Abstract
Genetic fate-mapping approaches provide a unique opportunity to assess differentiation pathways under physiological conditions. We have recently employed a lineage tracing approach to define hematopoietic differentiation pathways in relation to expression of the tyrosine kinase receptor Flk2.1 Based on our examination of reporter activity across all stem, progenitor and mature populations in our Flk2-Cre lineage model, we concluded that all mature blood lineages are derived through a Flk2+ intermediate, both at steady-state and under stress conditions. Here, we re-examine in depth our initial conclusions and perform additional experiments to test alternative options of lineage specification. Our data unequivocally support the conclusion that onset of Flk2 expression results in loss of self-renewal but preservation of multilineage differentiation potential. We discuss the implications of these data for defining stem cell identity and lineage potential among hematopoietic populations.
Keywords: hematopoietic stem cells, progenitor cell, cell fate decision, Flk2, Flt3, self-renewal, differentiation pathways, transplantation, lineage tracing, Cre/loxP, hematopoiesis
Introduction
Understanding the mechanisms that drive multipotent stem cells to self-renew or to commit to specific cell fates is a central goal of regenerative medicine. Accurate maps of differentiation pathways are not only critical for directed differentiation of pluripotent and multipotent cells for therapeutic use, but also for understanding disease pathogenesis and enabling targeting of the cells and molecules that are at the core of aberrant behavior. The hematopoietic system can be considered a model paradigm for dissecting stem cell differentiation pathways, as it has been established that a single, multipotent hematopoietic stem cell (HSC) can both self-renew and give rise to all mature blood cell types. Furthermore, progressively restricted progenitor cells capable of giving rise to unilineage-committed precursors and, ultimately, mature cells have been identified. Our knowledge of hematopoietic differentiation has benefitted greatly from an array of assays capable of measuring the lineage potential of defined cell populations both in vitro and in vivo. Unfortunately, recent advances in technical capability combined with development of more sensitive assays have generated more confusion than consensus. Previously defined cell populations have been further subdivided, and the lineage potential of both myeloid and lymphoid populations has been contested in iterations of classical and novel assays.
Transplantation assays have long been considered the highest standard for measuring the functional capacity of phenotypically distinct populations. Most in vivo reconstitution experiments are based on CD45 allelic discrimination between host- and donor-derived cells. Because the mature megakaryocyte/erythroid (MegE) cells, platelets (Plt) and red blood cells (RBC) do not express CD45, many studies on hematopoietic lineage potential, including early identification of “multipotent” populations capable of giving rise to granulocytes/macrophages (GM), B and T cells, did not include analysis of in vivo MegE potential.2-4 Many studies have instead relied heavily on in vitro assays to assess whether defined progenitor populations give rise to MegE cells. Interestingly, in vitro differentiation assays have reported both lack and gain of lineage potential compared with readout from in vivo transplantation experiments (reviewed in refs. 5 and 6). While it is clear that the assay conditions can have a profound impact on the outcome, it is unclear which assays are insufficiently sensitive and what conditions induce lineage readout that does not normally occur. Thus, the true role of several distinct progenitor populations in development of mature hematopoietic cells remains uncertain.
To enable interrogation of hematopoietic differentiation pathways under unperturbed, physiological conditions, we recently established a Cre/lox-based lineage tracing model (Fig. 1A).1 We found two properties of fate mapping models particularly appealing: the irreversibility of the genetic excision of the floxed locus and the opportunity to examine steady-state hematopoiesis. We reasoned that steady-state differentiation pathways would enable us to determine the physiological relevance of specific differentiation steps, and that the irreversible change in reporter expression would provide definitive information on the hierarchical relationship between distinct cell populations. In addition, inducing stress and performing transplantations would enable us to determine whether steady-state paths change under different conditions.
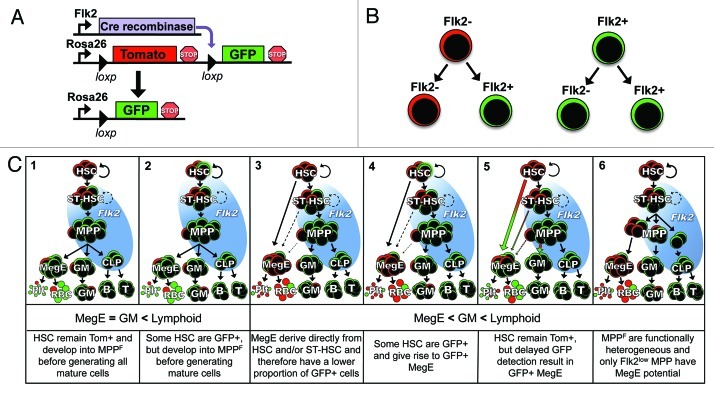
Figure 1. Modeling hematopoiesis with Flk2-Cre lineage tracing. (A) Flk2-Cre mice were crossed to mT/mG dual-color reporter mice to generate FlkSwitch mice. (B) Simplified model of lineage tracing strategy for FlkSwitch mice. (C) Six possible outcomes from FlkSwitch lineage tracing. Flk2 expression is indicated by the blue background circle. The second row indicates the relative proportion of GFP+ cells in MegE, GM and lymphoid lineages. The third row describes a hypothetical scenario for each model. In Models 1, 3, 5 and 6, all HSC remain Tomato+. In Models 2 and 4, Flk2 expression is initiated in a fraction of HSC resulting in reporter floxing and GFP expression. Although additional versions of these models can be drawn, only Model 1 fits the results from lineage tracing studies. HSC, hematopoietic stem cell; ST-HSC, short-term hematopoietic stem cell; MPP, multipotent progenitor; MegE, megakaryocyte/erythrocyte progenitor; GM, granulocyte/myelomonocyte; Plt, platelet; RBC, red blood cell; B, B cell; T, T cell.
