Abstract
The replication checkpoint signaling network monitors the presence of replication-induced lesions to DNA and coordinates an elaborate cellular response that includes ample transcriptional reprogramming. Recent work has established two major groups of replication stress-induced genes in Saccharomyces cerevisiae, the DNA damage response (DDR) genes and G1/S cell cycle (CC) genes. In both cases, transcriptional activation is mediated via checkpoint-dependent inhibition of a transcriptional repressor (Crt1 for DDR and Nrm1 for CC) that participates in negative feedback regulation. This repressor-mediated regulation enables transcription to be rapidly repressed once cells have dealt with the replication stress. The recent finding of a new class of CC genes, named “switch genes,” further uncovers a mode of transcription regulation that prevents overexpression of replication stress induced genes during G1. Collectively, these findings highlight the need for mechanisms that tightly control replication stress-induced transcription, allowing rapid transcriptional activation during replication stress but also avoiding long-term hyperaccumulation of the induced protein product that may be detrimental to cell proliferation.
Keywords: replication stress, replication checkpoint, yeast, G1-S transcription, Rad53, Nrm1, SBF and MBF, switch genes, checkpoint effectors, dosage-sensitive genes
Introduction
Stress generated during the process of DNA replication is arguably one of the most important challenges that proliferating cells need to cope with in order to ensure cell survival and the maintenance of genomic integrity.1-5 Every cell going through S-phase experiences a certain level of replication stress generated either through the collisions of the replication fork with transcriptional machineries localized in highly transcribed regions of the DNA (e.g., rDNA and tRNA genes), through the replication of “hard-to-replicate” repetitive DNA sequences, by limited supplies of dNTPs, or when the replication fork encounters DNA templates damaged by endogenous or exogenous factors (e.g., reactive oxygen species and UV radiation, respectively).6-12 Transcriptional activation of selected genes is an important part of the cellular response that allows cells to better cope with DNA replication stress. However, this transcriptional activation needs to be tightly regulated, as overexpression of some proteins may adversely affect the ability of cells to progress in the cell cycle once the stress ceases.
Here we discuss our current understanding of the molecular circuitries that control transcription levels of DNA replication stress-induced genes. Our recent work, published in back-to-back papers with the Wittenberg group in The EMBO Journal, establishes two major groups of replication stress-induced genes in Saccharomyces cerevisiae, the DNA damage response (DDR) genes and G1/S cell cycle (CC) genes. In both cases, transcriptional activation is mediated via checkpoint-dependent inhibition of a transcriptional repressor (Crt1 for DDR and Nrm1 for CC) that participates in negative feedback regulation.13,14 This repressor-mediated regulation enables transcription to be rapidly repressed once cells have dealt with the replication stress and shares similarities with transcriptional repression mechanisms found in bacteria and mammals.15,16 Our recent discovery of a select group of CC promoter genes that undergo a transcriptional factor switch during the G1-to-S transition (which we named the “switch genes”) further illustrates the need for tight control of replication stress-induced genes.
Sensing Replication Stress via DNA Damage Checkpoint Kinases
Replication stress-induced transcriptional responses are mostly regulated by phosphorylation-mediated signals emanated by DNA damage checkpoint kinases. DNA damage checkpoint kinases are evolutionarily conserved proteins that constantly monitor DNA structure during any stage of the cell cycle.17,18 The involvement of checkpoint kinases in specifically monitoring DNA structures during DNA replication is often referred to as the replication checkpoint. Many reviews specifically focus on the DNA checkpoint kinases and their multiple roles in the DNA damage response, and we refer to them for more detailed information about their role in controlling transcription-independent responses.19-21 At the heart of the DNA damage checkpoint in human cells lays the phosphatidyl-inositol-3-kinase-like sensor kinases ATR/ATM and the downstream effector kinases CHK2/CHK1. ATR and ATM are at the top of the signaling cascade and act partially redundantly to phosphorylate common substrates at S/T-Q motifs.22 ATR and ATM phosphorylate and control the activity of CHK1 and CHK2.17,23-26 ATM and CHK2 are mainly activated by DNA double-strand breaks (DSBs), whereas ATR and CHK1 respond to a wide variety of replication-induced lesions that lead to the accumulation of single-stranded DNA (ssDNA). Although there is some degree of crosstalk between the two kinases, ATR has a preponderant role in replication checkpoint signaling. In mammalian cells, ATR and CHK1 are essential kinases that phosphorylate an extremely complex network of substrates; however, how these phosphorylations regulate the replication stress response is not fully understood.27
Budding yeast is a very useful system to study the replication checkpoint due to a high degree of conservation of the main checkpoint kinases. In addition to its powerful genetics, the use of budding yeast allows the deletion of the functional counterparts of human ATR and CHK1 (Mec1 and Rad53, respectively) in the presence of a rescuing deletion of the ribonucleotide reductase inhibitor Sml1.28 In contrast, deletion of these essential kinases leads to lethality in most model organisms including human tissue culture cells. The architecture of the pathway in budding yeast and humans is strikingly similar. In the same way to ATR and CHK1, Mec1 and Rad53 are central to the replication stress response, and dysfunctions in these kinases cause hypersensitivity to replication stress and induce genomic instability.17,18,24,29-31 Mec1 and Rad53-dependent signaling is crucial to mediate stabilization and repair of replication forks, inhibition of late origin firing, increased production of deoxyribonucleotides (dNTPs), among other responses.1,9,10,14,28,31-38 While the dramatic loss of viability observed in cells lacking Mec1 and/or Rad53 exposed to replication stress is related to translation-independent functions, the identification of multiple functional connections between Mec1/Rad53 signaling and transcription highlight the central importance of regulating transcription during replication stress.30,31,39,40
