Abstract
The mammalian LIN complex (LINC) plays important roles in regulation of cell cycle genes. LIN54 is an essential core subunit of the LINC and has a DNA binding region (CHC domain), which consists of two cysteine-rich (CXC) domains separated by a short spacer. We generated various LIN54 mutants, such as CHC deletion mutant, and investigated their subcellular localizations and effects on cell cycle. Wild-type LIN54 was predominantly localized in the nucleus. We identified two nuclear localization signals (NLSs), both of which were required for nuclear localization of LIN54. Interestingly, deletion of one CXC domain resulted in an increased cytoplasmic localization. The cytoplasmic LIN54 mutant accumulated in the nucleus after leptomycin B treatment, suggesting CRM1-mediated nuclear export of LIN54. Point mutations (C525Y and C611Y) in conserved cysteine residues of CXC domain that abolish DNA binding activity also increased cytoplasmic localization. These data suggest that DNA binding activity of LIN54 is required for its nuclear retention. We also found that LIN54C525Y and LIN54C611Y inhibited cell cycle progression and led to abnormal nuclear morphology. Other CXC mutants also induced similar abnormalities in cell cycle progression. LIN54C525Y led to a decreased expression of some G2/M genes, whose expressions are regulated by LINC. This cell cycle inhibition was partially restored by overexpression of wild-type LIN54. These results suggest that abnormal cellular localization of LIN54 may have effects on LINC activity.
Keywords: LIN54, LINC, CXC domain, cell cycle, nuclear localization signal, nuclear export
Introduction
A cell cycle regulation is essential for proper proliferation and differentiation. Aberrations in the cell cycle elicit dire consequences for the cell, such as aneuploidy, genomic instability and oncogenic transformation. A retinoblastoma tumor suppressor protein (pRB) and two related pocket proteins (p107 and p130) are the most important proteins for regulating the cell cycle related genes and function together with different subsets of E2F transcription factors.1-4 p107 and p130 specifically interact with E2F4 and E2F5 and function as transcriptional repressors in quiescent cells.1,5-7
Recently, a new complex that interacts with p130/E2F4 in G0/G1 phase of the cell cycle was identified.8-10 This complex, named DREAM (Drosophila RBF, dE2F2 and dMyb-interacting proteins), includes a core module that consists of LIN9, LIN37, LIN52, LIN54 and RBBP4. This core module is termed LIN complex (LINC). Similar complexes have been identified in Drosophila and Caenorhabditis elegans.11-13 Human LINC in quiescent stage of cultured cells interacts with p130, E2F4 and DP1. This complex (DREAM) functions as a transcriptional repressor by binding to promoters of E2F target genes that regulate G1/S transition. In S phase, LINC dissociates from p130/E2F4/DP1 and associates with a transcription factor B-MYB. LINC-B-MYB complex binds to promoters of G2/M genes and activates their transcriptions.8,9,14-16
LIN54, which is one of the essential core subunits of LINC, contains two cysteine-rich (CXC) domains separated by a short spacer called hinge. It has been reported that CXC-hinge-CXC (CHC) domain functions as a DNA binding domain, and conserved cysteine residues in two CXC domains are essential for DNA binding.17 Therefore, it is considered that LINC binds to the target promoters through the CHC domain of LIN54. In fact, depletion of LIN54 in human and mouse cells leads to a decreased expression of G2/M genes and an inhibition of cell cycle progression.17-19
LIN54 homologs are conserved in many species, such as plants, invertebrates and mammals. In mammals, in addition to LIN54, another CXC family protein, tesmin, has been identified.20 Tesmin exhibits testis-specific expression and contains a CHC domain similarly to that of LIN54.20,21 We have previously demonstrated that tesmin exhibits dynamic changes in subcellular localization during spermatogenesis. Tesmin is localized in the cytoplasm of early to late spermatocytes and then translocates into the nucleus just before meiotic division. After meiosis, tesmin relocates into the cytoplasm of spermatids.22
It is known that functions of some nuclear proteins are regulated by changing the subcellular localization. Among the components of DREAM, E2F4 and p130 have been shown to exhibit subcellular localization changes in a cell cycle-dependent manner.23,24 E2F4 is primarily localized to the nucleus in quiescent cells, and it is exported from the nucleus in a CRM1-dependent manner during entry into S phase.25 It is possible that the components of LINC also exhibit nucleo-cytoplasmic shuttling; however, it remains unclear about their subcellular localization.
In this study, we generated various LIN54 mutants and investigated their subcellular localization and effects on cell cycle. We identified two nuclear localization signals (NLSs) that are essential for the nuclear localization of LIN54. In addition to NLSs, we found that CHC domain is required for the nuclear retention. LIN54 harboring a mutated CHC domain was efficiently exported from nucleus to cytoplasm, although LIN54 contains no region homologous to the known nuclear export signals (NESs). We also found that LIN54 CHC mutants led to a decreased expression of G2/M genes and inhibited cell cycle progression. We propose a possibility that LIN54 harboring a mutated CHC domain may act as a dominant-negative mutant.
Results
Expression of Lin54 in various mouse tissues
We have shown previously that tesmin, which is a member of the CHC family, is exclusively expressed in the testis.20 LIN54 also belongs to the CHC family, but its expression pattern in most tissues is not known. To investigate whether Lin54 exhibits tissue-specific expression, reverse transcription-PCR (RT-PCR) was performed with various mouse tissues. The expression of Lin54 was detected in all tissues tested (Fig. 1). In lung, brain, colon and testis, Lin54 expression was abundant compared with other tissues. On the other hand, Lin54 expression was low in non-proliferative tissues such as skeletal muscle.
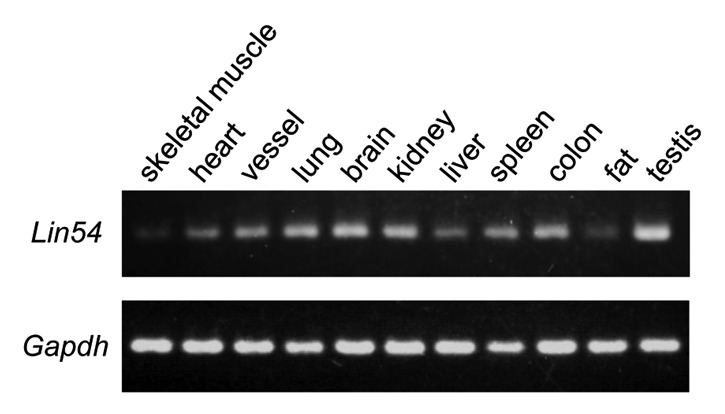
Figure 1. Expression of Lin54 in various mouse tissues was analyzed by RT-PCR. All tissues were excised from 7-week-old male ICR mouse. Gapdh was used as an internal control.