We are particularly interested in whether fate decisions are made by stem cells themselves or deferred until later differentiation steps. Surface expression of the tyrosine kinase receptor Flk2 (Flt3) within the “KLS” (cKithi, Lin-, Sca1+) stem and progenitor cell compartment corresponds with loss of long-term self-renewal ability2,3 and therefore separates KLS cells into multipotent, self-renewing HSC and non-self-renewing, multipotent progenitor cells (MPPF). Flk2 expression is sustained during lymphoid differentiation but is not detectable on myeloid-committed progenitors or mature cells (Fig. 1C). This off-on-off expression pattern provides an excellent opportunity to determine whether Flk2+ progenitors represent an obligatory or optional stage in myeloid differentiation, a question that has been debated in particular for the MegE lineage.7 Thus, we established an in vivo lineage tracing model that enabled us to determine the relative contribution of Flk2+ cells to the distinct hematopoietic lineages during steady-state hematopoiesis.1 Our group used a mouse model expressing Cre under Flk2 promoter elements8 combined with a dual-fluorescent reporter mouse model,9 whereby Flk2-Cre expression resulted in reporter switching from Tomato to GFP expression (“FlkSwitch” mice; Fig. 1A). Due to the permanent removal of the Tomato allele induced by Cre expression, a cell’s Flk2 expression history can be determined in FlkSwitch mice (Fig. 1B). In our previous report, we made the simple observation that mature cells of all major hematopoietic lineages, including MegE lineages, switch from Tomato to GFP expression when Cre recombinase is driven by the Flk2 locus.1 Because HSC do not express detectable cell surface levels of Flk22,3 and remain Tomato+ in the FlkSwitch model, we concluded that all hematopoietic lineages differentiate through a Flk2+ stage.1 Here, we consider alternative explanations to the conclusions drawn in our previous report, provide additional experiments to test these alternative options and discuss our new insights in the context of related studies in the field.
Results and Discussion
Both the reporter and Cre transgenes are expressed as intended and do not alter endogenous Flk2 expression or population size.
A basic requirement of lineage tracing models is that the Cre transgene is expressed in the intended pattern. Several different methods to regulate transgene expression can be utilized, including targeted gene replacement, knock-ins, fusion proteins or randomly integrated genes driven by small or large regulatory elements. Each approach has specific advantages, but no strategy guarantees perfect recapitulation of the intended expression pattern. For cell-ubiquitous reporter expression, we utilized a previously characterized mT/mG dual-color cassette inserted into the Rosa26 locus.9 The Rosa26 locus is a well-characterized and very commonly used site for targeted transgene integration with no effects on hematopoietic function. The Flk2-Cre mice were generated by BAC transgenic methods,8 thereby leaving both endogenous Flk2 loci intact, yet allowing regulation of the Cre transgene by a very large region of the Flk2 locus. We previously demonstrated that Cre expression parallels endogenous Flk2 expression: MPPF display high levels of both Flk2 and Cre expression, whereas the Flk2-negative HSC and CMP do not express Cre.1 These analyses were greatly facilitated by the fact that Flk2 is a surface marker, allowing FACS isolation of clearly Flk2+ and Flk2- populations. To also test for residual Cre activity in the myeloid lineage, the fraction of common myeloid progenitors (CMP) that remained Tom+ in FlkSwitch mice were isolated and subjected to in vitro and in vivo differentiation. Reporter switching (e.g., from Tomato to GFP expression) was not observed in progeny derived from in vitro differentiated or transplanted Tom+ CMP, indicating that neither Cre expression nor activity persist in myeloid progenitors.1
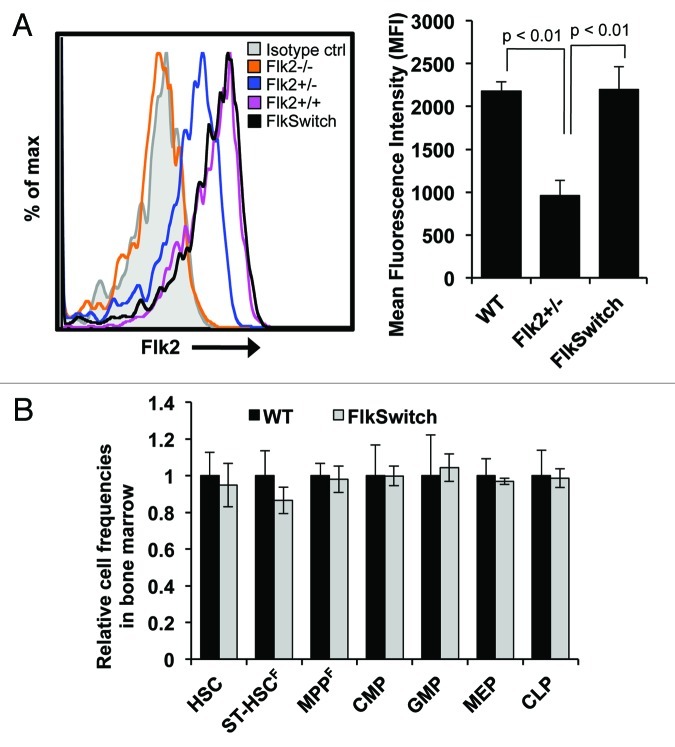
To be confident in our conclusions regarding lineage specification in the FlkSwitch model, we must also be certain that expression of endogenous Flk2 is unchanged by insertion of the Flk2-Cre transgene. Since BAC transgenesis was used to integrate the Flk2-Cre construct randomly into the genome, both of the endogenous Flk2 loci remain intact. However, it is possible that the Flk2-Cre construct(s) may alter endogenous Flk2 expression by other mechanisms, such as promoter competition for limiting factors, subsequently affecting hematopoiesis. We therefore compared Flk2 surface levels in KLS cells derived from FlkSwitch, Flk2‑/-, Flk2+/- and wt mice. Flk2 cell surface levels on FlkSwitch KLS cells were equal to the levels on wt KLS but significantly higher compared with KLS from Flk2-/- and Flk2+/- mice (Fig. 2A). Additionally, we also found no differences in the cell frequency of stem and progenitor cell populations in the bone marrow of wt and FlkSwitch mice (Fig. 2B). Together, these data show that both the reporter and Cre transgenes are expressed as intended without effects on Flk2 expression levels or lineage specification.
Figure 2. Flk2 expression and lineage specification is not altered in Flk2-Cre reporter mice. (A) Flow cytometric analysis (left) showing Flk2 expression within the KLS compartment of Flk2−/−, Flk2+/−, Flk2+/+ (wt) and FlkSwitch (Flk2+/+) mice. Isotype control is shown in gray. Mean fluorescence intensity for Flk2 surface levels from multiple mice (right). (B) Cell frequencies of hematopoietic stem and progenitors in FlkSwitch bone marrow relative to wt mice. n = 3–5 mice per experiment.