Checkpoint-Dependent Transcriptional Regulation in Yeast
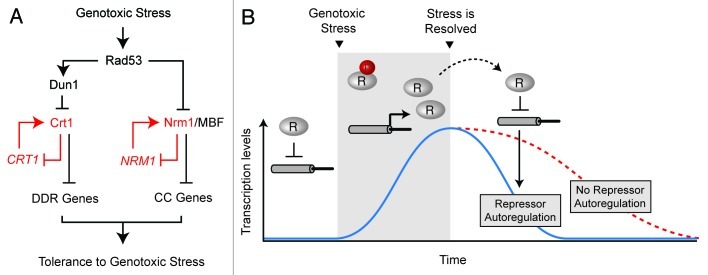
Since the very early work in the 80s monitoring gene expression in response to DNA damage, it was known that genotoxicity leads to repression and activation of different subsets of genes.41-45 This became clear in the late 90s with the use of high-throughput microarray technologies.46 Genome-wide expression analysis in yeast has revealed that in response to replication stress induced by methyl methane sulphonate (MMS), ionizing radiation (IR), hydroxyurea (HU) and UV radiation, hundreds of genes are either induced or repressed.46-48 Consistent with the notion that the DNA damage checkpoint is a central trigger for most of the transcriptional responses to replication stress, the vast majority of the observed replication-specific changes in expression were indeed found to be dependent on the Mec1/Rad53 kinase-signaling pathway.47,49 This pathway can be divided into two major branches, one mediated by the most downstream checkpoint protein kinase Dun1, a kinase that is directly phosphorylated and activated by Rad53, while the other branch is Dun1-independent (Fig. 1A).13,14,50,51
Figure 1. (A) Checkpoint-dependent transcriptional pathways in budding yeast. Upon genotoxic stress, Rad53 de-represses transcription of DNA damage response (DDR) and G1/S cell cycle (CC) genes via Dun1/Crt1 and Nrm1/MBF pathways, respectively. Crt1 and Nrm1 auto-regulation is indicated. (B) Crt1 and Nrm1 activity is regulated by a negative feedback loop. Upon DNA replication stress, Crt1 and Nrm1 repressors (indicated by the letter “R”) are phosphorylated in a checkpoint dependent manner. Phosphorylation releases Crt1 and Nrm1 from target promoters including their own promoters, leading to transcription activation. Increased transcription results in a continues production of unphosphorylated Crt1 and Nrm1 during the stress response, which allow for rapid downregulation of transcription after genotoxic stress is resolved.
Transcriptional Regulation of DNA Damage Response (DDR) Genes
The first set of co-regulated genes induced as part of the DNA replication and damage checkpoint response was identified in S. cerevisiae and named the DNA damage response (DDR) genes. This small group of genes includes genes that encode for the subunits of ribonucleotide reductase (RNR2, RNR3 and RNR4) and additional genes whose functions are not well understood, such as HUG1, FSH3, YLR345W and NTH2.14,52 The initial study, performed in the Elledge lab, showed that the mechanism of DDR transcriptional regulation involves Dun1-dependent phosphorylation and inactivation of the transcriptional repressor Crt1. Crt1 is bound to the promoters of DDR genes, as well as to its own promoter, and is released upon phosphorylation. Therefore, Dun1-dependent phosphorylation of Crt1 results in de-repression and consequent activation of DDR genes.14 The Dun1-Crt1 regulatory circuit is not conserved in higher eukaryotes, and in budding yeast it represents only a small part of the checkpoint-dependent transcriptional response.
Transcriptional Regulation of G1/S Cell Cycle (CC) Genes during Replication Stress
Recent work revealed that the transcription of a large group of cell cycle-dependent genes known as G1/S genes is also induced as part of the DNA replication stress transcriptional response in S. cerevisiae.13,51 Under unperturbed conditions, the transcription of G1/S cell cycle genes peaks during the transition from G1 to S-phase, and as cells progress into S-phase the transcription of G1/S cell cycle genes is inactivated (for review on G1/S genes, see ref. 53). Under replication stress, a number of G1/S genes exhibited sustained transcription in S phase. This group of replication stress-induced G1/S genes includes close to 100 genes and consequently represents one of the largest single groups of co-regulated targets among DNA-damage induced genes.13,51 It is well established that G1/S transcription depends on the SBF and MBF transcription factor complexes that regulate over 200 cell cycle genes.53,54 SBF and MBF are comprised of the common regulatory component Swi6 and a specific DNA binding protein, Swi4 or Mbp1, respectively.53,55 Swi4 and Mbp1 recognize distinct DNA sequence elements named SCB and MCB, respectively, which define SBF and MBF target promoters. Whereas the majority of G1/S targets are regulated by either SBF or MBF, a small subset is thought to be regulated by both.56-58 SBF has been shown to be required for activating transcription during G1, while MBF is involved in repressing G1/S transcription outside of G1, thereby confining expression to the G1 phase of the cell cycle.56,59,60
Our recent work, published in The EMBO Journal, revealed that CC genes that depend on MBF for inactivation once cells progress into S phase are transcriptionally induced upon replication stress. These two studies show that MBF targets, but not SBF targets, are upregulated via checkpoint-dependent inactivation of the Nrm1 co-repressor. The regulation of Nrm1 was found to be Rad53-dependent but Dun1-independent, and is clear that it represents a pathway that is parallel to the Rad53-Dun1-Crt1-DDR pathway downstream of Rad53. Interestingly, similarly to Crt1, Nrm1 also targets its own promoter and represses its expression.
Checkpoint Interference of Negative Feedback Loops
As described above, during DNA replication stress, part of the G1/S cell cycle transcription program is maintained in an active state by interference of a negative feedback-loop involving the transcriptional repressor Nrm1.61,62 Interestingly, the repressor Crt1, involved in regulation of the DNA damage-inducible genes also participates in a negative feedback response. The capacity of both Nrm1 and Crt1 to repress their own expression allows rapid downregulation of the transcription of their target after cells recovery from a DNA replication arrest (Fig. 1B). Overall, interference of transcriptional repressors that participate in a negative feedback, as part of the checkpoint transcriptional response, appears to be conserved throughout evolution. In E. coli, LexA mediates feedback regulation during the SOS response and mammalian Mdm2, a negative regulator of p53, mediates feedback regulation of transcription during recovery from the DNA damage checkpoint response.15,16 The conservation of this particular network wiring shows that the transcriptional response initiated by DNA damage and replication stress needs to be rapidly repressed once those problems have been rectified.