Nuclear and cytoplasmic localization of LIN54 protein
To investigate the subcellular localization of LIN54, LIN54-EGFP fusion construct was generated and expressed in some mammalian cultured cells (HeLa, COS-7, HEK 293, NIH3T3, GC-1 and GC-2) with PolyFect Transfection Reagent. The chimeric protein was visualized by direct fluorescence. However, very few EGFP-positive cells were detected. Similar results were observed when cells were transiently transfected with the FLAG epitope-tagged LIN54 constructs. Therefore, we used an adenovirus expression system to transiently express LIN54. GC-1 cells were infected with recombinant adenoviruses and the expression of the EGFP fusion protein was visualized using fluorescence microscopy. The green fluorescence of the LIN54-EGFP protein was detected in the most infected cells and predominantly localized in the nucleus (Fig. 2A), which is consistent with its role in transcriptional regulation. Interestingly, it was observed that in some GC-1 cells, LIN54-EGFP was localized to both the nucleus and the cytoplasm, suggesting that LIN54 may be a nucleo-cytoplasmic shuttling protein (Fig. 2B). Furthermore, the cytoplasmic LIN54-EGFP accumulated in the nucleus after treatment with leptomycin B, a specific inhibitor of CRM1/exportin 1 (Fig. 2B).26 This result suggests that the cytoplasmic localization of LIN54-EGFP is due to a nuclear export by CRM1/exportin 1. We also found that LIN54-EGFP was equally distributed in both the nucleus and the cytoplasm when the cells were cultured in serum-starved condition (Fig. 2C). After readdition of serum the cytoplasmic LIN54 was translocated into the nucleus. LIN54 may change the subcellular localization in a cell cycle-dependent manner.
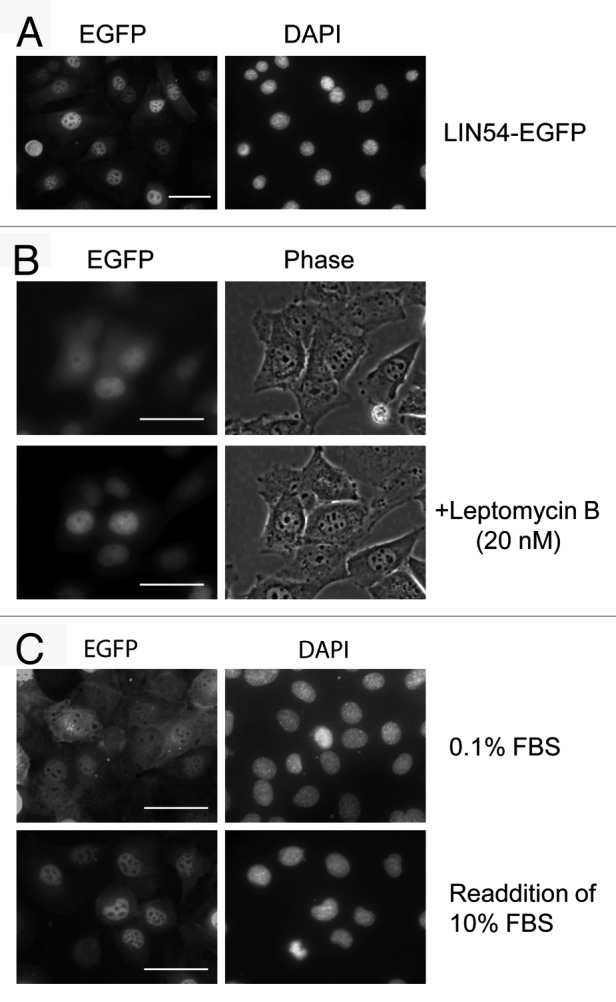
Figure 2. Subcellular localization of LIN54. (A) GC-1 cells were infected with recombinant adenovirus expressing wild-type LIN54-EGFP. At 24 h after infection, the cells were fixed and EGFP-fused proteins were detected using fluorescent microscope. Nuclei were stained with DPAI. (B) The upper panels show examples of cytoplasmic localizations of LIN54. The lower panels are the result of observing the same field of view shown in the upper panels after leptomycin B treatment. Leptomycin B treatment (20 nM) was performed for 4 h. The phase panels show the corresponding field visualized by phase-contrast microscopy. (C) GC-1 cells were cultured in 0.1% FBS for 48 h and then infected with the recombinant adenovirus. After infection, cells were cultured in 0.1% FBS for 24 h and then LIN54-EGFP was detected. Serum-starved cells were stimulated with serum for 10 h. The scale bar is 50 μM.
Role of the putative NLSs in nuclear targeting of LIN54
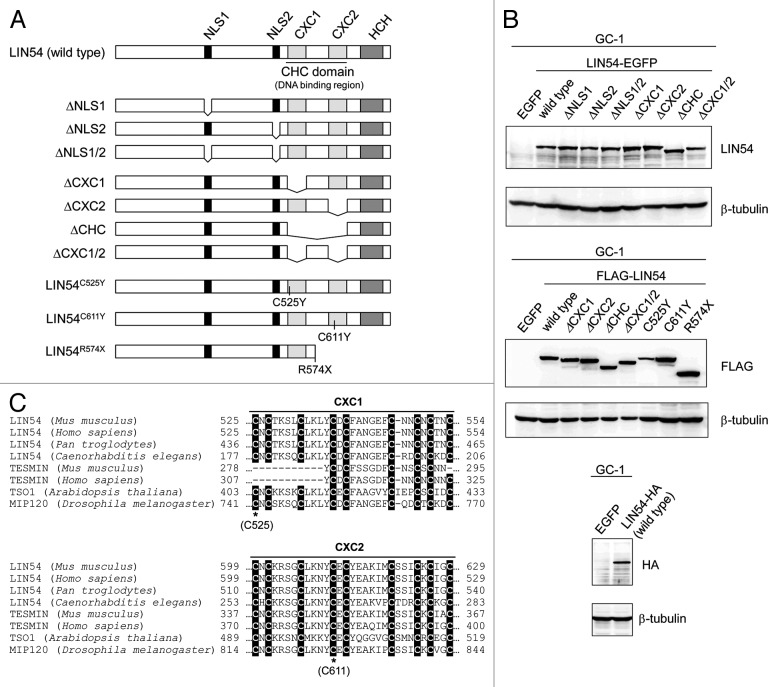
Two putative NLSs were identified in LIN54 by using PSORT II Prediction (www.psort.hgc.jp/form2.html) (Fig. 3A). The amino acid sequences of NLS1, 231-KKPR-234, correspond to the classical NLS consensus sequence K-K/R-X-K/R for monopartite NLSs. By contrast, the amino acid sequences of NLS2, 520-RPRK-523, are partially conserved. In order to investigate whether each of the putative NLSs are functional in the nuclear import of LIN54, NLS-deletion mutants of LIN54 were generated and expression of the EGFP fusion proteins was confirmed by western blotting with anti-LIN54 antibody (Fig. 3A and B). GC-1 cells were infected with recombinant adenoviruses and the NLS-deletion mutants of LIN54-EGFP were visualized using fluorescence microscopy. Deletion of the NLS1 or NLS2 resulted in an increased cytoplasmic localization compared with the wild-type LIN54 distribution (Fig. 4). Furthermore, LIN54 lacking both NLSs was predominantly localized to the cytoplasm. These results indicate that two NLSs play a major role in the nuclear import of LIN54.