Theoretically, the ideal lineage tracing model is one in which 100% of parent cells are unfloxed, and 100% of Cre-expressing cells and their progeny are floxed (Fig. 1B). Unexpectedly, however, we found that the “imperfect,” highly variable floxing efficiency between different individual mice was an advantageous feature of the FlkSwitch model.1 While mice with high reporter-switching at the MPPF stage have a “ceiling effect” that prevents additional reporter switching, low-floxing mice allow for the detection of differences in reporter activity downstream of MPPF. In our model, low-floxing mice enabled us to demonstrate that both B and T cells utilized multiple Flk2+ developmental stages.1 GM and MegE lineages, in contrast, did not exhibit increased reporter switching compared with MPPF, as illustrated in Model 1 (Fig. 1C). It is important to note that although the floxing efficiency between different mice was highly variable, the Tom/GFP ratios of different populations within individual mice always followed the same pattern. This unintended but highly beneficial feature of the FlkSwitch model enabled a more quantitative assessment of the relationship between different lineages than an all-or-none floxing response, as illustrated below.
All functional HSC remain Tomato+.
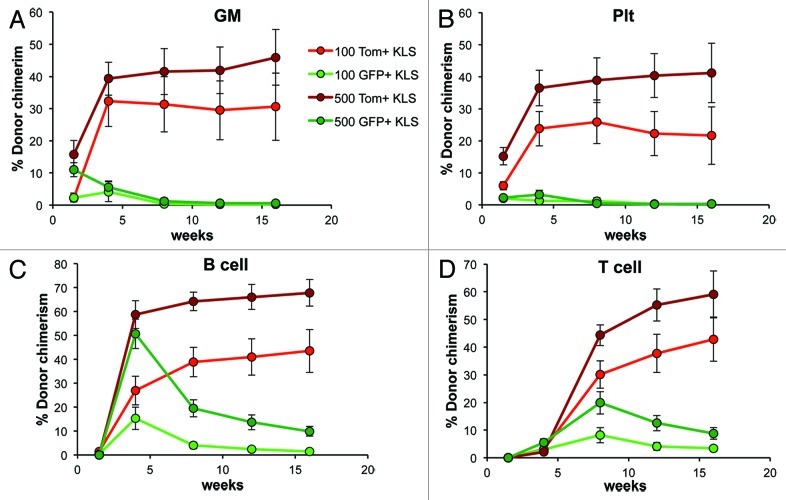
While it is clear that the vast majority of mouse HSC lack detectable Flk2 cell surface protein,2,3 it is possible that Flk2 (and therefore Cre) expression occurs in HSC prior to display of the protein on the cell surface (Models 2 and 4, Fig. 1C). Low-level Cre expression in HSC could lead to Tomato excision and result in GFP+ progeny without differentiation through a truly Flk2+ stage (Model 4, Fig. 1C). Such “premature” excision could mean that our conclusion that all lineages derive through a Flk2+ stage is erroneous. In our previous analyses, we were unable to detect Cre mRNA in HSC, and we also showed that all phenotypic HSC are Tom+ in FlkSwitch mice.1 However, phenotypic analysis does not exclude the possibility that HSC also exist in the GFP+ fraction in FlkSwitch mice. We therefore tested this directly by subfractionating KLS cells from FlkSwitch mice based on Tomato and GFP expression and measuring their functional capacity upon transplantation into recipient mice. Long-term, multilineage reconstitution was observed when either 100 or 500 Tomato+ KLS cells were transplanted (Fig. 3). In contrast, contribution from GFP+ KLS cells to B-cells (Fig. 3C) and T-cells (Fig. 3D) peaked at week 4 and 8, after which readout steadily declined. Similarly, Plt and GM production from GFP+ KLS cells declined to undetectable levels by week 8, consistent with retention of multilineage potential but loss of self-renewal ability in GFP+ cells (Fig. 3A and B). Thus, transplantation of either 100 or 500 GFP+ KLS cells was unable to sustain myeloid (Fig. 3A and B) or lymphoid (Fig. 3C and D) output for the longterm, even when 500 cells were transplanted. Consistent with the lack of detectable Cre expression in phenotypic HSC, these data demonstrate that all, or virtually all, long-term reconstituting HSC in FlkSwitch mice remain Tomato+, and therefore that GFP expression observed in mature lineages cannot be attributed to direct generation from a GFP+ HSC.
Figure 3.HSC are contained within the Tomato+ fraction in FlkSwitch mice. Percent donor chimerism of (A) GM, (B) platelet, (C) B cell and (D) T cell lineages in the peripheral blood from mice injected with 100 or 500 Tomato+ or GFP+ KLS cells from FlkSwitch mice. Data are from 12–15 recipients per cell type and dose from three independent experiments.
The possibility of delayed floxing has to be applied equivalently to all progeny.
We then considered the possibility that a high proportion of myeloid cells are GFP+ due to delayed detection of floxing (Model 5, Fig. 1C). In this model, a precursor cell expresses Cre but remains Tom+ due to the lag between Cre expression, excision of the first reporter and transcription/translation of the second reporter. If low levels of Cre led to initiation of floxing in HSC, they could remain GFP-negative (and Tom+) due to the delay in GFP protein detection. In this scenario, all HSC progeny could turn GFP+, even if these progeny do not express Cre themselves. Such delayed detection of floxing could lead us to erroneously conclude that the Flk2- myeloid cells—or maybe more likely, only the contested MegE lineage of the myeloid branch—are derived through a Flk2+ stage, while in reality they are derived directly from HSC.
When evaluating reporter expression in MegE progeny only, it is difficult to rule out the possibility that GFP expression is due to delayed floxing in HSC. When considering this in the bigger context of other hematopoietic lineages, however, it is clear that this explanation is highly unlikely. First, cell populations of the MegE and GM lineages have the same proportion of GFP+ cells, not only on average, but in every individual mouse.1 If delayed floxing results in MegE cells that have an increased proportion of GFP+ cells compared with their immediate progeny, analogous increases in GFP+ proportion should also be observed for the GM lineage (Model 5, Fig. 1C). Because MPPF possess strong and undisputed GM potential, delayed floxing predicts that GM populations would exhibit a higher proportion of GFP+ cells than MPPF, in addition to a higher GFP+ proportion than MegE cells (Model 5, Fig. 1C). This is not what we observe. Furthermore, the consistently higher proportion of GFP+ cells in lymphoid populations demonstrates that a “ceiling” of possible floxing efficiency has not been reached in GM cells.1 The fact that neither MegE nor GM lineages express Flk2 (or Cre) and yet exhibit equivalent levels of floxing indicates that they are either derived from the same progenitor population or from two different populations with equivalent floxing efficiencies. The identity of these precursor populations is discussed in the next section.