“Switch Genes:” A New Class of G1/S Cell Cycle Genes
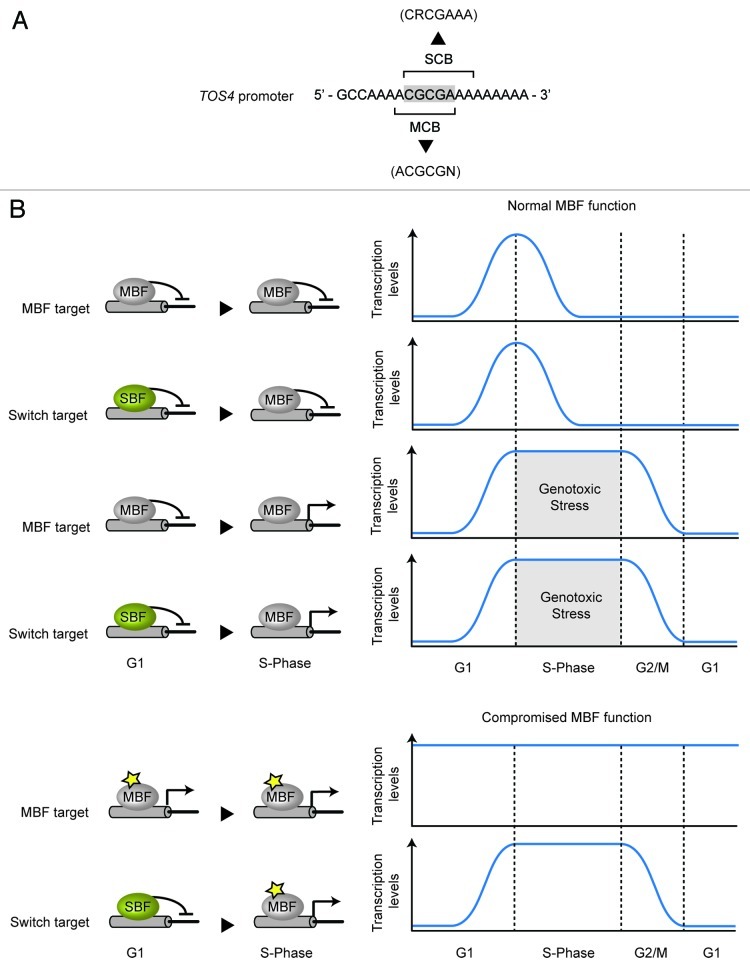
Our work established that only G1/S genes that depend on MBF for repression during S phase are induced by replication stress. Interestingly, we found that this set of genes included not only G1/S genes solely regulated by MBF throughout the cell cycle, but also a new subset of G1/S genes regulated by SBF during G1 and by MBF during S-phase.13 This subset of genes was named “switch genes” based on the SBF-to-MBF switch at their promoters during the G1-to-S transition. The SBF-to-MBF switch was first established for the G1/S gene TOS4. Since Tos4 protein accumulates in response to replication stress, like MBF-dependent genes, but was previously identified as a target of SBF, detailed analysis was performed to establish its transcriptional mode of regulation. This revealed that peak expression of TOS4 during G1 depends on SBF, whereas repression outside of G1 requires MBF. Chromatin immunoprecipitation (ChIP) experiments showed that SBF binds the TOS4 promoter during G1 but is subsequently replaced by MBF once cells progress into S phase. Analysis of the TOS4 promoter revealed that it contains an overlapping MCB/SCB motif that maintains both consensus-binding sequences (Fig. 2A). Further analysis established that in the absence of Swi4, Mbp1 is able to further bind the TOS4 promoter during G1, indicating that Swi4 competes for binding at the overlapping site during G1. Since Swi4 leaves G1/S target promoters once cells progress into S phase, in a Clb/CDK dependent manner, Mbp1 is able to bind the TOS4 promoter at the overlapping MCB/SCB site. Analysis of SBF and MBF target promoters identified in three genome-wide binding studies, resulted in a list of 44 genes containing an overlapping MCB/SCB site, which were annotated as putative switch genes (Table 1).54,63,64 Additional analysis of these genes confirmed that the majority are cell cycle-regulated, induced in response to HU treatment and display an SBF-to-MBF switch at their promoters. But what is the biological relevance of this SBF-to-MBF switch mechanism? We propose that G1/S genes that need to be induced in response to replication stress, but whose constitutive expression is detrimental to cell cycle progression, would benefit from an SBF-to-MBF switch regulation. In circumstances where MBF function is compromised, this switch regulation would work as a “fail-safe” mechanism, allowing the switch-genes to behave as a replication stress-inducible gene during S phase but, differently from a MBF-only regulated gene, will avoid constitutive high expression and hyper-accumulation (Fig. 2B). Consistent with this notion, switch genes are significantly enriched for dosage-sensitive genes reported to result in cell cycle delay and/or growth defects when overexpressed.65,66 Moreover, analysis of transcription induction upon replication stress showed that this group of genes is enriched for genes that are upregulated during replication stress conditions.13,51
Figure 2. (A) The prototypical SBF (SCB) and MBF (MCB) overlapping motif identified in the TOS4 promoter (-232 to -214 from the ATG start codon). Grey shade represents SCB/MCB overlapping region. (B) SBF-to-MBF switch mechanism avoids protein accumulation outside of S-phase. During replication stress, G1/S genes regulated by MBF or SBF-to-MBF switch are repressed during G1 and being kept active during S-phase. However, in case of MBF malfunction the SBF-to-MBF switch on G1/S promoters provides a fail-safe mechanism that allows gene expression during S-phase but prevents loss of periodicity and hyper-accumulation of dosage sensitive genes outside of S-phase. Star represents MBF malfunction.