Figure 3. Constructs of various LIN54 mutants. (A) Schematic representation of LIN54 mutants. Fragments of LIN54 were ligated in frame with one of various epitope tags (EGFP, FLAG and HA). EGFP and HA epitope tag were fused to the C terminus and FLAG epitope tag was fused to the N terminus. DNA constructs encoding these mutants were inserted into adenoviral vectors. (B) GC-1 cells were infected with each recombinant adenovirus and then LIN54 and mutant proteins were detected by western blot with indicated antibodies. β-tubulin is a loading control. (C) Alignment of the amino acid sequence of CXC domains with those of other CHC family proteins. Conserved cysteine residues are shaded in black. The asterisks represent the point-mutated cysteine residues (C525 and C611).
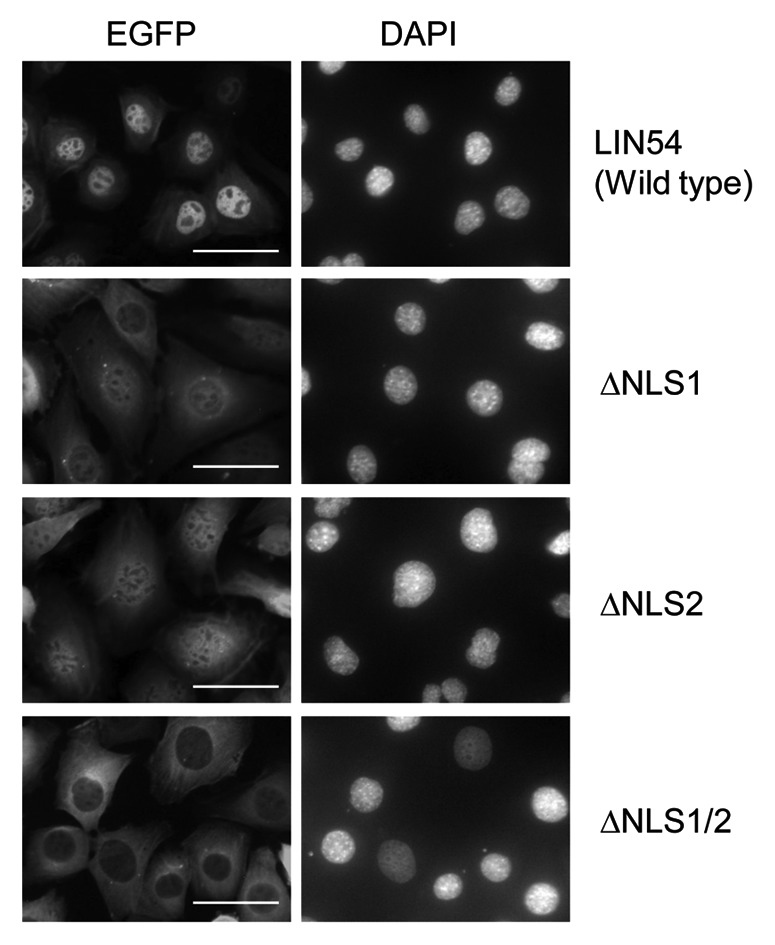
Figure 4. Subcellular localization of LIN54 NLS mutants. GC-1 cells were infected with recombinant adenoviruses expressing the indicated constructs. At 24 h after infection, the cells were fixed and EGFP-fused proteins were detected using fluorescent microscope. Nuclei were stained with DPAI. The scale bar is 50 μM.
Effects of mutated CHC domain on LIN54 localization
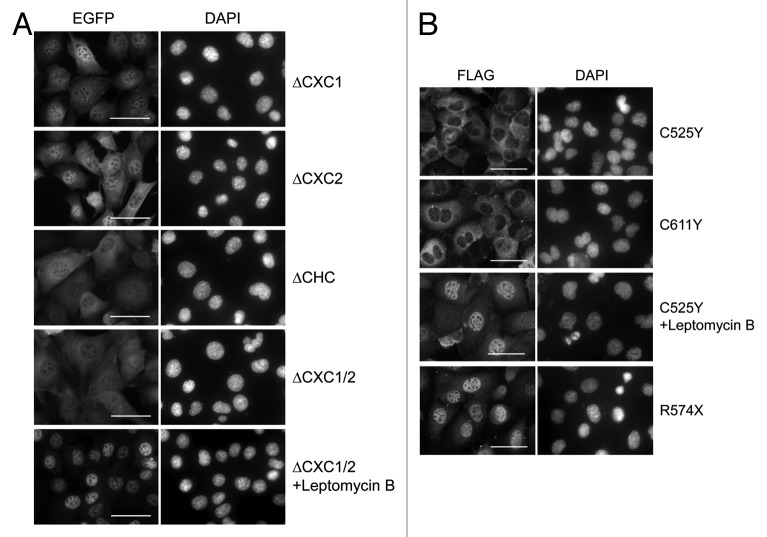
LIN54 contains two cysteine-rich CXC domains, which are highly conserved among LIN54 homologs (Fig. 3C). Recently, it was reported that the CXC domain of LIN54 was a DNA-binding domain.17 To investigate more features of the CXC domain, various CXC domain deletion mutants were constructed and transfected into GC-1 cells with adenoviruses (Fig. 3A and B). We first examined the subcellular localizations of CXC deletion mutants. LIN54(ΔCXC1)-EGFP and LIN54(ΔCXC2)-EGFP exhibited an increased cytoplasmic localization compared with wild-type LIN54 (Fig. 5A). Furthermore, LIN54 lacking two CXC domains, LIN54(ΔCHC)-EGFP and LIN54(ΔCXC1/2)-EGFP, were primarily localized to the cytoplasm. These results indicate that both CXC domains of LIN54 were required for nuclear localization. We also examined subcellular localization of FLAG epitope-tagged LIN54 and its derivatives. FLAG-LIN54 constructs were transiently expressed in GC-1 cells by adenoviral vectors and immunostained with anti-FLAG monoclonal antibody. Similar to EGFP fusion proteins, wild-type FLAG-LIN54 was localized to the nucleus and a deletion of CXC domain resulted in an increased cytoplasmic localization (data not shown).
Figure 5. Subcellular localization of LIN54 harboring mutations in CHC domain. (A) GC-1 cells were infected with recombinant adenoviruses expressing the indicated constructs. At 24 h after infection, the cells were fixed, and EGFP-fused proteins were detected using fluorescent microscope. Leptomycin B treatment (20 nM) was performed for 4 h prior to fixation. Nuclei were stained with DPAI. (B) FLAG-tagged LIN54 mutants were visualized by immunostaining with anti-FLAG antibody. The scale bar is 50 μM.
Next, to test whether conserved cysteines in the CXC domain were required for the nuclear localization, we introduced a point mutation into the CXC domain (Cys525 or Cys611) and transiently expressed the constructs in GC-1 cells. It has been reported that mutations of the cysteine residues corresponding to Cys525 and Cys611 in TSO1, which is a LIN54 homolog in Arabidopsis thaliana, cause abnormal flower development and defects in cell division in floral meristem cells.27-29 LIN54 with Cys525 substituted with tyrosine (LIN54C525Y) was predominately localized to the cytoplasm (Fig. 5B). A similar result was observed when Cys611 was mutated to tyrosine (LIN54C611Y). These results suggest that the conserved cysteine residues are required for the nuclear localization of LIN54. Furthermore, point-mutated LIN54 accumulated in the nucleus after treatment with leptomycin B, suggesting that the steady-state cytoplasmic distributions of LIN54C525Y and LIN54C611Y reflect the dominance of CRM1-medimated nuclear export mechanisms. Cytoplasmic LIN54(ΔCXC1/2) also accumulated in the nucleus after treatment with leptomycin B (Fig. 5A). The CHC domain presumably plays a role in the nuclear retention of LIN54.