Another argument against the possibility of delayed floxing in our model stems from the fact that HSC are exclusively Tomato+. Due to self-renewal, HSC are also their own progeny. If all progeny turn GFP+ due to initiation of floxing in pre-existing HSC, newly derived HSC would also be GFP+. This is not what we observe at steady-state or even after hematopoietic stress. We have shown that all HSC remain Tom+ after stress induced by irradiation or transplantation into irradiated recipients. These conditions induce HSC proliferation but do not lead to detection of GFP+ HSC.1 Furthermore, since floxing is irreversible, we would expect that low-level Cre activity in HSC would lead to an accumulation of GFP+ HSC over time. Analysis of aging FlkSwitch mice revealed that all phenotypic BM HSC remained Tom+ in mice as old as 22 mo, even with GFP+ MPPF proportions as high as 92% (data not shown). Combined with the lack of detectable Cre mRNA in HSC and lack of HSC activity in GFP+ KLS fraction, we are compelled to conclude that Flk2 and Cre are only expressed at functionally significant levels upon differentiation, but not in HSC themselves. These results strongly support our conclusion that all hematopoietic lineages differentiate through a Flk2+ stage (Model 1, Fig. 1C).
Flk2+ KLS cells must be MegE progenitors.
Having concluded that all myeloid lineages must differentiate through a progenitor that expresses Flk2, thereby excluding Flk2- HSC as an immediate precursor, we revisit our conclusion that MPPF is the Flk2+ intermediate stage for all hematopoietic lineages. In part, this conclusion was based on our observation that the proportion of GFP+ cells within all myeloid populations was strikingly similar to that of MPPF in each individual mouse across a wide range of floxing efficiencies (6–97% in MPPF).1 Thus, two distinct populations, for example MPPF and MEP, each consisted of the same proportion of Tom:GFP cells. Clearly, this does not in itself necessarily mean one population gives rise to the other. Since we had already established that HSC do not express Flk2 or exhibit reporter-switching whereas downstream progenitors upregulate Flk2, the options were quickly narrowed down to Flk2+ cells within the KLS fraction: no other Flk2+ cell type with MegE potential has been identified. Quite contrary, increased Flk2 expression has been proposed to correlate with loss of MegE capability.10,11 Thus, the remaining challenge is to determine whether Flk2+ progenitors are multipotent at the population and/or clonal levels, and whether the MegE and GM lineages derive from different or overlapping populations within this fraction.
Multipotency at the clonal vs. population level: Heterogeneity of cell populations.
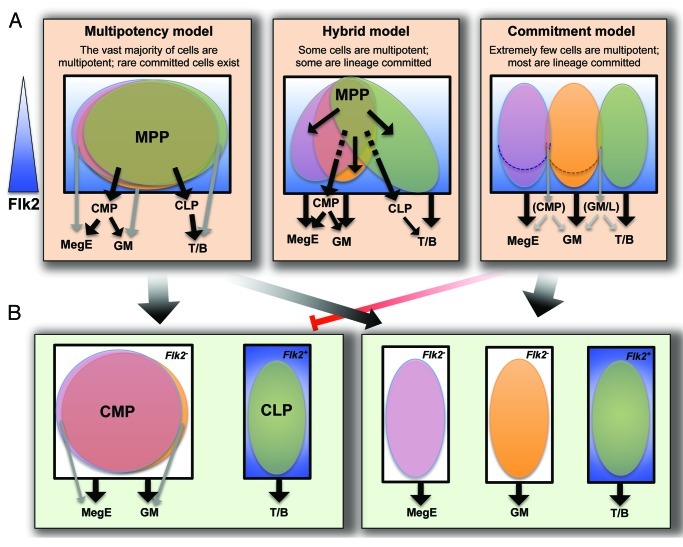
To determine which Flk2+ KLS cells give rise to the MegE lineage and whether these same cells also are GM progenitors, we considered three main models representing complete population overlap to no overlap at all. Figure 4 illustrates the range of possibilities using, for simplicity, only the three main lineages (GM, MegE and lymphoid) discussed in this report. In the “multipotentcy” model, the vast majority of Flk2+ KLS cells are multipotent at the single cell level. Because any one cell can give rise to any lineage, this model represents complete overlap between MegE, GM and lymphoid progenitors. At the opposite end, there is very little or no overlap in the “commitment” model, in which most cells have committed to a specific lineage. Here, the “MPP” compartment consists of three lineage-committed cell populations despite sharing the defining Flk2+ cell surface phenotype. The “hybrid” model consolidates the two extremes by including both true multipotent cells as well as substantial numbers of oligopotent and lineage-committed cells.
Figure 4. Models for lineage potential at the clonal or population level. (A) The phenotypic MPPF compartment (Flk2low-high KLS cells, with Flk2 levels indicated by a gradually intensifying blue background) may consist of cells with clonal multilineage potential (multipotentcy model), a mix of phenotypically similar cells that are lineage committed (Commitment model), or a mix of clonally multipotent and lineage biased cells (Hybrid model). If different cells within the phenotypic MPPF fraction give rise to MegE and GM progeny, these two cell populations must have equivalent levels of Flk2 (dotted lines in Commitment model to delineate lower Flk2 levels) as MegE and GM lineages have the same proportion of floxed cells. (B) Lineage-committed progenitor populations may consist of cells with clonal, oligopotent potential (left), or a mix of lineage biased cells (right). If lineage decisions are made by MPPF (or by HSC prior to differentiating into MPPF) as depicted in the Commitment model, it is unlikely that clonal, oligopotent progenitor cells such as CMP exist, as indicated by the red inhibition arrow. Cell populations within a black box share defining cell surface markers, whereas cell populations in different black boxes are phenotypically distinct. Major differentiation paths are indicated by black arrows, whereas alternative, infrequently used paths are shown in gray.