Table 1. List of putative switch genes.
| # | ORF (SGD) | Switch Gene | Function | Up in HU? | Overexpression (Phenotype) | Cell Cycle Gene? | |||
|---|---|---|---|---|---|---|---|---|---|
| |
|
|
|
de Oliveira 2012 |
Travesa 2012 |
|
Travesa 2012 |
Spellman 1998 |
Orlando 2008 |
| 1 |
YBR070C |
ALG14 |
Glycosylation |
√ |
√ |
not observed |
√ |
√ |
√ |
| 2 |
YBR071W |
YBR071W |
Unknown |
- |
√ |
vegetative growth: decreased rate |
√ |
√ |
√ |
| 3 |
YBR162C |
TOS1 |
Unknown |
X |
- |
not observed |
√ |
X |
√ |
| 4 |
YCR065W |
HCM1 |
Transcription factor |
√ |
√ |
not observed |
√ |
√ |
√ |
| 5 |
YDL003W |
MCD1 |
Cohesin subunit |
√ |
√ |
vegetative growth: decreased rate |
√ |
√ |
√ |
| 6 |
YDL055C |
PSA1 |
Cell wall biosynthesis |
X |
√ |
not observed |
√ |
√ |
X |
| 7 |
YDL127W |
PCL2 |
Cyclin |
X |
- |
not observed |
√ |
√ |
√ |
| 8 |
YDR222W |
YDR222W |
Unknown |
√ |
X |
vegetative growth: decreased rate |
√ |
X |
√ |
| 9 |
YDR224C |
HTB1 |
Histone H2B |
- |
X |
vegetative growth: decreased rate |
√ |
√ |
√ |
| 10 |
YDR225W |
HTA1 |
Histone H2A |
- |
X |
vegetative growth: decreased rate |
√ |
√ |
√ |
| 11 |
YDR501W |
PLM2 |
Homolog of TOS4 |
- |
- |
vegetative growth: decreased rate |
√ |
√ |
√ |
| 12 |
YER001W |
MNN1 |
Glycosylation |
- |
√ |
not observed |
√ |
√ |
√ |
| 13 |
YER111C |
SWI4 |
Subunit of the SBF complex |
√ |
√ |
abnormal cell cycle progression in G2 |
√ |
X |
√ |
| 14 |
YER112W |
LSM4 |
RNA processing |
- |
X |
not observed |
√ |
X |
X |
| 15 |
YGL096W |
TOS8 |
Putative transcription factor |
X |
- |
vegetative growth: decreased rate |
√ |
X |
X |
| 16 |
YGL179C |
TOS3 |
Kinase: activates Snf1p |
√ |
√ |
not observed |
√ |
X |
√ |
| 17 |
YGR140W |
CBF2 |
Kinetochore protein |
√ |
X |
vegetative growth: decreased rate |
√ |
√ |
√ |
| 18 |
YGR151C |
YGR151C |
Unknown |
- |
√ |
not observed |
√ |
√ |
X |
| 19 |
YGR153W |
YGR153W |
Unknown |
- |
- |
not observed |
X |
X |
X |
| 20 |
YIL123W |
SIM1 |
Unknown |
√ |
- |
not observed |
√ |
X |
√ |
| 21 |
YIL141W |
YIL141W |
Unknown |
√ |
- |
not observed |
√ |
√ |
X |
| 22 |
YJL187C |
SWE1 |
Kinase: inhibits Cdc28 |
√ |
√ |
vegetative growth: decreased rate |
√ |
√ |
√ |
| 23 |
YJR054W |
ERM6 |
Vacuolar protein |
X |
- |
vegetative growth: decreased rate |
√ |
√ |
√ |
| 24 |
YKL008C |
LAC1 |
Ceramide synthesis |
√ |
X |
not observed |
√ |
√ |
√ |
| 25 |
YKL102C |
YKL102C |
Unknown |
X |
- |
not observed |
√ |
X |
X |
| 26 |
YLR183C |
TOS4 |
Checkpoint effector, HDAC regulator |
√ |
√ |
abnormal cell cycle progression in G1/S |
√ |
√ |
√ |
| 27 |
YLR332W |
MID2 |
Cell wall integrity |
- |
- |
vegetative growth: decreased rate |
√ |
X |
X |
| 28 |
YMR144W |
YMR144W |
Unknown |
- |
√ |
not observed |
√ |
√ |
√ |
| 29 |
YNL231C |
PDR16 |
Lipid homeostasis |
√ |
√ |
vegetative growth: decreased rate |
√ |
√ |
√ |
| 30 |
YNL278W |
CAF120 |
Transcription factor |
√ |
X |
vegetative growth: decreased rate |
√ |
X |
√ |
| 31 |
YNL283C |
WSC2 |
Cell wall integrity |
√ |
- |
vegetative growth: decreased rate |
√ |
√ |
√ |
| 32 |
YNL301C |
RPL18B |
Ribosomal protein |
X |
- |
not observed |
√ |
X |
X |
| 33 |
YOL007C |
CSI2 |
Unknown |
X |
√ |
vegetative growth: decreased rate |
√ |
√ |
√ |
| 34 |
YOR247W |
SRL1 |
Cell wall integrity |
√ |
X |
vegetative growth: decreased rate |
√ |
√ |
√ |
| 35 |
YOR342C |
YOR342C |
Unknown |
X |
- |
not observed |
√ |
X |
√ |
| 36 |
YPL126W |
NAN1 |
Ribosomal protein |
- |
- |
not observed |
√ |
X |
X |
| 37 |
YPL127C |
HHO1 |
Histone H1 |
√ |
√ |
abnormal cell cycle progression in G1 |
√ |
√ |
√ |
| 38 |
YPR204W |
YPR204W |
DNA helicase |
√ |
- |
not observed |
√ |
X |
X |
| 39 |
YKL007W |
CAP1 |
Actin polymerisation inhibitor |
- |
- |
not observed |
√ |
X |
√ |
| 40 |
YPL267W |
ACM1 |
APC inhibitor |
- |
√ |
vegetative growth: decreased rate |
√ |
√ |
√ |
| 41 |
YBR161W |
CSH1 |
Glycosylation |
- |
- |
vegetative growth: decreased rate |
√ |
√ |
√ |
| 42 |
YGL027C |
GLS1 |
Glycosylation |
- |
- |
vegetative growth: decreased rate |
√ |
√ |
X |
| 43 |
YGR221C |
TOS2 |
Budding |
- |
√ |
not observed |
√ |
√ |
√ |
| 44 | YNL300W | TOS6 | GPI-dependent cell wall protein | - | √ | not observed | √ | √ | √ |
Switch Genes as Dosage-Sensitive Checkpoint Effectors
TOS genes.