Since we failed to find any known nuclear export signals (NESs) in LIN54, it is suspected that the nuclear export of LIN54 is mediated by other NES-containing proteins. It has been reported that LIN54 contains a helix-coil-helix (HCH) domain, which is required for the interaction with p130 and B-MYB.17 To investigate whether HCH domain is involved in nuclear export of LIN54, we introduced a nonsense mutation into the hinge region (Arg574). The resulted construct (LIN54R574X) is lacking one CXC domain and HCH domain (Fig. 3A and B). In spite of the lack of one CXC domain, LIN54R574X was predominantly localized to the nucleus, suggesting a failure of nuclear export. p130 or B-MYB may contribute to the nuclear export of LIN54 by associating through the HCH domain.
LIN54 harboring a mutated CHC domain impedes cell cycle progression
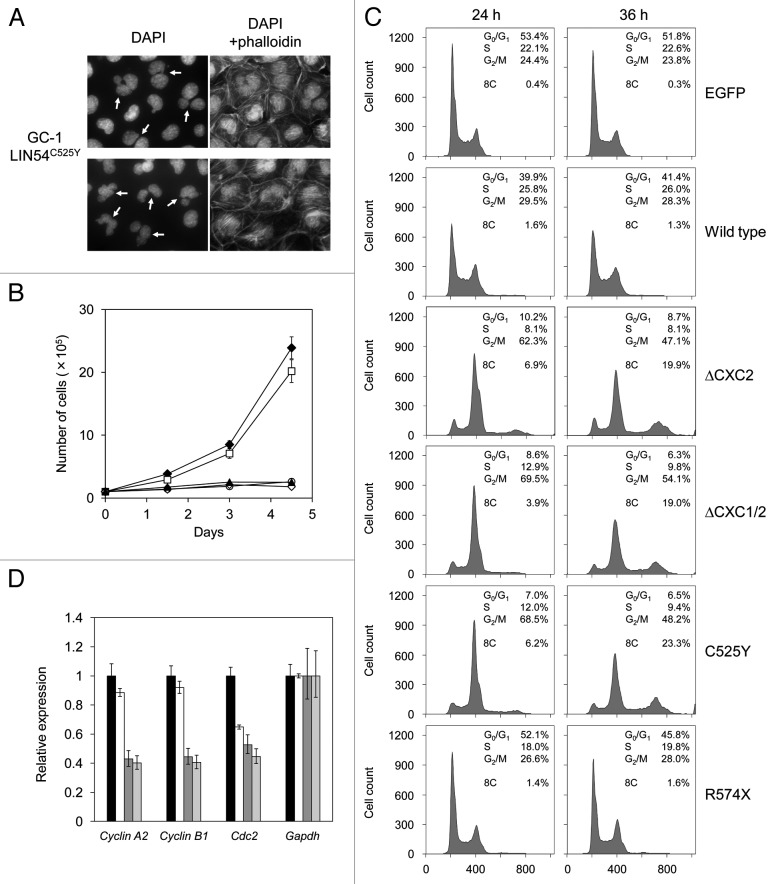
When various LIN54 mutants were transiently expressed in GC-1 cells, we found that the proportion of abnormal nuclear morphology was increased in cells expressing LIN54 derivatives with mutations in CHC domains (Table 2). Even in wild-type LIN54, its overexpression slightly caused abnormal nuclear morphology. These abnormal cells had aberrant nuclei, such as binucleation, micronucleation and nuclei with multiple lobes (Fig. 6A). Furthermore, LIN54(ΔCXC1/2), LIN54C525Y and LIN54C611Y seriously inhibited cell proliferation (Fig. 6B). We next performed cell cycle analysis using flow cytometry. LIN54C525Y caused a marked reduction of cells in G1 phase and accumulation of cells in G2/M phase compared with control EGFP and wild-type LIN54-expressing cells (Fig. 6C). Interestingly, cells with an 8C DNA content were observed in LIN54C525Y-expressing cells. Similar result was observed when LIN54(ΔCXC2) and LIN54(ΔCXC1/2) were transiently expressed in GC-1 cells (Fig. 6C). We also analyzed cell cycle profiles of HeLa cells expressing LIN54 mutants. LIN54C525Y and LIN54(ΔCXC1/2) led to a decrease in G1 cells and an increased in G2/M cells (data not shown). These data suggest that the expression of LIN54 CHC mutants induce mitotic defects and impede cell cycle progression.
Table 2. Nuclear aberrations in GC-1 cells expressing LIN54 mutants.
| Nuclear morphology |
||
|---|---|---|
| Normal | Aberration | |
|
EGFP |
97.4% |
2.6% |
|
LIN54 |
89.6% |
10.4% |
|
LIN54(ΔCXC1) |
55.7% |
44.3% |
|
LIN54(ΔCXC2) |
51.2% |
48.8% |
|
LIN54(ΔCXC1/2) |
55.8% |
44.2% |
|
LIN54C525Y |
65.4% |
34.6% |
| LIN54C611Y | 64.1% | 35.9% |
More than 200 cells were counted from multiple random frames.
Figure 6. LIN54 CHC mutants lead to nuclear aberrations and impede cell cycle progression. (A) Examples of nuclear abnormalities in GC-1 cells expressing FLAG-LIN54C525Y. At 36 h after infection, cells were fixed and stained with DAPI and FITC-phalloidin that binds specifically to F-actin. The arrows indicate cells with abnormal nuclear morphology. (B) GC-1 cells expressing EGFP (filled diamond), FLAG-LIN54 (open square), FLAG-LIN54(ΔCXC1/2) (open diamond), FLAG-LIN54C525Y (open circle) or FLAG-LIN54C611Y (filled triangle) were seeded at 1 x 105 cells per dish in triplicate and counted at indicated time points. The experiment was performed three times. One representative experiment is shown. Error bars represent standard deviation. (C) GC-1 cells were infected with recombinant adenoviruses expressing the indicated constructs. At 24 or 36 h after infection, the cells were fixed and stained with PI. The cell cycle profiles were determined by flow cytometry. (D) GC-1 cells were infected with recombinant adenoviruses expressing EGFP (black bars), FLAG-LIN54 (white bars), FLAG-LIN54(ΔCXC1/2) (dark gray bars), or FLAG-LIN54C525Y (light gray bars). At 24 h after infection, cyclin A2, cyclin B1 and Cdc2 expression were analyzed by quantitative RT-PCR. Relative expression was normalized to Gapdh. Error bars represent standard deviation.