The distinction between these models may depend on both the phenotypic definition and functional heterogeneity of the cell populations involved. It is clear from our previous analyses that Tom/GFP expression does not segregate functionally distinct MPPF:Tom+ and GFP+ MPPF gave rise to similar levels of Plt, GM, B and T cells in the PB upon transplantation as well as to similar numbers of CFU-S colonies.1 Analogous results were obtained for Tom+ and GFP+ CMP. Thus, if these populations consist of a mix of lineage-committed cells, their relative numbers are roughly equivalent between Tom+ and GFP+ fractions in both MPPF and CMP.
To address the impact of the phenotypic definition of cell populations, we previously tested whether alternatively defined myeloid progenitor populations12 had different Tom/GFP proportions that correlated with commitment toward MegE or GM lineages. Regardless of how the myeloid progenitor fraction (cKit+Lin-Sca1- cells) was subdivided, the different populations consisted of similar proportions of Tom+ and GFP+ cells. These data demonstrated that Tom/GFP reporter expression does not correlate with commitment into MegE vs. GM lineages, consistent with the unipotent (EP) or mature (Plt and GM) cells of each lineage displaying similar levels of floxing.
In contrast to the myeloid progenitor pool, dividing the KLS fraction in different ways results in clear differences in Tom/GFP proportions. This is consistent with the variable Flk2 (and Cre) expression levels in KLS. Therefore, slicing the KLS fraction in different ways results in different proportions of Tom/GFP cells in different populations, with implications on progenitor-progeny relationships. One model has proposed that MegE potential is lost in cells with the highest levels of Flk2 expression.10 Do MPP that are less Flk2+ have more MegE potential than MPP that express high levels of Flk2? Because GM potential is retained in the highly Flk2+ fraction, adherence to this model would result in higher proportions of GFP+ cells in GM compared with MegE populations (Model 6, Fig. 1C). This is not what we observe. Thus, it seems that if MPPF are functionally heterogeneous with regards to MegE and GM potential, these distinct MegE and GM precursor populations have the same proportion of GFP:Tom cells, suggesting that they cannot be distinguished by Flk2 expression levels alone.
Time as a factor in floxing efficiency.
So far, we have focused on the level of Flk2 expression as the major factor of floxing efficiency. However, it is also highly likely that time spent within a Flk2+ stage also significantly influences the extent of floxing. The consistently higher GFP+ proportion of lymphoid cells, known to differentiate through additional Flk2+ stages, provides strong evidence for this. In addition, the floxing efficiency of MPPF after irradiation- or transplantation-induced stress is decreased.1 The stress-induced increase in proliferation rates likely causes cells to transition faster through the Flk2+ stage, resulting in a reduction of overall floxing efficiency. Thus, if one considers the “time spent” parameter in addition to the “Flk2 level” parameter, it is conceivable that MegE cells could derive from a KLS cell with lower Flk2 levels than the KLS that give rise to GM. This scenario would be consistent with models proposing loss of MegE, but retention of GM, potential in highly Flk2+ progenitors (Models 3 and 4, Fig. 1C). However, if Flk2 levels are lower in the KLS population that give rise to MegE than in KLS cells generating GM, the time MegE precursors spend at this Flk2low stage must be prolonged to achieve the equivalent levels of reporter switching observed for these two lineages. Although it is difficult to measure whether MegE precursors spend more time at a Flk2+ stage, the notion seems inconsistent with the very rapid appearance of Plt and EP after HSC transplantation.7 It is also important to note that the ratio of Tom:GFP cells in MPPF and all myeloid populations were similar, and equally reduced, after stress.1 Thus, if MegE- and GM-destined progenitor cells spend a differential amount of time at a Flk2+ stage during differentiation, this time is equally reduced upon two different stress conditions. We find it much more likely that all MegE and GM cells follow a shared path of differentiation through the MPPF stage, with subsequent divergence into lineage committed progenitor cells.
Segregation into lineage committed cells.
Our conclusions in favor of the multipotency model based on our FlkSwitch data are supported by several lines of additional evidence. Using the same Flk2-Cre model coupled to a single-color reporter (Rosa26-eYFP), Buza-Vidas and colleagues demonstrated high reporter activity in MegE progenitors, consistent with a significant contribution of Flk2+ progenitors to MegE lineages.13 Although there were some differences between their study and ours (see below), both reports demonstrated that GM and MegE progenitors had similar frequencies of reporter activity. As described above, these data support the existence of common myeloid progenitors (CMP), originally isolated based on combined MegE and GM potential in functional assays in vitro and in vivo.14 Because phenotypic MPPF have much higher burst size relative to downstream progenitors, and the timing of CFU-S or mature cell readout in PB is distinctly different between MPPF and CMP/MEP,7 MPPF cannot simply consist of a mix of CMP, MEP, GMP and CLP, but rather precursors to such populations.
The multipotency model has been challenged by subfractionation of MPP and other progenitor populations into phenotypically distinct populations by use of both cell surface and intracellular markers. However, identification of novel phenotypic subpopulations has not led to clean separation of functional activity in vivo. Quite the contrary, subfractionation of cells using new marker combinations has supported the existence of cell populations with oligo- or bilineage potential, sometimes also demonstrated clonally.12,15,16 These data highlight the notion that differentiation is gradual, and that marker heterogeneity is not synonymous with functional heterogeneity. A simple example is a protein expressed in a cell cycle-dependent pattern; both Ki67- and Ki67+ MPP are still MPP, even though the marker distinction may be accompanied by transient differences in functional readout.17 We therefore find little evidence for a strict commitment model at the level of MPP. If the commitment model is nonetheless correct, we expect that new marker combinations will very soon enable clear separation of the functionally distinct subsets within current cell compartments.