Among the genes found to be regulated by a SBF-to-MBF switch, seven were previously identified in the Iyer ChIP-chip study as targets of SBF (TOS1, 2, 3, 4, 6 and 8).54 Why these genes containing MCB/SCB overlapping motifs were previously identified as SBF targets is still unclear. However, it is well established that the enrichment for promoter DNA in ChIP analysis of MBF targets when pulling down Mbp1 is much lower than that of SCB target promoters when pulling down Swi4. This is assumed to be a reflection of the cross-linking methodology, rather than a reflection of the actual binding. Since TOS1-4, 6 and 8 have both SCBs and MCBs, they were only annotated as SBF genes after the genome-wide ChIP analysis, probably because signals for SBF were much higher than for MBF pulldowns. Of the Tos proteins, TOS4 was found to encode for a Forkhead-associated (FHA) domain-containing protein. Interestingly, upon replication stress, Tos4 accumulates in a checkpoint-dependent manner and interacts with the Hos2 and Rpd3 histone deacetylases (HDAC) subunits, which are part of two major HDAC complexes.13 Interaction between Tos4 and the HDACs is mediated by its FHA-domain, and this interaction is critical for Tos4 function. Genetic evidence suggests that Tos4 is important for cell viability during replication stress and functions in a parallel pathway to that of the checkpoint protein kinase Dun1.13 Although the role of Tos4 during replication stress response is still unclear, we speculate that in response to genotoxic stress, Tos4 might be involved in either promoting or inhibiting recruitment of HDACs to certain gene promoters or modulating the activity of certain HDACs. The function of the TOS genes remains largely unknown. However, whereas the TOS genes share no sequence homology, they are part of a very select group of genes linked by their expression profile and the mechanism of transcriptional control both during the cell cycle and in response to replication stress. Based on this, it is anticipated that the proteins encoded by these genes have a specific role outside of the G1 phase of the cell cycle and in response to checkpoint activation.
MCD1, SWE1 and ACM1.
Among the genes controlled by the SBF-to-MBF switch, we also identified MCD1, a gene that encodes a subunit of the yeast cohesin complex, which is required for cohesion, DSB repair and recombination between sister chromatids during mitosis. Previous work has shown that Mcd1 is a checkpoint target, and the checkpoint-dependent phosphorylation of Mcd1 was shown to be important for cohesion during G2/M.67 Like MCD1, SWE1 is another switch gene that also plays an important role during G2/M transition. SWE1 encodes a protein kinase involved in the inhibition of the cyclin-dependent kinase (CDK) Cdc28. Swe1-dependent inhibition of CDK is a central mechanism to regulate cell cycle transition during G2/M.68 The ACM1 switch gene plays a role in a later cell cycle transition, namely the exit of mitosis, by inhibiting the anaphase-promoting complex (APC) via competitive inhibition. The APC coactivator Cdh1, which is involved in the recruitment of target substrates to the APC, is inhibited from binding its targets by Cdc28-dependent stabilization of Acm1.69
TOS4, MCD1, SWE1 and ACM1 are cell cycle-regulated, and, corroborating our hypothesis that switch genes expression needs to be tightly regulated, overexpression of these genes was linked to growth defect and/or cell cycle arrest.65,66 Further analysis of the other switch genes, such as the TOS genes and eight putative ORFs, will show their potential role as checkpoint effectors.
Conclusion
With the discovery of the SBF-to-MBF switch in budding yeast we have identified a group of genes involved in the checkpoint response that require timely inactivation of transcription. The switch mechanism not only allows the upregulation of genes that are important for the checkpoint response during S-phase, but it also prevents the overexpression of these dosage-sensitive genes outside of G1 in case of MBF malfunction. Furthermore, preliminary work from the de Bruin lab in fission yeast indicates that, whereas G1/S transcription levels are maintained at a high level in response to genotoxic stress, repression outside of G1 is required to maintain genome stability (C. Caetano and RdB). Together with the Nrm1 negative feedback mechanism of transcriptional repression, which rapidly shuts off transcription of MBF targets, the SBF-to-MBF switch provides an important regulatory mechanism to tightly control protein accumulation not only during S-phase, but also throughout the whole cell cycle. Consequently, switch genes are enriched for cell cycle-regulated, dosage-dependent and replication stress-induced genes. These features could direct the future characterization of this select group of co-regulated genes, many of which are of unknown function. In addition, future research will shed light on how widely used this mode of regulation is and the importance of this particular network wiring for cell survival in response to stress and during evolution.
Acknowledgments
The authors acknowledge support by Research Scholar Grant # RSG-11-146-01-DMC from the American Cancer Society to M.B.S., Cornell Fleming Research Fellowship to FMBdO and by RdB’s MRC Career Development Award (G0800297).