LINC are required for activation of some G2/M genes such as Cdc2 and cyclin B1 that are involved in G2/M transition.8,14,15 We next investigated whether the mitotic defects of these cells are caused by downregulation of G2/M genes. We analyzed expressions of some G2/M genes by quantitative RT-PCR, whose transcriptional activation is responsible for LINC. The expression of cyclin A2, cyclin B1 and Cdc2 were decreased in GC-1 cells expressing LIN54C525Y compared with the cells expressing EGFP (Fig. 6D). Wild-type LIN54 slightly decreased expression of these genes, which is consistent with the result that wild-type LIN54 induced a slight increase in cells with abnormal nuclear morphology. These results suggest that the downregulation of G2/M genes caused the mitotic defects. Since it has been reported that these three genes are downregulated by depletion of LIN components, such as LIN9 and LIN54,8,14,19 it is possible that LIN54 CHC mutants such as LIN54C525Y and LIN54C611Y may disturb the endogenous LIN54 function.
LIN54C525Y and LIN54C611Y are dominant-negative mutants
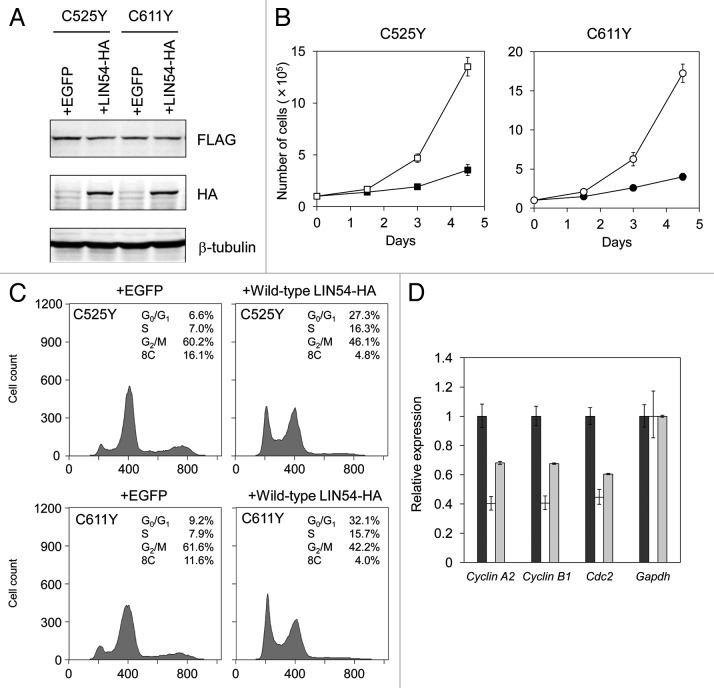
The phenotypes of GC-1 cells expressing LIN54C525Y, LIN54C611Y or other CHC mutants were similar to that of LIN54-depleted cells, such as a decreased proportion of G1 cells and an accumulation of G2/M cells.16,17 Therefore, to examine whether LIN54C525Y and LIN54C611Y act as dominant-negative mutants, we overexpressed wild-type LIN54 in GC-1 cells expressing LIN54C525Y or LIN54C611Y. Expression of hemagglutinin (HA)-tagged wild-type LIN54 and FLAG-tagged LIN54C525Y and LIN54C611Y were confirmed by western blotting with anti-HA and anti-FLAG antibodies, respectively (Fig. 7A). Overexpression of wild-type LIN54 recovered the proliferation and partially suppressed the decrease in G1 cells and the accumulation of G2/M cells (Fig. 7B and C). A population of cells with an 8C DNA content was also decreased by overexpression of wild-type LIN54. Quantitative RT-PCR analysis showed that decreased expression of G2/M genes were partially recovered by wild-type LIN54 expression (Fig. 7D). Interestingly, in spite of the lack of one CXC domain, LIN54R574X had no influence on cell cycle progression, although LIN54(ΔCXC2) seriously inhibited cell cycle progression (Fig. 6C). LIN54R574X may fail to impede LINC formation by competing with endogenous LIN54 because of the lack of HCH domain. These results suggest that LIN54C525Y and LIN54C611Y inhibit the endogenous LIN54 in a dominant-negative manner, and as a result, LINC becomes dysfunctional.
Figure 7. Wild-type LIN54 partially restores the cell cycle progression. (A) FLAG-LIN54C525Y, FLAG-LIN54C611Y and LIN54-HA expression were confirmed by western blotting with indicated antibodies. β-tubulin is a loading control. (B) GC-1 cells expressing FLAG-LIN54C525Y +EGFP (filled square), FLAG-LIN54C525Y +LIN54-HA (open square), FLAG-LIN54C611Y +EGFP (filled circle) or FLAG-LIN54C611Y +LIN54-HA (open circle) were seeded at 1 x 105 cells per dish in triplicate and counted at the indicated time points. The experiment was performed three times. One representative experiment is shown. Error bars represent standard deviation. (C) GC-1 cells were coinfected with recombinant adenoviruses expressing the indicated constructs. At 36 h after infection, the cells were fixed and stained with PI. The cell cycle profiles were determined by flow cytometry. (D) GC-1 cells were infected with recombinant adenoviruses expressing EGFP (black bars), FLAG-LIN54C525Y +EGFP (white bars) or FLAG-LIN54C525Y +LIN54-HA (gray bars). At 24 h after infection, Cyclin A2, Cyclin B1, and Cdc2 expression were analyzed by quantitative RT-PCR. Relative expression was normalized to Gapdh. Error bars represent standard deviation.
Discussion
LINC is involved in cell cycle progression by binding to p130/E2F4/DP1 in G0/G1 and to B-MYB in S phase. It was reported that LIN54, one of the components of LINC, is an essential core subunit to bind to the target gene promoter.17 However, the intracellular transport of LIN54 remains unclear. In this study, we found that LIN54 was primarily localized in the nucleus. Furthermore, in some cells, LIN54 was localized to both the nucleus and the cytoplasm and the cytoplasmic LIN54 accumulated in the nucleus after treatment with leptomycin B. These results suggest that LIN54 is a nucleo-cytoplasmic shuttling protein, and the nuclear export of LIN54 is probably dependent on CRM1. It is possible that LIN54 changes the subcellular localization in a cell cycle-dependent manner.
LIN54 possesses two putative classical monopartite nuclear localization signals (NLSs), and we have revealed that both two NLSs are required for an efficient nuclear import of LIN54. We have also demonstrated that not only two NLSs but also a CHC domain, which consists of two cysteine-rich (CXC) domains and functions as a DNA binding domain, are needed for LIN54 to localize to the nucleus. Deletion of the CHC domain resulted in increased cytoplasmic localization of LIN54. It has been reported that a DNA binding domain of some transcription factors serves as a NLS.30-32 However, it is not clear whether CHC domain acts as a NLS. LIN54R574X was primarily localized in the nucleus, although it has a deficient CHC domain. It indicates that the CXC domains are not essential for nuclear import of LIN54. Interestingly, LIN54C525Y and LIN54C611Y, harboring a point mutation that abolishes DNA binding activity of LIN54, were predominantly localized to the cytoplasm. This suggests that DNA binding activity of LIN54 is important for its nuclear retention. Furthermore, LIN54 CHC mutants accumulated in the nucleus by leptomycin B treatment, suggesting that cytoplasmic localization of CHC-deficient mutants reflects the dominance of nuclear export mechanisms. Recently, crystal structures of the RanGTP-CRM1 complex were revealed and nuclear export signal (NES) consensus was redefined.33 We searched for the NES consensus in LIN54 amino acid sequence, but such a sequence did not exist. It is possible that a nuclear export of LIN54 is mediated by other NES-containing proteins. Since LIN54R574X was localized in the nucleus, HCH domain that is required for the interaction with p130 and B-MYB seems to be involved in a nuclear export of LIN54. The retinoblastoma-related pocket protein p130 is known to shuttle between the nucleus and cytoplasm.24 A cytoplasmic translocation of LIN54 may be dependent on an interaction with p130.