Instead of MPP being comprised of cells with clearly distinct functional capabilities, MPP may consist of two or more populations of lineage-biased cells. Lineage bias and recent claims of novel lineage combinations, such as GM/T- or GM/B-restricted progenitors, seem to support the hybrid model (Fig. 4). However, novel lineage combination claims have relied heavily on in vitro assays and have been rapidly refuted.1,6,7,16,18-20 A recent IL7R-Cre fate mapping study provided particularly convincing evidence for a shared T/B cell differentiation path and clear separation of myeloid and lymphoid lineages,6,19 as proposed upon isolation of CLP and CMP.14,21 Our own data, as well as the results from Buza-Vidas et al.,13 also support this model, as all myeloid-committed cells have similar Flk2-Cre-mediated floxing efficiencies that are lower than those of lymphoid-committed cells. These studies also show that the contribution of putative Flk2+ myeloid restricted progenitors, proT cells or “lymphoid-biased” MPP10,11,22,23 to the total GM output is either minimal or includes equivalent MegE production. Because in vitro assays can both underestimate and induce lineage potential (reviewed in refs. 5 and 6), we are strongly inclined to favor models based on genetic lineage tracing experiments performed under unperturbed, physiological conditions. Reassuringly, the three fate mapping studies described above1,6,13 all support a closer relationship between GM and MegE than between GM and T or GM and B cell lineages.
Flk2 expression delineates loss of self-renewal but is compatible with multipotency.
We previously established that all phenotypic HSC in the FlkSwitch model are Tom+ and therefore must be derived from a Flk2- precursor.1 In this report, we established that both the reporter and Cre transgenes are inert, without effects on endogenous Flk2 expression or the size or function of populations defined by Flk2 expression (Fig. 2). Using an unbiased approach of transplanting Tom+ or GFP+ KLS cells, we further demonstrated that all self-renewing cells with long-term, multilineage capacity do not express Cre or Flk2 (Fig. 3). In contrast to our findings, Buza-Vidas and colleagues reported that a fraction of HSC exhibited reporter activity, as many as 23% of phenotypic HSC (CD150+CD48- KLS cells) were reporter-positive.13 However, a serial transplantation assay demonstrated that the self-renewal capability of the floxed KLS cells was far inferior to that of unfloxed KLS.13 Combined with our transplantation data (Fig. 3), it is clear that all cells expressing Flk2 and therefore Cre and GFP in FlkSwitch mice, have lost the ability to self-renew. These results also show that addition of Flk2 to the KLS CD150/CD48 marker combination eliminates a fraction of cells that have lost self-renewal potential. Thus, in the mouse, Flk2 is a valuable marker for isolation of highly pure functional HSC.
Conversely, while upregulation of Flk2 marks loss of self-renewal, Flk2 expression does not result in loss of multilineage potential. Data from our previous transplantation experiments in which we quantitatively assessed multilineage reconstitution, including MegE generation, across multiple stem and progenitor populations support a model in which multilineage capability is retained as Flk2- HSC develop into Flk2+ MPPF.7 Despite this clear demonstration of MegE potential, it remained possible that MPPF only give rise to MegE cells under conditions of acute stress in transplantation models, whereas the physiological role of MPPF under normal conditions is to promote lymphoid development. The FlkSwitch model clearly demonstrates multilineage differentiation through a Flk2+ intermediate under physiological conditions, and that Flk2 expression does not restrict lineage potential. Expression of Flk2 in human HSC provides additional evidence that Flk2 expression is compatible with multilineage potential.24,25
Definitive conclusions and questions that remain.
Our fate mapping data allows us to definitively conclude that Flk2 is not expressed during HSC development or maintenance, but that a Flk2+ stage is obligatory for differentiation into all lineages at both steady-state and under stress conditions. However, despite the strong evidence for a shared Flk2+ progenitor for all lineages, definitive demonstration for this is still imperative. An essential missing clue is what proportion of MPP are multipotent at the clonal level. Importantly, conclusions regarding multipotency of one population have clear implications for the lineage potential of its progeny: if the “MPP” compartment in reality is composed of several unilineage-committed cell types that happen to share a handful of surface markers (commitment model, Fig. 4A), it seems highly unlikely that clonal, oligopotent CMP exist (Fig. 4B). Likewise, if lineage commitment is made by HSC, the argument for the existence of clonal, multipotent MPP is severely undermined. While a growing number of reports have established multilineage, long-term readout from transplanted single HSC,26-30 recent evidence also points toward significant lineage bias between different types of HSC.31-36 For MPP, we believe that the most convincing data converge on a slightly imperfect version of the multipotency model (Fig. 4A). Cells with exclusive GM/T or GM/B potential may exist, but our data indicate that their numbers are vanishingly small and likely of little physiological relevance. Establishment of assays capable of efficiently measuring physiologically accurate lineage potential at the clonal level seems necessary to resolve conflicting views. This will be extremely challenging, but strikes us as an issue of high significance to the field as the very definition of a stem cell—multilineage and self-renewal potential at the single-cell level—is under debate.
Materials and Methods
Mice.
Mice were maintained by the UCSC animal facility according to approved protocols. Flk2-/- mice have been described previously.37 Flk2-Cre mice8 were crossed to mT/mG9 mice to generate FlkSwitch mice.1 FlkSwitch mice with high floxing efficiency (> 80% GFP in myeloid cells) were used for all experiments.
Flow cytometry to determine cell frequencies and Flk2 surface levels.
Whole bone marrow samples were stained with antibody cocktails, as described previously1,38,39 to identify HSC (Lin-Sca1+cKit+CD48-Slam1+Flk2-), ST-HSCF (Lin-Sca1+cKit+Flk2intermediate), MPPF (Lin-Sca1+cKit+CD48+Slam1-Flk2+), CMP (Lin-Flk2-Sca1-cKit+FcgRmidCD34mid), GMP (Lin-Flk2-Sca1-cKit+FcgRhiCD34hi), MEP (Lin-Flk2-Sca1-cKit+FcgRloCD34lo) and CLP (Lin-Sca1midcKitmidFlk2+IL7Ra+).
Transplantation assays.
100 or 500 Tomato-positive and GFP-positive KLS cells were double sorted from cKit-enriched bone marrow from individual FlkSwitch mice and transplanted by retroorbital transfer into sublethally irradiated mice. Peripheral blood samples from recipient mice were analyzed by flow cytometry, as described previously,1,38 after staining with B220, CD3, Gr1, Mac1 and Ter119 antibodies. GM (Ter119-, B220-, CD3-, Mac1+, Gr1hi), Plt (SSClo, CD61+, Ter119-), B-cell (Ter119-, B220+, CD3-, Mac1-, Gr1-), T-cell (Ter119-, B220-, CD3+, Mac1-, Gr1-).