Footnotes
Previously published online: www.landesbioscience.com/journals/cc/article/21197
References
- 1.Segurado M, Tercero JA. The S-phase checkpoint: targeting the replication fork. Biol Cell. 2009;101:617–27. doi: 10.1042/BC20090053. [DOI] [PubMed] [Google Scholar]
- 2.Branzei D, Foiani M. The DNA damage response during DNA replication. Curr Opin Cell Biol. 2005;17:568–75. doi: 10.1016/j.ceb.2005.09.003. [DOI] [PubMed] [Google Scholar]
- 3.Putnam CD, Jaehnig EJ, Kolodner RD. Perspectives on the DNA damage and replication checkpoint responses in Saccharomyces cerevisiae. DNA Repair (Amst) 2009;8:974–82. doi: 10.1016/j.dnarep.2009.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zegerman P, Diffley JF. DNA replication as a target of the DNA damage checkpoint. DNA Repair (Amst) 2009;8:1077–88. doi: 10.1016/j.dnarep.2009.04.023. [DOI] [PubMed] [Google Scholar]
- 5.Friedel AM, Pike BL, Gasser SM. ATR/Mec1: coordinating fork stability and repair. Curr Opin Cell Biol. 2009;21:237–44. doi: 10.1016/j.ceb.2009.01.017. [DOI] [PubMed] [Google Scholar]
- 6.Branzei D, Foiani M. Maintaining genome stability at the replication fork. Nat Rev Mol Cell Biol. 2010;11:208–19. doi: 10.1038/nrm2852. [DOI] [PubMed] [Google Scholar]
- 7.Ivessa AS, Lenzmeier BA, Bessler JB, Goudsouzian LK, Schnakenberg SL, Zakian VA. The Saccharomyces cerevisiae helicase Rrm3p facilitates replication past nonhistone protein-DNA complexes. Mol Cell. 2003;12:1525–36. doi: 10.1016/S1097-2765(03)00456-8. [DOI] [PubMed] [Google Scholar]
- 8.Torres JZ, Bessler JB, Zakian VA. Local chromatin structure at the ribosomal DNA causes replication fork pausing and genome instability in the absence of the S. cerevisiae DNA helicase Rrm3p. Genes Dev. 2004;18:498–503. doi: 10.1101/gad.1154704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lopes M, Cotta-Ramusino C, Pellicioli A, Liberi G, Plevani P, Muzi-Falconi M, et al. The DNA replication checkpoint response stabilizes stalled replication forks. Nature. 2001;412:557–61. doi: 10.1038/35087613. [DOI] [PubMed] [Google Scholar]
- 10.Tercero JA, Diffley JF. Regulation of DNA replication fork progression through damaged DNA by the Mec1/Rad53 checkpoint. Nature. 2001;412:553–7. doi: 10.1038/35087607. [DOI] [PubMed] [Google Scholar]
- 11.Bermejo R, Lai MS, Foiani M. Preventing replication stress to maintain genome stability: resolving conflicts between replication and transcription. Mol Cell. 2012;45:710–8. doi: 10.1016/j.molcel.2012.03.001. [DOI] [PubMed] [Google Scholar]
- 12.Branzei D, Foiani M. The checkpoint response to replication stress. DNA Repair (Amst) 2009;8:1038–46. doi: 10.1016/j.dnarep.2009.04.014. [DOI] [PubMed] [Google Scholar]
- 13.Bastos de Oliveira FM, Harris MR, Brazauskas P, de Bruin RA, Smolka MB. Linking DNA replication checkpoint to MBF cell-cycle transcription reveals a distinct class of G1/S genes. EMBO J. 2012;31:1798–810. doi: 10.1038/emboj.2012.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huang M, Zhou Z, Elledge SJ. The DNA replication and damage checkpoint pathways induce transcription by inhibition of the Crt1 repressor. Cell. 1998;94:595–605. doi: 10.1016/S0092-8674(00)81601-3. [DOI] [PubMed] [Google Scholar]
- 15.Wu X, Bayle JH, Olson D, Levine AJ. The p53-mdm-2 autoregulatory feedback loop. Genes Dev. 1993;7(7A):1126–32. doi: 10.1101/gad.7.7a.1126. [DOI] [PubMed] [Google Scholar]
- 16.Butala M, Zgur-Bertok D, Busby SJ. The bacterial LexA transcriptional repressor. Cell Mol Life Sci. 2009;66:82–93. doi: 10.1007/s00018-008-8378-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rouse J, Jackson SP. Interfaces between the detection, signaling, and repair of DNA damage. Science. 2002;297:547–51. doi: 10.1126/science.1074740. [DOI] [PubMed] [Google Scholar]
- 18.Kolodner RD, Putnam CD, Myung K. Maintenance of genome stability in Saccharomyces cerevisiae. Science. 2002;297:552–7. doi: 10.1126/science.1075277. [DOI] [PubMed] [Google Scholar]
- 19.Paulsen RD, Cimprich KA. The ATR pathway: fine-tuning the fork. DNA Repair (Amst) 2007;6:953–66. doi: 10.1016/j.dnarep.2007.02.015. [DOI] [PubMed] [Google Scholar]
- 20.López-Contreras AJ, Fernandez-Capetillo O. The ATR barrier to replication-born DNA damage. DNA Repair (Amst) 2010;9:1249–55. doi: 10.1016/j.dnarep.2010.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bartek J, Lukas C, Lukas J. Checking on DNA damage in S phase. Nat Rev Mol Cell Biol. 2004;5:792–804. doi: 10.1038/nrm1493. [DOI] [PubMed] [Google Scholar]
- 22.Kim ST, Lim DS, Canman CE, Kastan MB. Substrate specificities and identification of putative substrates of ATM kinase family members. J Biol Chem. 1999;274:37538–43. doi: 10.1074/jbc.274.53.37538. [DOI] [PubMed] [Google Scholar]
- 23.Sun Z, Fay DS, Marini F, Foiani M, Stern DF. Spk1/Rad53 is regulated by Mec1-dependent protein phosphorylation in DNA replication and damage checkpoint pathways. Genes Dev. 1996;10:395–406. doi: 10.1101/gad.10.4.395. [DOI] [PubMed] [Google Scholar]
- 24.Sanchez Y, Bachant J, Wang H, Hu F, Liu D, Tetzlaff M, et al. Control of the DNA damage checkpoint by chk1 and rad53 protein kinases through distinct mechanisms. Science. 1999;286:1166–71. doi: 10.1126/science.286.5442.1166. [DOI] [PubMed] [Google Scholar]
- 25.Matsuoka S, Huang M, Elledge SJ. Linkage of ATM to cell cycle regulation by the Chk2 protein kinase. Science. 1998;282:1893–7. doi: 10.1126/science.282.5395.1893. [DOI] [PubMed] [Google Scholar]