We found that LIN54 harboring mutations in CHC domains induced mitotic errors and inhibited cell proliferation. FACS analysis showed a decreased proportion of LIN54C525Y-expressing cells in G1 and accumulation of cells in G2/M. Furthermore, some G2/M genes were downregulated in LIN54C525Y-expressing cells. Since these phenotypes are strikingly similar to those of LIN54- or LIN9-depleted cells,8,14,16,17,19 it seems that LIN54C525Y inhibits the function of the endogenous wild-type LIN54 in a dominant-negative manner. Indeed, in LIN54C525Y-expressing cells, overexpression of wild-type LIN54 partially recovered the proliferation and suppressed a decrease in G1 phase cells and an increase in G2/M phase cells. However, it is not clear how the cytoplasmic LIN54C525Y inhibits the function of intranuclear LIN54. LIN54R574X, which is defect in one CXC domain and HCH domain, had no influence on cell cycle progression, suggesting that the HCH domain is required to act as a dominant-negative mutant. As mentioned above, the HCH domain is essential for the binding of LIN54 to p130 and B-MYB. LIN54C525Y may interact with these proteins in cytoplasm and inhibit their nuclear import. If LIN54 is transported together with other components of LINC, such as LIN9 and LIN52, by interacting through the HCH domain, LIN54C525Y may also inhibit their nuclear localizations, which may cause a decrease in intranuclear LINC. However, further studies are needed to elucidate this proposed mechanism.
Point mutations of conserved cysteine residues in CXC domain of TSO1, which is homolog of LIN54 in Arabidopsis thaliana, result in abnormal flower development and defects in cell division in floral meristem cells including large nuclei that are aberrant in shape and have high DNA content.27-29 On the other hand, tso1–3 mutant that contains a nonsense mutation (R453X) and lacks one of the two CXC domains and C-terminal region corresponding to HCH domain has little effect on flower development. The tso1–3 mutant plants exhibit normal flower formation and defects only in ovules.34 This is consistent with our data that LIN54C525Y and LIN54C611Y have strong effects on cell cycle progression, but LIN54R574X does not. A weak phenotype of tso1–3 mutant is presumably due to loss of TSO1 interaction with other proteins through HCH domain.
In this study, we found that LIN54 harboring a point mutation of the conserved cysteine residues in CXC domain most likely act as dominant-negative mutants and inhibit cell cycle progression. Thus, the presence of a heterozygous point-mutated allele may result in decreased transcriptional activation of several LINC target genes and defects in mitosis. Lin9 heterozygous mice are more susceptible to lung tumorigenesis induced by the c-Raf oncogene than wild-type mice.18 It is suspected that decreased Lin9 expression reduces LINC formation and induces genomic instability by causing aberrant mitosis. Point mutations of conserved cysteine residues in CXC domain of LIN54 may also contribute to oncogene-induced tumorigenesis.
Materials and Methods
Cell culture
GC-1 cells, a mouse-derived spermatogonia line, were purchased from ATCC (ATCC number: CRL-2053) and cultivated in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum (Gibco) at 37°C and 5% CO2.
Antibodies
The LIN54 polyclonal antibodies which were raised in rabbits against the peptide (DSNTTVKPAFPSGLQK) were purified by Melon Gel Monoclonal IgG Purification Kit (Pierce). Monoclonal antibodies used were anti-FLAG M2 (Sigma), anti-FLAG M2-Peroxidase (Sigma), anti-HA (Nacalai Tesque) and anti-β-tubulin (Sigma). Secondary antibodies for immunoblotting were anti-rabbit IgG, HRP-linked whole Ab donkey (Amersham) and goat anti-mouse IgG-HRP: sc-2302 (Santa Cruz Biotechnology). Secondary antibodies for immunofluorescence were Alexa 488-conjugated rabbit anti-mouse IgG (Molecular Probes) and Alexa 555-conjugated goat anti-mouse IgG (Molecular Probes).
RNA isolation, reverse transcription PCR, quantitative real-time PCR
Total RNA was extracted with QIAGEN RNeasy Mini Kit (QIAGEN). Seven-weeks-old male ICR mouse tissues were used to investigate the expression pattern of Lin54. Extracted total RNA was treated with TURBO DNA-free Kit (Ambion). RT-PCR was performed with the QIAGEN One-Step RT-PCR Kit (QIAGEN). PCR without reverse transcription reaction was performed to confirm no detectable DNA contamination. RT-PCR product was analyzed by 3% Agarose-LM Sieve gel (Nacalai Tesque) electrophoresis. One microgram of RNA was used for reverse transcription reaction with Transcriptor high-fidelity cDNA synthesis kit (Roche). Quantitative real-time PCR was performed with SYBR Premix Ex Taq (TaKaRa) and Thermal Cycler Dice Real Time System TP800 (TaKaRa). Primer sequences used are described in Table 1.
Table 1. Primers used in this study.
| Primer name |
Sequence |
|---|---|
| Site-directed mutagenesis | |
| Lin54-NLS1-F |
ACAGTACAGATTGCTACACCAACCTCGGGT |
| Lin54-NLS1-R |
ACCCGAGGTTGGTGTAGCAATCTGTACTGT |
| Lin54-NLS2-F |
TCAGAGTCTACCAGTCCCTGTAACTGTACA |
| Lin54-NLS2-R |
TGTACAGTTACAGGGACTGGTAGACTCTGA |
| Lin54-C525Y-F |
CCAGAAAACCCTATAACTGTACAAA |
| Lin54-C525Y-R |
TTTGTACAGTTATAGGGTTTTCTGG |
| Lin54-C611Y-F |
TTAAAAACTACTACGAGTGCTATGA |
| Lin54-C611Y-R |
TCATAGCACTCGTAGTAGTTTTTAA |
| Lin54-R574X-F |
GCATGCCTTGATTGAAATCCGGAAG |
| Lin54-R574X-R |
CTTCCGGATTTCAATCAAGGCATGC |
| Lin54-CXC1-F |
CGAACGTCTCACCTACAACAATTTAGAGCACGAG |
| Lin54-CXC1-R |
ATTTCGTCTCATAGGGTTTTCTGGGCCGACTGG |
| Lin54-CXC2-F |
CGAACGTCTCCGCAAGAATTTTGAAGAAAGCCC |
| Lin54-CXC2-R |
TTTTCGTCTCCTTGCCTTTGCTGTGCCGCCGATC |
| Lin54-CHC-F |
CGAACGTCTCCCCAAGAATTTTGAAGAAAGCCC |
| Lin54-CHC-R | ATTTCGTCTCATTGGGTTTTCTGGGCCGACTG |
| RT-PCR | |
|---|---|
| Lin54 RT-F |
ATACTACAACTCAGCCCTCT |
| Lin54 RT-R |
TCTGTCCTGTAACTGCTTTG |
| Cyclin A2 RT-F |
GTCTGTGTTAAGAGGGAAGC |
| Cyclin A2 RT-R |
ACTAGGTGCTCCATTCTCAG |
| Cyclin B1 RT-F |
GCCTGAGCCTGAACCTGAAC |
| Cyclin B1 RT-R |
GCAGGCGCACATCCAGATG |
| Cdc2 RT-F |
AGGACTCCAGGCTGTATCTC |
| Cdc2 RT-R |
ATTCCCTGGAGGATTTGGTG |
| Gapdh RT-F |
GAGTATGTCGTGGAGTCTA |
| Gapdh RT-R | TCACACCCATCACAAACA |
Constructions of expression plasmid for LIN54-EGFP, FLAG-LIN54 and LIN54-HA
A mouse Lin54 cDNA (ID: AK134320) was obtained from DNAFORM corporation. The entire open reading frame without stop codon was amplified by PCR using KOD -plus- DNA polymerase (TOYOBO) and a pair of primers; 5′-ATCCTCGAGCTCTAGAAGCGGTGCGATCATG-3′ and 5′-CGAGGATCCCGGCAGTGCATGGCACAGGGGTC-3′. The PCR products were subcloned into pEGFP-N1 (Clontech) in frame with enhanced green fluorescent protein (EGFP). For FLAG epitope-tagged LIN54, Lin54 cDNA was digested with XbaI and subcloned into pFLAG-CMV2 (Sigma) in frame with FLAG peptide. Lin54 fragment in frame with the hemagglutinin (HA) epitope was cloned into pcDNA3.1/Hyg expression vector (Invitrogen).