Acknowledgments
We thank Drs. Bleul and Boehm for generously sharing the Flk2-Cre mice. We thank Dr. Fernando Ugarte for insightful comments on the manuscript and Bertram Linderkamp for animal care. This work was supported by a California Institute for Regenerative Medicine (CIRM) New Faculty Award to E.C.F., by CIRM Shared Stem Cell Facilities (FA1–00617–1) and Major Facilities (FA1–00617–1) awards to UCSC. S.W.B. is supported by NIH training grant 2T32GM008646. A.E.B. is supported by CIRM Training grant TG2–01157. The authors declare no conflicts of interest.
Glossary
Abbreviations:
- BM
bone marrow
- HSC
hematopoietic stem cell
- MPP
multipotent progenitor
- CMP
common myeloid progenitor
- CLP
common lymphoid progenitor
- MegE
megakaryocyte/erythroid
- EP
erythroid progenitor
- GM
granulocyte/macrophage
- PB
peripheral blood
- GFP
green fluorescent protein
Footnotes
Previously published online: www.landesbioscience.com/journals/cc/article/21279
References
- 1.Boyer SW, Schroeder AV, Smith-Berdan S, Forsberg EC. All hematopoietic cells develop from hematopoietic stem cells through Flk2/Flt3-positive progenitor cells. Cell Stem Cell. 2011;9:64–73. doi: 10.1016/j.stem.2011.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Adolfsson J, Borge OJ, Bryder D, Theilgaard-Mönch K, Astrand-Grundström I, Sitnicka E, et al. Upregulation of Flt3 expression within the bone marrow Lin(-)Sca1(+)c-kit(+) stem cell compartment is accompanied by loss of self-renewal capacity. Immunity. 2001;15:659–69. doi: 10.1016/S1074-7613(01)00220-5. [DOI] [PubMed] [Google Scholar]
- 3.Christensen JL, Weissman IL. Flk-2 is a marker in hematopoietic stem cell differentiation: a simple method to isolate long-term stem cells. Proc Natl Acad Sci USA. 2001;98:14541–6. doi: 10.1073/pnas.261562798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Morrison SJ, Wandycz AM, Hemmati HD, Wright DE, Weissman IL. Identification of a lineage of multipotent hematopoietic progenitors. Development. 1997;124:1929–39. doi: 10.1242/dev.124.10.1929. [DOI] [PubMed] [Google Scholar]
- 5.Ichii M, Shimazu T, Welner RS, Garrett KP, Zhang Q, Esplin BL, et al. Functional diversity of stem and progenitor cells with B-lymphopoietic potential. Immunol Rev. 2010;237:10–21. doi: 10.1111/j.1600-065X.2010.00933.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schlenner SM, Rodewald HR. Early T cell development and the pitfalls of potential. Trends Immunol. 2010;31:303–10. doi: 10.1016/j.it.2010.06.002. [DOI] [PubMed] [Google Scholar]
- 7.Forsberg EC, Serwold T, Kogan S, Weissman IL, Passegué E. New evidence supporting megakaryocyte-erythrocyte potential of flk2/flt3+ multipotent hematopoietic progenitors. Cell. 2006;126:415–26. doi: 10.1016/j.cell.2006.06.037. [DOI] [PubMed] [Google Scholar]
- 8.Benz C, Martins VC, Radtke F, Bleul CC. The stream of precursors that colonizes the thymus proceeds selectively through the early T lineage precursor stage of T cell development. J Exp Med. 2008;205:1187–99. doi: 10.1084/jem.20072168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Muzumdar MD, Tasic B, Miyamichi K, Li L, Luo L. A global double-fluorescent Cre reporter mouse. Genesis. 2007;45:593–605. doi: 10.1002/dvg.20335. [DOI] [PubMed] [Google Scholar]
- 10.Adolfsson J, Månsson R, Buza-Vidas N, Hultquist A, Liuba K, Jensen CT, et al. Identification of Flt3+ lympho-myeloid stem cells lacking erythro-megakaryocytic potential a revised road map for adult blood lineage commitment. Cell. 2005;121:295–306. doi: 10.1016/j.cell.2005.02.013. [DOI] [PubMed] [Google Scholar]
- 11.Nutt SL, Metcalf D, D’Amico A, Polli M, Wu L. Dynamic regulation of PU.1 expression in multipotent hematopoietic progenitors. J Exp Med. 2005;201:221–31. doi: 10.1084/jem.20041535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pronk CJ, Rossi DJ, Månsson R, Attema JL, Norddahl GL, Chan CK, et al. Elucidation of the phenotypic, functional, and molecular topography of a myeloerythroid progenitor cell hierarchy. Cell Stem Cell. 2007;1:428–42. doi: 10.1016/j.stem.2007.07.005. [DOI] [PubMed] [Google Scholar]
- 13.Buza-Vidas N, Woll P, Hultquist A, Duarte S, Lutteropp M, Bouriez-Jones T, et al. FLT3 expression initiates in fully multipotent mouse hematopoietic progenitor cells. Blood. 2011;118:1544–8. doi: 10.1182/blood-2010-10-316232. [DOI] [PubMed] [Google Scholar]
- 14.Akashi K, Traver D, Miyamoto T, Weissman IL. A clonogenic common myeloid progenitor that gives rise to all myeloid lineages. Nature. 2000;404:193–7. doi: 10.1038/35004599. [DOI] [PubMed] [Google Scholar]
- 15.Arinobu Y, Mizuno S, Chong Y, Shigematsu H, Iino T, Iwasaki H, et al. Reciprocal activation of GATA-1 and PU.1 marks initial specification of hematopoietic stem cells into myeloerythroid and myelolymphoid lineages. Cell Stem Cell. 2007;1:416–27. doi: 10.1016/j.stem.2007.07.004. [DOI] [PubMed] [Google Scholar]
- 16.Karsunky H, Inlay MA, Serwold T, Bhattacharya D, Weissman IL. Flk2+ common lymphoid progenitors possess equivalent differentiation potential for the B and T lineages. Blood. 2008;111:5562–70. doi: 10.1182/blood-2007-11-126219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Passegué E, Wagers AJ, Giuriato S, Anderson WC, Weissman IL. Global analysis of proliferation and cell cycle gene expression in the regulation of hematopoietic stem and progenitor cell fates. J Exp Med. 2005;202:1599–611. doi: 10.1084/jem.20050967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Richie Ehrlich LI, Serwold T, Weissman IL. In vitro assays misrepresent in vivo lineage potentials of murine lymphoid progenitors. Blood. 2011;117:2618–24. doi: 10.1182/blood-2010-05-287102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schlenner SM, Madan V, Busch K, Tietz A, Läufle C, Costa C, et al. Fate mapping reveals separate origins of T cells and myeloid lineages in the thymus. Immunity. 2010;32:426–36. doi: 10.1016/j.immuni.2010.03.005. [DOI] [PubMed] [Google Scholar]
