logic of TGFbeta signaling. FEBS Lett. 2006;580:2811–2820. doi: 10.1016/j.febslet.2006.04.033. [DOI] [PubMed] [Google Scholar]
- Massagué J, Seoane J, Wotton D. Smad transcription factors. Genes Dev. 2005;19:2783–2810. doi: 10.1101/gad.1350705. [DOI] [PubMed] [Google Scholar]
- Mikkelsen TS, Ku M, Jaffe DB, Issac B, Lieberman E, Giannoukos G, Alvarez P, Brockman W, Kim TK, Koche RP, et al. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature. 2007;448:553–560. doi: 10.1038/nature06008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morsut L, Yan KP, Enzo E, Aragóna M, Soligo SM, Wendling O, Mark M, Khetchoumian K, Bressan G, Chambon P, et al. Negative control of Smad activity by ectodermin/Tif1gamma patterns the mammalian embryo. Development. 2010;137:2571–2578. doi: 10.1242/dev.053801. [DOI] [PubMed] [Google Scholar]
- Mullen AC, Orlando DA, Newman JJ, Loven J, Kumar RM, Bilodeau S, Reddy J, Guenther MG, DeKoter RP, Young RA. Master transcription factors determine cell-type-specific responses to TGF-beta signaling. Cell. 2011;147:565–576. doi: 10.1016/j.cell.2011.08.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murry CE, Keller G. Differentiation of embryonic stem cells to clinically relevant populations: lessons from embryonic development. Cell. 2008;132:661–680. doi: 10.1016/j.cell.2008.02.008. [DOI] [PubMed] [Google Scholar]
- Ozato K, Shin DM, Chang TH, Morse HC., 3rd TRIM family proteins and their emerging roles in innate immunity. Nat Rev Immunol. 2008;8:849–860. doi: 10.1038/nri2413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ransom DG, Bahary N, Niss K, Traver D, Burns C, Trede NS, Paffett-Lugassy N, Saganic WJ, Lim CA, Hersey C, et al. The zebrafish moonshine gene encodes transcriptional intermediary factor 1gamma, an essential regulator of hematopoiesis. PLoS Biol. 2004;2:E237. doi: 10.1371/journal.pbio.0020237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reissmann E, Jornvall H, Blokzijl A, Andersson O, Chang C, Minchiotti G, Persico MG, Ibanez CF, Brivanlou AH. The orphan receptor ALK7 and the Activin receptor ALK4 mediate signaling by Nodal proteins during vertebrate development. Genes Dev. 2001;15:2010–2022. doi: 10.1101/gad.201801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross S, Cheung E, Petrakis TG, Howell M, Kraus WL, Hill CS. Smads orchestrate specific histone modifications and chromatin remodeling to activate transcription. Embo J. 2006;25:4490–4502. doi: 10.1038/sj.emboj.7601332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruthenburg AJ, Li H, Patel DJ, Allis CD. Multivalent engagement of chromatin modifications by linked binding modules. Nat Rev Mol Cell Biol. 2007;8:983–994. doi: 10.1038/nrm2298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sapkota G, Alarcón C, Spagnoli FM, Brivanlou AH, Massagué J. Balancing BMP signaling through integrated inputs into the Smad1 linker. Mol Cell. 2007;25:441–454. doi: 10.1016/j.molcel.2007.01.006. [DOI] [PubMed] [Google Scholar]
- Schier AF. Nodal signaling in vertebrate development. Annu Rev Cell Dev Biol. 2003;19:589–621. doi: 10.1146/annurev.cellbio.19.041603.094522. [DOI] [PubMed] [Google Scholar]
- Silvestri C, Narimatsu M, von Both I, Liu Y, Tan NB, Izzi L, McCaffery P, Wrana JL, Attisano L. Genome-wide identification of Smad/Foxh1 targets reveals a role for Foxh1 in retinoic acid regulation and forebrain development. Dev Cell. 2008;14:411–423. doi: 10.1016/j.devcel.2008.01.004. [DOI] [PubMed] [Google Scholar]
- Singh PB. HP1 proteins--what is the essential interaction? Genetika. 2011;46:1424–1429. [PubMed] [Google Scholar]
- Tang Y, Katuri V, Dillner A, Mishra B, Deng CX, Mishra L. Disruption of transforming growth factor-beta signaling in ELF beta-spectrin-deficient mice. Science. 2003;299:574–577. doi: 10.1126/science.1075994. [DOI] [PubMed] [Google Scholar]