Site-directed mutagenesis
NLS deletion and point mutation constructs were generated by PCR-directed mutagenesis using the primers listed in Table 1. To construct CXC deletion mutants, Lin54 cDNA without CXC domain coding region was amplified by PCR using the primers listed in Table 1. The PCR products were digested with BsmBI and self-ligated. All constructs were verified by DNA sequencing.
Constructions of recombinant adenoviruses
Adenoviruses were generated by using Adenovirus Expression Vector Kit (TaKaRa). All cDNAs were cloned into pAxCAwtit under the CAG promoter. The recombinant cosmids were transfected into HEK 293 cells using PolyFect Transfection Reagent (QIAGEN). Adenovirus purifications and propagations were performed according to the manuals.
Cell proliferation assay
Cells were infected with recombinant adenoviruses at a multiplicity of infection (MOI) of 50 and seeded at 1 x 105 cells per dish in triplicate. For recovery assay, infection of recombinant adenovirus expressing EGFP or wild-type HA tagged LIN54 (50 MOI) was followed by infection of recombinant adenovirus expressing FLAG tagged LIN54C525Y or LIN54C611Y (50 MOI). The infected cells were seeded at 1 × 105 cells per dish in triplicate. The cells were counted at the indicated time points.
Flow cytometry
Cells were infected with recombinant adenoviruses at an MOI of 50 and seeded at 2 x 106 cells per dish. Coinfection of adenoviruses was performed according to the methods described above. The cells were fixed at different time points in 70% ethanol at -20°C, washed with PBS and resuspended in PBS containing 150 μg/ml RNase A. DNA was stained with 50 μg/ml propidium iodide (PI) for 20 min at room temperature. DNA content was then analyzed by FACS analysis on a Becton Dickinson FACSCanto.
Financial Support
Financial support for this study was provided by Tokushima Bunri University.
Glossary
Abbreviations:
- LINC
LIN complex
- DREAM
Drosophila RBF, dE2F2, and dMyb-interacting proteins
- CHC
CXC-hinge-CXC
- HCH
helix-coil-helix
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Footnotes
Previously published online: www.landesbioscience.com/journals/cc/article/21569
References
- 1.Cobrinik D. Pocket proteins and cell cycle control. Oncogene. 2005;24:2796–809. doi: 10.1038/sj.onc.1208619. [DOI] [PubMed] [Google Scholar]
- 2.DeGregori J, Johnson DG. Distinct and overlapping roles for E2F family members in transcription, proliferation and apoptosis. Curr Mol Med. 2006;6:739–48. doi: 10.2174/1566524010606070739. [DOI] [PubMed] [Google Scholar]
- 3.Krek W, Livingston DM, Shirodkar S. Binding to DNA and the retinoblastoma gene product promoted by complex formation of different E2F family members. Science. 1993;262:1557–60. doi: 10.1126/science.8248803. [DOI] [PubMed] [Google Scholar]
- 4.Sardet C, Vidal M, Cobrinik D, Geng Y, Onufryk C, Chen A, et al. E2F-4 and E2F-5, two members of the E2F family, are expressed in the early phases of the cell cycle. Proc Natl Acad Sci USA. 1995;92:2403–7. doi: 10.1073/pnas.92.6.2403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Beijersbergen RL, Kerkhoven RM, Zhu L, Carlée L, Voorhoeve PM, Bernards R. E2F-4, a new member of the E2F gene family, has oncogenic activity and associates with p107 in vivo. Genes Dev. 1994;8:2680–90. doi: 10.1101/gad.8.22.2680. [DOI] [PubMed] [Google Scholar]
- 6.Ginsberg D, Vairo G, Chittenden T, Xiao ZX, Xu G, Wydner KL, et al. E2F-4, a new member of the E2F transcription factor family, interacts with p107. Genes Dev. 1994;8:2665–79. doi: 10.1101/gad.8.22.2665. [DOI] [PubMed] [Google Scholar]
- 7.Hijmans EM, Voorhoeve PM, Beijersbergen RL, van ’t Veer LJ, Bernards R. E2F-5, a new E2F family member that interacts with p130 in vivo. Mol Cell Biol. 1995;15:3082–9. doi: 10.1128/mcb.15.6.3082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schmit F, Korenjak M, Mannefeld M, Schmitt K, Franke C, von Eyss B, et al. LINC, a human complex that is related to pRB-containing complexes in invertebrates regulates the expression of G2/M genes. Cell Cycle. 2007;6:1903–13. doi: 10.4161/cc.6.15.4512. [DOI] [PubMed] [Google Scholar]
- 9.Litovchick L, Sadasivam S, Florens L, Zhu X, Swanson SK, Velmurugan S, et al. Evolutionarily conserved multisubunit RBL2/p130 and E2F4 protein complex represses human cell cycle-dependent genes in quiescence. Mol Cell. 2007;26:539–51. doi: 10.1016/j.molcel.2007.04.015. [DOI] [PubMed] [Google Scholar]
- 10.Pilkinton M, Sandoval R, Colamonici OR. Mammalian Mip/LIN-9 interacts with either the p107, p130/E2F4 repressor complex or B-Myb in a cell cycle-phase-dependent context distinct from the Drosophila dREAM complex. Oncogene. 2007;26:7535–43. doi: 10.1038/sj.onc.1210562. [DOI] [PubMed] [Google Scholar]
- 11.Korenjak M, Taylor-Harding B, Binné UK, Satterlee JS, Stevaux O, Aasland R, et al. Native E2F/RBF complexes contain Myb-interacting proteins and repress transcription of developmentally controlled E2F target genes. Cell. 2004;119:181–93. doi: 10.1016/j.cell.2004.09.034. [DOI] [PubMed] [Google Scholar]
