Abstract
Comment on: Travesa A, et al. EMBO J 2012; 31:1811-22.
Keywords: checkpoint, DNA damage, DNA replication, G1/S transcription, MBF
Genomic stability is crucial to maintain cell viability and avoid errors in control of proliferation that often lead to disease. Cell cycle checkpoints have evolved as molecular mechanisms to prevent this problem. DNA damage or DNA replication stress during S phase activates the intra-S phase checkpoint, which ensures faithful DNA replication by multiple mechanisms, including inducing expression of genes involved in DNA repair and DNA replication and blocking cell cycle progression until the damage has been resolved.1,2
Until recently, only a small number of genes, primarily those having the DNA damage signature, have been shown to be induced in response to DNA damage in budding yeast.3 However, the use of asynchronous populations in those studies obscured the induction of gene expression that occurs only during specific cell cycle phases. To address that problem, we studied the effect of DNA damage and DNA replication stress by genome-wide expression analysis using budding yeast cells synchronously traversing the cell division cycle.4
Cells were treated with three well-studied genotoxins: methyl methane sulfonate (MMS), camptothecin (CPT) or hydroxyurea (HU). MMS and CPT both generate DNA damage. However, nucleotide base methylation induced by MMS is sensed during S phase, whereas the double-strand breaks that ultimately result from treatment with CPT are thought to be sensed and repaired during G2 phase.5,6 Unlike MMS and CPT, HU generates replication stress by depleting the dNTP pools.7 Thus, each of these three genotoxic agents has distinct effects, enabling us to study the transcriptional response to three different forms of genotoxic stress.
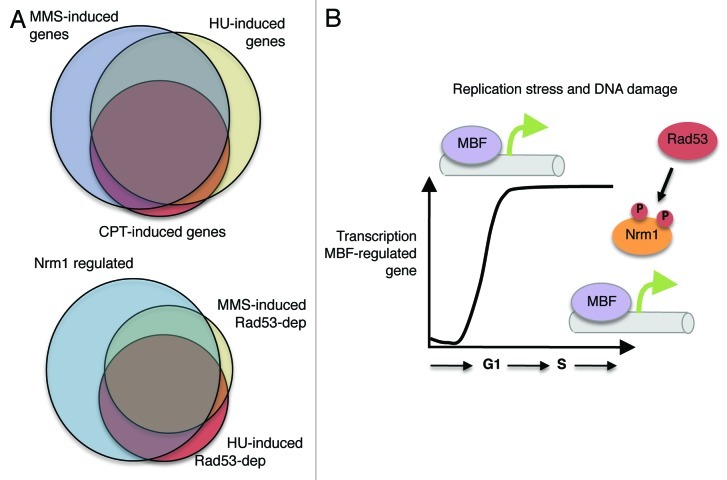
Interestingly, cells responding to MMS-induced DNA damage or HU-induced DNA replication stress lead to induction of a subset of G1/S genes during S phase, an interval during which those genes are normally repressed (Fig. 1A).4 The overlap between the genes induced by each of those treatments is greater than 75%. In contrast, treatment with CPT leads to induction of only about a quarter of the G1/S genes. Yet, despite the smaller number, nearly all of the genes that are induced by CPT are common with those induced by the other treatments, suggesting that the damage induced by CPT generates DNA replication stress to an extent sufficient to promote a response during S phase. Furthermore, the large degree of overlap between the genes induced by each of these treatments suggests that expression of a common set of genes is generally beneficial to cells responding to genotoxic insults (Fig. 1A).
Figure 1. (A) Relationship between genes induced by genotoxins. Venn diagrams showing the overlap between genes activated in response to MMS, HU and CPT (top diagram) and between the checkpoint-dependent genes induced by HU, MMS and Nrm1-regulated genes (bottom diagram). (B) Regulation of G1/S transcription by the checkpoint. Rad53 phosphorylates Nrm1, blocking its binding to MBF and, thus, allowing the MBF-regulated transcription to remain active during S phase.
To establish whether the induction of genes in response to the genotoxic agents is a consequence of checkpoint activation, we evaluated the genome-wide expression profile in cells deficient in the Rad53 protein kinase, the central effector checkpoint kinase in S. cerevisiae. Approximately half of the genes induced by these genotoxic agents are dependent upon a functional checkpoint. Furthermore, they are enriched for genes encoding functions related to cell cycle, DNA replication and DNA repair. The checkpoint-dependent genes that are induced in response to HU and MMS exhibit greater overlap (70%, Fig. 1A) than those that are checkpoint-independent (less than 50%).
G1/S genes are expressed under the control of two transcription factors, MBF and SBF. Whereas, MBF-regulated genes encode many proteins involved in DNA replication and repair, those regulated by SBF more often encode proteins involved in cell cycle timing and morphogenesis. Importantly, almost all the genes that are induced in a checkpoint-dependent manner are regulated by MBF and its transcriptional corepressor Nrm1 (Fig. 1A). This suggests that a large portion of the genes induced in response to DNA replication stress are regulated through a common pathway.
Indeed, using biochemical and genetic approaches, we show that the Rad53 protein kinase directly phosphorylates Nrm1 in response to activation of the checkpoint, thereby inducing MBF-regulated transcription (Fig. 1B). Those findings are reinforced by a companion report, reviewed here by Smolka, et al., 2012.8,9 Taken together, these observations elucidate a previously uncharacterized branch of the DNA replication checkpoint pathway that is responsible for induction of the larger of the two clusters of checkpoint-regulated genes. This checkpoint regulation of G1/S gene expression is conserved in the distantly related fungi, Schizosaccharomyces pombe.10 By inducing many genes involved in DNA replication and repair, this newly characterized pathway enhances genomic stability in the face of a broad range of genotoxic stresses.
Acknowledgments
The work described was supported by U.S. Public Health Service Grant R01 GM59441 to C.W.
Glossary
Abbreviations:
- MMS
methyl methane sulfonate
- CPT
camptothecin
- HU
hydroxyurea
- SBF
SCB binding factor
- MBF
MCB binding factor
- DNA
deoxyribonucleic acid
- dNTP
deoxynucleoside triphosphate
Footnotes
Previously published online: www.landesbioscience.com/journals/cc/article/21587
References
- 1.Hanahan D, et al. Cell. 2000;100:57–70. doi: 10.1016/S0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- 2.Hartwell LH, et al. Science. 1989;246:629–34. doi: 10.1126/science.2683079. [DOI] [PubMed] [Google Scholar]
- 3.Gasch AP, et al. Mol Biol Cell. 2001;12:2987–3003. doi: 10.1091/mbc.12.10.2987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Travesa A, et al. EMBO J. 2012;31:1811–22. doi: 10.1038/emboj.2012.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tercero JA, et al. Nature. 2001;412:553–7. doi: 10.1038/35087607. [DOI] [PubMed] [Google Scholar]
- 6.Redon C, et al. EMBO Rep. 2003;4:678–84. doi: 10.1038/sj.embor.embor871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lopes M, Cotta-Ramusino C, Pellicioli A, Liberi G, Plevani P, Muzi-Falconi M, et al. Nature. 2001;412:557–61. doi: 10.1038/35087613. [DOI] [PubMed] [Google Scholar]
- 8.Bastos de Oliveira FM, et al. EMBO J. 2012;31:1798–810. doi: 10.1038/emboj.2012.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Smolka MB, et al. Cell Cycle. 2012 doi: 10.4161/cc.22933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.de Bruin RA, et al. Proc Natl Acad Sci USA. 2008;105:11230–5. doi: 10.1073/pnas.0801106105. [DOI] [PMC free article] [PubMed] [Google Scholar]