- 26.Sanchez Y, Desany BA, Jones WJ, Liu Q, Wang B, Elledge SJ. Regulation of RAD53 by the ATM-like kinases MEC1 and TEL1 in yeast cell cycle checkpoint pathways. Science. 1996;271:357–60. doi: 10.1126/science.271.5247.357. [DOI] [PubMed] [Google Scholar]
- 27.Matsuoka S, Ballif BA, Smogorzewska A, McDonald ER, 3rd, Hurov KE, Luo J, et al. ATM and ATR substrate analysis reveals extensive protein networks responsive to DNA damage. Science. 2007;316:1160–6. doi: 10.1126/science.1140321. [DOI] [PubMed] [Google Scholar]
- 28.Zhao X, Muller EG, Rothstein R. A suppressor of two essential checkpoint genes identifies a novel protein that negatively affects dNTP pools. Mol Cell. 1998;2:329–40. doi: 10.1016/S1097-2765(00)80277-4. [DOI] [PubMed] [Google Scholar]
- 29.Myung K, Chen C, Kolodner RD. Multiple pathways cooperate in the suppression of genome instability in Saccharomyces cerevisiae. Nature. 2001;411:1073–6. doi: 10.1038/35082608. [DOI] [PubMed] [Google Scholar]
- 30.Allen JB, Zhou Z, Siede W, Friedberg EC, Elledge SJ. The SAD1/RAD53 protein kinase controls multiple checkpoints and DNA damage-induced transcription in yeast. Genes Dev. 1994;8:2401–15. doi: 10.1101/gad.8.20.2401. [DOI] [PubMed] [Google Scholar]
- 31.Tercero JA, Longhese MP, Diffley JF. A central role for DNA replication forks in checkpoint activation and response. Mol Cell. 2003;11:1323–36. doi: 10.1016/S1097-2765(03)00169-2. [DOI] [PubMed] [Google Scholar]
- 32.Segurado M, Diffley JF. Separate roles for the DNA damage checkpoint protein kinases in stabilizing DNA replication forks. Genes Dev. 2008;22:1816–27. doi: 10.1101/gad.477208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cobb JA, Schleker T, Rojas V, Bjergbaek L, Tercero JA, Gasser SM. Replisome instability, fork collapse, and gross chromosomal rearrangements arise synergistically from Mec1 kinase and RecQ helicase mutations. Genes Dev. 2005;19:3055–69. doi: 10.1101/gad.361805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pasero P, Shimada K, Duncker BP. Multiple roles of replication forks in S phase checkpoints: sensors, effectors and targets. Cell Cycle. 2003;2:568–72. doi: 10.4161/cc.2.6.577. [DOI] [PubMed] [Google Scholar]
- 35.Santocanale C, Diffley JFAA. Mec1- and Rad53-dependent checkpoint controls late-firing origins of DNA replication. Nature. 1998;395:615–8. doi: 10.1038/27001. [DOI] [PubMed] [Google Scholar]
- 36.Choi DH, Oh YM, Kwon SH, Bae SH. The mutation of a novel Saccharomyces cerevisiae SRL4 gene rescues the lethality of rad53 and lcd1 mutations by modulating dNTP levels. J Microbiol. 2008;46:75–80. doi: 10.1007/s12275-008-0013-6. [DOI] [PubMed] [Google Scholar]
- 37.Zhao X, Rothstein R. The Dun1 checkpoint kinase phosphorylates and regulates the ribonucleotide reductase inhibitor Sml1. Proc Natl Acad Sci USA. 2002;99:3746–51. doi: 10.1073/pnas.062502299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhao X, Chabes A, Domkin V, Thelander L, Rothstein R. The ribonucleotide reductase inhibitor Sml1 is a new target of the Mec1/Rad53 kinase cascade during growth and in response to DNA damage. EMBO J. 2001;20:3544–53. doi: 10.1093/emboj/20.13.3544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sidorova JM, Breeden LL. Rad53-dependent phosphorylation of Swi6 and down-regulation of CLN1 and CLN2 transcription occur in response to DNA damage in Saccharomyces cerevisiae. Genes Dev. 1997;11:3032–45. doi: 10.1101/gad.11.22.3032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kiser GL, Weinert TA. Distinct roles of yeast MEC and RAD checkpoint genes in transcriptional induction after DNA damage and implications for function. Mol Biol Cell. 1996;7:703–18. doi: 10.1091/mbc.7.5.703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Maga JA, McClanahan TA, McEntee K. Transcriptional regulation of DNA damage responsive (DDR) genes in different rad mutant strains of Saccharomyces cerevisiae. Mol Gen Genet. 1986;205:276–84. doi: 10.1007/BF00430439. [DOI] [PubMed] [Google Scholar]
- 42.Barker DG, White JH, Johnston LH. The nucleotide sequence of the DNA ligase gene (CDC9) from Saccharomyces cerevisiae: a gene which is cell-cycle regulated and induced in response to DNA damage. Nucleic Acids Res. 1985;13:8323–37. doi: 10.1093/nar/13.23.8323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ruby SW, Szostak JW. Specific Saccharomyces cerevisiae genes are expressed in response to DNA-damaging agents. Mol Cell Biol. 1985;5:75–84. doi: 10.1128/mcb.5.1.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Elledge SJ, Davis RW. DNA damage induction of ribonucleotide reductase. Mol Cell Biol. 1989;9:4932–40. doi: 10.1128/mcb.9.11.4932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Treger JM, Heichman KA, McEntee K. Expression of the yeast UB14 gene increases in response to DNA-damaging agents and in meiosis. Mol Cell Biol. 1988;8:1132–6. doi: 10.1128/mcb.8.3.1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jelinsky SA, Samson LD. Global response of Saccharomyces cerevisiae to an alkylating agent. Proc Natl Acad Sci USA. 1999;96:1486–91. doi: 10.1073/pnas.96.4.1486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gasch AP, Huang M, Metzner S, Botstein D, Elledge SJ, Brown PO. Genomic expression responses to DNA-damaging agents and the regulatory role of the yeast ATR homolog Mec1p. Mol Biol Cell. 2001;12:2987–3003. doi: 10.1091/mbc.12.10.2987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Benton MG, Somasundaram S, Glasner JD, Palecek SP. Analyzing the dose-dependence of the Saccharomyces cerevisiae global transcriptional response to methyl methanesulfonate and ionizing radiation. BMC Genomics. 2006;7:305. doi: 10.1186/1471-2164-7-305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.De Sanctis V, Bertozzi C, Costanzo G, Di Mauro E, Negri R. Cell cycle arrest determines the intensity of the global transcriptional response of Saccharomyces cerevisiae to ionizing radiation. Radiat Res. 2001;156:379–87. doi: 10.1667/0033-7587(2001)156[0379:CCADTI]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- 50.Fu Y, Pastushok L, Xiao W. DNA damage-induced gene expression in Saccharomyces cerevisiae. FEMS Microbiol Rev. 2008;32:908–26. doi: 10.1111/j.1574-6976.2008.00126.x. [DOI] [PubMed] [Google Scholar]