- 12.Lewis PW, Beall EL, Fleischer TC, Georlette D, Link AJ, Botchan MR. Identification of a Drosophila Myb-E2F2/RBF transcriptional repressor complex. Genes Dev. 2004;18:2929–40. doi: 10.1101/gad.1255204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Harrison MM, Ceol CJ, Lu X, Horvitz HR. Some C. elegans class B synthetic multivulva proteins encode a conserved LIN-35 Rb-containing complex distinct from a NuRD-like complex. Proc Natl Acad Sci USA. 2006;103:16782–7. doi: 10.1073/pnas.0608461103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Osterloh L, von Eyss B, Schmit F, Rein L, Hübner D, Samans B, et al. The human synMuv-like protein LIN-9 is required for transcription of G2/M genes and for entry into mitosis. EMBO J. 2007;26:144–57. doi: 10.1038/sj.emboj.7601478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pilkinton M, Sandoval R, Song J, Ness SA, Colamonici OR. Mip/LIN-9 regulates the expression of B-Myb and the induction of cyclin A, cyclin B, and CDK1. J Biol Chem. 2007;282:168–75. doi: 10.1074/jbc.M609924200. [DOI] [PubMed] [Google Scholar]
- 16.Knight AS, Notaridou M, Watson RJAA. Lin-9 complex is recruited by B-Myb to activate transcription of G2/M genes in undifferentiated embryonal carcinoma cells. Oncogene. 2009;28:1737–47. doi: 10.1038/onc.2009.22. [DOI] [PubMed] [Google Scholar]
- 17.Schmit F, Cremer S, Gaubatz S. LIN54 is an essential core subunit of the DREAM/LINC complex that binds to the cdc2 promoter in a sequence-specific manner. FEBS J. 2009;276:5703–16. doi: 10.1111/j.1742-4658.2009.07261.x. [DOI] [PubMed] [Google Scholar]
- 18.Reichert N, Wurster S, Ulrich T, Schmitt K, Hauser S, Probst L, et al. Lin9, a subunit of the mammalian DREAM complex, is essential for embryonic development, for survival of adult mice, and for tumor suppression. Mol Cell Biol. 2010;30:2896–908. doi: 10.1128/MCB.00028-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kittler R, Pelletier L, Heninger AK, Slabicki M, Theis M, Miroslaw L, et al. Genome-scale RNAi profiling of cell division in human tissue culture cells. Nat Cell Biol. 2007;9:1401–12. doi: 10.1038/ncb1659. [DOI] [PubMed] [Google Scholar]
- 20.Sugihara T, Wadhwa R, Kaul SC, Mitsui Y. A novel testis-specific metallothionein-like protein, tesmin, is an early marker of male germ cell differentiation. Genomics. 1999;57:130–6. doi: 10.1006/geno.1999.5756. [DOI] [PubMed] [Google Scholar]
- 21.Matsuura T, Kawasaki Y, Miwa K, Sutou S, Ohinata Y, Yoshida F, et al. Germ cell-specific nucleocytoplasmic shuttling protein, tesmin, responsive to heavy metal stress in mouse testes. J Inorg Biochem. 2002;88:183–91. doi: 10.1016/S0162-0134(01)00377-4. [DOI] [PubMed] [Google Scholar]
- 22.Sutou S, Miwa K, Matsuura T, Kawasaki Y, Ohinata Y, Mitsui Y. Native tesmin is a 60-kilodalton protein that undergoes dynamic changes in its localization during spermatogenesis in mice. Biol Reprod. 2003;68:1861–9. doi: 10.1095/biolreprod.102.005603. [DOI] [PubMed] [Google Scholar]
- 23.Lindeman GJ, Gaubatz S, Livingston DM, Ginsberg D. The subcellular localization of E2F-4 is cell-cycle dependent. Proc Natl Acad Sci USA. 1997;94:5095–100. doi: 10.1073/pnas.94.10.5095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chestukhin A, Litovchick L, Rudich K, DeCaprio JA. Nucleocytoplasmic shuttling of p130/RBL2: novel regulatory mechanism. Mol Cell Biol. 2002;22:453–68. doi: 10.1128/MCB.22.2.453-468.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gaubatz S, Lees JA, Lindeman GJ, Livingston DM. E2F4 is exported from the nucleus in a CRM1-dependent manner. Mol Cell Biol. 2001;21:1384–92. doi: 10.1128/MCB.21.4.1384-1392.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fornerod M, Ohno M, Yoshida M, Mattaj IW. CRM1 is an export receptor for leucine-rich nuclear export signals. Cell. 1997;90:1051–60. doi: 10.1016/S0092-8674(00)80371-2. [DOI] [PubMed] [Google Scholar]
- 27.Liu Z, Running MP, Meyerowitz EM. TSO1 functions in cell division during Arabidopsis flower development. Development. 1997;124:665–72. doi: 10.1242/dev.124.3.665. [DOI] [PubMed] [Google Scholar]
- 28.Song JY, Leung T, Ehler LK, Wang C, Liu Z. Regulation of meristem organization and cell division by TSO1, an Arabidopsis gene with cysteine-rich repeats. Development. 2000;127:2207–17. doi: 10.1242/dev.127.10.2207. [DOI] [PubMed] [Google Scholar]
- 29.Hauser BA, He JQ, Park SO, Gasser CS. TSO1 is a novel protein that modulates cytokinesis and cell expansion in Arabidopsis. Development. 2000;127:2219–26. doi: 10.1242/dev.127.10.2219. [DOI] [PubMed] [Google Scholar]
- 30.Kuwahara J, Watanabe Y, Kayasuga T, Itoh K. Zn finger and nuclear localization of transcription factor Sp1. Nucleic Acids Symp Ser. 2000;44:265–6. doi: 10.1093/nass/44.1.265. [DOI] [PubMed] [Google Scholar]
- 31.Ito T, Azumano M, Uwatoko C, Itoh K, Kuwahara J. Role of zinc finger structure in nuclear localization of transcription factor Sp1. Biochem Biophys Res Commun. 2009;380:28–32. doi: 10.1016/j.bbrc.2008.12.165. [DOI] [PubMed] [Google Scholar]
- 32.Quadrini KJ, Bieker JJ. Krüppel-like zinc fingers bind to nuclear import proteins and are required for efficient nuclear localization of erythroid Krüppel-like factor. J Biol Chem. 2002;277:32243–52. doi: 10.1074/jbc.M205677200. [DOI] [PubMed] [Google Scholar]
- 33.Güttler T, Madl T, Neumann P, Deichsel D, Corsini L, Monecke T, et al. NES consensus redefined by structures of PKI-type and Rev-type nuclear export signals bound to CRM1. Nat Struct Mol Biol. 2010;17:1367–76. doi: 10.1038/nsmb.1931. [DOI] [PubMed] [Google Scholar]
- 34.Hauser BA, Villanueva JM, Gasser CS. Arabidopsis TSO1 regulates directional processes in cells during floral organogenesis. Genetics. 1998;150:411–23. doi: 10.1093/genetics/150.1.411. [DOI] [PMC free article] [PubMed] [Google Scholar]