- Taverna SD, Li H, Ruthenburg AJ, Allis CD, Patel DJ. How chromatin-binding modules interpret histone modifications: lessons from professional pocket pickers. Nat Struct Mol Biol. 2007;14:1025–1040. doi: 10.1038/nsmb1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trompouki E, Bowman TV, Lawton LN, Fan ZP, Wu DC, DiBiase A, Martin CS, Cech JN, Sessa AK, Leblanc JL, et al. Lineage regulators direct BMP and Wnt pathways to cell-specific programs during differentiation and regeneration. Cell. 2011;147:577–589. doi: 10.1016/j.cell.2011.09.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai WW, Wang Z, Yiu TT, Akdemir KC, Xia W, Winter S, Tsai CY, Shi X, Schwarzer D, Plunkett W, et al. TRIM24 links a non-canonical histone signature to breast cancer. Nature. 2010;468:927–932. doi: 10.1038/nature09542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukazaki T, Chiang TA, Davison AF, Attisano L, Wrana JL. SARA, a FYVE domain protein that recruits Smad2 to the TGFbeta receptor. Cell. 1998;95:779–791. doi: 10.1016/s0092-8674(00)81701-8. [DOI] [PubMed] [Google Scholar]
- Varelas X, Samavarchi-Tehrani P, Narimatsu M, Weiss A, Cockburn K, Larsen BG, Rossant J, Wrana JL. The Crumbs complex couples cell density sensing to Hippo-dependent control of the TGF-beta-SMAD pathway. Dev Cell. 2010;19:831–844. doi: 10.1016/j.devcel.2010.11.012. [DOI] [PubMed] [Google Scholar]
- Vincent DF, Yan KP, Treilleux I, Gay F, Arfi V, Kaniewski B, Marie JC, Lepinasse F, Martel S, Goddard-Leon S, et al. Inactivation of TIF1gamma cooperates with Kras to induce cystic tumors of the pancreas. PLoS Genet. 2009;5:e1000575. doi: 10.1371/journal.pgen.1000575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu MY, Hill CS. Tgf-beta superfamily signaling in embryonic development and homeostasis. Dev Cell. 2009;16:329–343. doi: 10.1016/j.devcel.2009.02.012. [DOI] [PubMed] [Google Scholar]
- Xi Q, He W, Zhang XH, Le HV, Massagué J. Genome-wide impact of the BRG1 SWI/SNF chromatin remodeler on the transforming growth factor beta transcriptional program. J Biol Chem. 2008;283:1146–1155. doi: 10.1074/jbc.M707479200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xi Q, Wang Z, Zaromytidou AI, Zhang XH, Chow-Tsang LF, Liu JX, Kim H, Barlas A, Manova-Todorova K, Kaartinen V, et al. A poised chromatin platform for TGF-beta access to master regulators. Cell. 2011;147:1511–1524. doi: 10.1016/j.cell.2011.11.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu L, Massagué J. Nucleocytoplasmic shuttling of signal transducers. Nat Rev Mol Cell Biol. 2004;5:209–219. doi: 10.1038/nrm1331. [DOI] [PubMed] [Google Scholar]
- Xu L, Yao X, Chen X, Lu P, Zhang B, Ip YT. Msk is required for nuclear import of TGF-{beta}/BMP-activated Smads. J Cell Biol. 2007;178:981–994. doi: 10.1083/jcb.200703106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamakawa N, Tsuchida K, Sugino H. The rasGAP-binding protein, Dok-1, mediates activin signaling via serine/threonine kinase receptors. EMBO J. 2002;21:1684–1694. doi: 10.1093/emboj/21.7.1684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao X, Chen X, Cottonham C, Xu L. Preferential utilization of Imp7/8 in nuclear import of Smads. J Biol Chem. 2008;283:22867–22874. doi: 10.1074/jbc.M801320200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yap KL, Zhou MM. Keeping it in the family: diverse histone recognition by conserved structural folds. Crit Rev Biochem Mol Biol. 2010;45:488–505. doi: 10.3109/10409238.2010.512001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeo C, Whitman M. Nodal signals to Smads through Cripto-dependent and Cripto-independent mechanisms. Mol Cell. 2001;7:949–957. doi: 10.1016/s1097-2765(01)00249-0. [DOI] [PubMed] [Google Scholar]
- Young RA. Control of the embryonic stem cell state. Cell. 2011;144:940–954. doi: 10.1016/j.cell.2011.01.032. [DOI] [PMC free article] [PubMed] [Google Scholar]