- 51.Travesa A, Kuo D, de Bruin RA, Kalashnikova TI, Guaderrama M, Thai K, et al. DNA replication stress differentially regulates G1/S genes via Rad53-dependent inactivation of Nrm1. EMBO J. 2012;31:1811–22. doi: 10.1038/emboj.2012.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zaim J, Speina E, Kierzek AM. Identification of new genes regulated by the Crt1 transcription factor, an effector of the DNA damage checkpoint pathway in Saccharomyces cerevisiae. J Biol Chem. 2005;280:28–37. doi: 10.1074/jbc.M404669200. [DOI] [PubMed] [Google Scholar]
- 53.Wittenberg C, Reed SI. Cell cycle-dependent transcription in yeast: promoters, transcription factors, and transcriptomes. Oncogene. 2005;24:2746–55. doi: 10.1038/sj.onc.1208606. [DOI] [PubMed] [Google Scholar]
- 54.Iyer VR, Horak CE, Scafe CS, Botstein D, Snyder M, Brown PO. Genomic binding sites of the yeast cell-cycle transcription factors SBF and MBF. Nature. 2001;409:533–8. doi: 10.1038/35054095. [DOI] [PubMed] [Google Scholar]
- 55.Breeden LL. Periodic transcription: a cycle within a cycle. Curr Biol. 2003;13:R31–8. doi: 10.1016/S0960-9822(02)01386-6. [DOI] [PubMed] [Google Scholar]
- 56.Bean JM, Siggia ED, Cross FR. High functional overlap between MluI cell-cycle box binding factor and Swi4/6 cell-cycle box binding factor in the G1/S transcriptional program in Saccharomyces cerevisiae. Genetics. 2005;171:49–61. doi: 10.1534/genetics.105.044560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Eser U, Falleur-Fettig M, Johnson A, Skotheim JM. Commitment to a cellular transition precedes genome-wide transcriptional change. Mol Cell. 2011;43:515–27. doi: 10.1016/j.molcel.2011.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ferrezuelo F, Colomina N, Futcher B, Aldea M. The transcriptional network activated by Cln3 cyclin at the G1-to-S transition of the yeast cell cycle. Genome Biol. 2010;11:R67. doi: 10.1186/gb-2010-11-6-r67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Amon A, Tyers M, Futcher B, Nasmyth K. Mechanisms that help the yeast cell cycle clock tick: G2 cyclins transcriptionally activate G2 cyclins and repress G1 cyclins. Cell. 1993;74:993–1007. doi: 10.1016/0092-8674(93)90722-3. [DOI] [PubMed] [Google Scholar]
- 60.de Bruin RA, Kalashnikova TI, Chahwan C, McDonald WH, Wohlschlegel J, Yates J, 3rd, et al. Constraining G1-specific transcription to late G1 phase: the MBF-associated corepressor Nrm1 acts via negative feedback. Mol Cell. 2006;23:483–96. doi: 10.1016/j.molcel.2006.06.025. [DOI] [PubMed] [Google Scholar]
- 61.de Bruin RA, Kalashnikova TI, Aslanian A, Wohlschlegel J, Chahwan C, Yates JR, 3rd, et al. DNA replication checkpoint promotes G1-S transcription by inactivating the MBF repressor Nrm1. Proc Natl Acad Sci USA. 2008;105:11230–5. doi: 10.1073/pnas.0801106105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.de Bruin RA, Wittenberg C. All eukaryotes: before turning off G1-S transcription, please check your DNA. Cell Cycle. 2009;8:214–7. doi: 10.4161/cc.8.2.7412. [DOI] [PubMed] [Google Scholar]
- 63.Simon I, Barnett J, Hannett N, Harbison CT, Rinaldi NJ, Volkert TL, et al. Serial regulation of transcriptional regulators in the yeast cell cycle. Cell. 2001;106:697–708. doi: 10.1016/S0092-8674(01)00494-9. [DOI] [PubMed] [Google Scholar]
- 64.Harbison CT, Gordon DB, Lee TI, Rinaldi NJ, Macisaac KD, Danford TW, et al. Transcriptional regulatory code of a eukaryotic genome. Nature. 2004;431:99–104. doi: 10.1038/nature02800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sopko R, Huang D, Preston N, Chua G, Papp B, Kafadar K, et al. Mapping pathways and phenotypes by systematic gene overexpression. Mol Cell. 2006;21:319–30. doi: 10.1016/j.molcel.2005.12.011. [DOI] [PubMed] [Google Scholar]
- 66.Yoshikawa K, Tanaka T, Ida Y, Furusawa C, Hirasawa T, Shimizu H. Comprehensive phenotypic analysis of single-gene deletion and overexpression strains of Saccharomyces cerevisiae. Yeast. 2011;28:349–61. doi: 10.1002/yea.1843. [DOI] [PubMed] [Google Scholar]
- 67.Ström L, Lindroos HB, Shirahige K, Sjögren C. Postreplicative recruitment of cohesin to double-strand breaks is required for DNA repair. Mol Cell. 2004;16:1003–15. doi: 10.1016/j.molcel.2004.11.026. [DOI] [PubMed] [Google Scholar]
- 68.Harvey SL, Charlet A, Haas W, Gygi SP, Kellogg DR. Cdk1-dependent regulation of the mitotic inhibitor Wee1. Cell. 2005;122:407–20. doi: 10.1016/j.cell.2005.05.029. [DOI] [PubMed] [Google Scholar]
- 69.Ostapenko D, Burton JL, Wang R, Solomon MJ. Pseudosubstrate inhibition of the anaphase-promoting complex by Acm1: regulation by proteolysis and Cdc28 phosphorylation. Mol Cell Biol. 2008;28:4653–64. doi: 10.1128/MCB.00055-08. [DOI] [PMC free article] [PubMed] [Google Scholar]