- 20.Serwold T, Ehrlich LI, Weissman IL. Reductive isolation from bone marrow and blood implicates common lymphoid progenitors as the major source of thymopoiesis. Blood. 2009;113:807–15. doi: 10.1182/blood-2008-08-173682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kondo M, Weissman IL, Akashi K. Identification of clonogenic common lymphoid progenitors in mouse bone marrow. Cell. 1997;91:661–72. doi: 10.1016/S0092-8674(00)80453-5. [DOI] [PubMed] [Google Scholar]
- 22.Bell JJ, Bhandoola A. The earliest thymic progenitors for T cells possess myeloid lineage potential. Nature. 2008;452:764–7. doi: 10.1038/nature06840. [DOI] [PubMed] [Google Scholar]
- 23.Wada H, Masuda K, Satoh R, Kakugawa K, Ikawa T, Katsura Y, et al. Adult T-cell progenitors retain myeloid potential. Nature. 2008;452:768–72. doi: 10.1038/nature06839. [DOI] [PubMed] [Google Scholar]
- 24.Doulatov S, Notta F, Eppert K, Nguyen LT, Ohashi PS, Dick JE. Revised map of the human progenitor hierarchy shows the origin of macrophages and dendritic cells in early lymphoid development. Nat Immunol. 2010;11:585–93. doi: 10.1038/ni.1889. [DOI] [PubMed] [Google Scholar]
- 25.Kikushige Y, Yoshimoto G, Miyamoto T, Iino T, Mori Y, Iwasaki H, et al. Human Flt3 is expressed at the hematopoietic stem cell and the granulocyte/macrophage progenitor stages to maintain cell survival. J Immunol. 2008;180:7358–67. doi: 10.4049/jimmunol.180.11.7358. [DOI] [PubMed] [Google Scholar]
- 26.Ema H, Sudo K, Seita J, Matsubara A, Morita Y, Osawa M, et al. Quantification of self-renewal capacity in single hematopoietic stem cells from normal and Lnk-deficient mice. Dev Cell. 2005;8:907–14. doi: 10.1016/j.devcel.2005.03.019. [DOI] [PubMed] [Google Scholar]
- 27.Kiel MJ, Yilmaz OH, Iwashita T, Yilmaz OH, Terhorst C, Morrison SJ. SLAM family receptors distinguish hematopoietic stem and progenitor cells and reveal endothelial niches for stem cells. Cell. 2005;121:1109–21. doi: 10.1016/j.cell.2005.05.026. [DOI] [PubMed] [Google Scholar]
- 28.Morita Y, Ema H, Nakauchi H. Heterogeneity and hierarchy within the most primitive hematopoietic stem cell compartment. J Exp Med. 2010;207:1173–82. doi: 10.1084/jem.20091318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Osawa M, Hanada K, Hamada H, Nakauchi H. Long-term lymphohematopoietic reconstitution by a single CD34-low/negative hematopoietic stem cell. Science. 1996;273:242–5. doi: 10.1126/science.273.5272.242. [DOI] [PubMed] [Google Scholar]
- 30.Wagers AJ, Sherwood RI, Christensen JL, Weissman IL. Little evidence for developmental plasticity of adult hematopoietic stem cells. Science. 2002;297:2256–9. doi: 10.1126/science.1074807. [DOI] [PubMed] [Google Scholar]
- 31.Benveniste P, Frelin C, Janmohamed S, Barbara M, Herrington R, Hyam D, et al. Intermediate-term hematopoietic stem cells with extended but time-limited reconstitution potential. Cell Stem Cell. 2010;6:48–58. doi: 10.1016/j.stem.2009.11.014. [DOI] [PubMed] [Google Scholar]
- 32.Challen GA, Boles NC, Chambers SM, Goodell MA. Distinct hematopoietic stem cell subtypes are differentially regulated by TGF-beta1. Cell Stem Cell. 2010;6:265–78. doi: 10.1016/j.stem.2010.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dykstra B, Kent D, Bowie M, McCaffrey L, Hamilton M, Lyons K, et al. Long-term propagation of distinct hematopoietic differentiation programs in vivo. Cell Stem Cell. 2007;1:218–29. doi: 10.1016/j.stem.2007.05.015. [DOI] [PubMed] [Google Scholar]
- 34.Weksberg DC, Chambers SM, Boles NC, Goodell MA. CD150- side population cells represent a functionally distinct population of long-term hematopoietic stem cells. Blood. 2008;111:2444–51. doi: 10.1182/blood-2007-09-115006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hock H. Some hematopoietic stem cells are more equal than others. J Exp Med. 2010;207:1127–30. doi: 10.1084/jem.20100950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schroeder T. Hematopoietic stem cell heterogeneity: subtypes, not unpredictable behavior. Cell Stem Cell. 2010;6:203–7. doi: 10.1016/j.stem.2010.02.006. [DOI] [PubMed] [Google Scholar]
- 37.Mackarehtschian K, Hardin JD, Moore KA, Boast S, Goff SP, Lemischka IR. Targeted disruption of the flk2/flt3 gene leads to deficiencies in primitive hematopoietic progenitors. Immunity. 1995;3:147–61. doi: 10.1016/1074-7613(95)90167-1. [DOI] [PubMed] [Google Scholar]
- 38.Smith-Berdan S, Nguyen A, Hassanein D, Zimmer M, Ugarte F, Ciriza J, et al. Robo4 cooperates with CXCR4 to specify hematopoietic stem cell localization to bone marrow niches. Cell Stem Cell. 2011;8:72–83. doi: 10.1016/j.stem.2010.11.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Smith-Berdan S, Schepers K, Ly A, Passegué E, Forsberg EC. Dynamic expression of the Robo ligand Slit2 in bone marrow cell populations. Cell Cycle. 2012;11:675–82. doi: 10.4161/cc.11.4.19146. [DOI] [PubMed] [Google Scholar]