Abstract
Uninterrupted replication across damaged DNA is critical to prevent replication fork collapse and resulting double-strand DNA breaks. Rad18-mediated PCNA ubiquitination is a crucial event that triggers a number of downstream pathways important for lesion bypass. Here, we report characterization of Spartan, an evolutionarily conserved protein containing a PCNA-interacting peptide motif, called a PIP box, and a UBZ4 ubiquitin-binding domain. Spartan is a nuclear protein and forms DNA damage-induced foci that colocalize with markers for stalled DNA replication. Focus formation of Spartan requires its PIP-box and the UBZ4 domain and is dependent on Rad18 and the PCNA ubiquitination site, indicating that Spartan is recruited to ubiquitinated PCNA. Spartan depletion results in increased mutagenesis during replication of UV-damaged DNA. Taken together, our data suggest that Spartan is recruited to sites of stalled replication via ubiquitinated PCNA and plays an important role to prevent mutations associated with replication of damaged DNA.
Keywords: C1orf124, DNA damage, PCNA, PIP box, Spartan, UBZ, mutagenesis, ubiquitin
Genomic DNA is under constant attack by environmental factors and endogenous metabolic processes. The resulting DNA lesions are normally eliminated by various DNA repair mechanisms; however, if not repaired before S phase, they could stall replicative DNA polymerases, leading to fork collapse, double-strand DNA breaks and cell death. To avoid such lethal events, cells have evolved a mechanism called DNA damage tolerance, which allows replication across the polymerase-blocking lesions without their actual repair.1
The ubiquitin ligase Rad18 is the master regulator of the DNA damage tolerance pathway. When DNA polymerases encounter DNA lesions, Rad18 induces PCNA monoubiquitination, a crucial step in DNA damage tolerance.2-4 Two subpathways are potentially activated via monoubiquitination of PCNA. One is translesion synthesis (TLS), in which specialized TLS polymerases directly replicate past lesions in a damaged DNA template. Monoubiquitinated PCNA recruits TLS polymerases via interactions with their PCNA-interacting motif, called a PIP box, and ubiquitin-binding domains.4-8 The TLS pathway is potentially error-prone, because it employs low-fidelity Y-family polymerases.9 In contrast, the second pathway, template switching (TS), is an error-free process. The TS pathway uses the newly synthesized, undamaged strand of the sister chromatid as template to bypass lesions.1 Although the underlying mechanism is still unclear, the TS pathway is activated through extension of K63-linked ubiquitin chains from monoubiquitinated PCNA by Rad5 (HLTF and SHPRH in human).10-12
Ubiquitin-binding domains play critical roles in DNA damage response.13,14 In particular, ubiquitin-binding zinc-finger 4 (UBZ4) is commonly found in proteins that localize to sites of DNA damage.15-26 To find potential regulators of DNA damage response, we performed a bioinformatics search for uncharacterized proteins containing UBZ4 domains. We noted that a human protein C1orf124 (NP_114407), which was recently named Spartan,27 contains a PIP box and UBZ4, similarly to TLS polymerases. In this paper, we report that Spartan is recruited to ubiquitinated PCNA via the PIP box and UBZ4 domain. At sites of stalled replication, Spartan plays an important role in suppressing mutagenesis associated with replication of damaged DNA.
Results
Spartan is an evolutionarily conserved protein
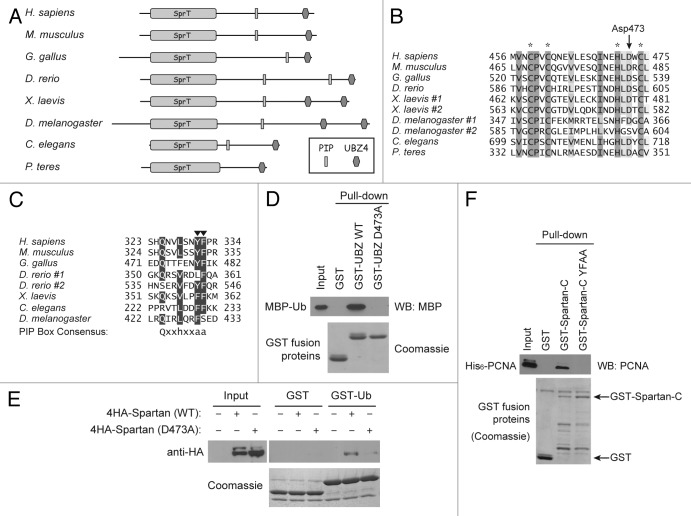
A homology search using the human Spartan protein sequence revealed putative Spartan orthologs in higher eukaryotes, including mouse (NP_001104611), chicken (XP_419571), fish (XP_696486), frog (AAI60677), fly (NP_573032) and worm (NP_505853) (Fig. 1A). Although Spartan is not widely conserved in fungus, its orthologs are found in several plant pathogens of fungal origin such as Pyrenophora teres. Spartan contains a conserved N-terminal domain that shares homology with an uncharacterized protein SprT from E. coli.28 The UBZ4 domain of Spartan is also highly conserved in eukaryotes and is duplicated in some species (Fig. 1A and B). We noted that human Spartan contains a potential PIP box that matches with the consensus sequence Qxxhxxaa (h: hydrophobic residue; a: aromatic residue; x: any amino acids)29 (Fig. 1A and C). Although some of the residues deviate from the consensus sequence, the PIP box appears to be conserved in Spartan orthologs in other species (Fig. 1C). This high degree of conservation suggests that Spartan plays a fundamental role in eukaryotes. Importantly, the Spartan orthologs in other organisms are mostly uncharacterized except for the C. elegans homolog T19B10.6, whose knockdown by RNA interference (RNAi) caused sterility.30,31
Figure 1. Spartan interacts with ubiquitin and PCNA. (A) Domain structure of Spartan and conservation across species. Putative homologs containing both SprT and UBZ4 domains are shown. (B) Multiple alignment of the UBZ4 domains from Spartan homologs in selected species. Zinc-coordinating residues are highlighted with asterisks. The conserved aspartate residue mutated in this study is indicated by an arrow. (C) Multiple alignment of the PIP boxes from Spartan homologs in selected species. Conserved aromatic residues that were mutated to alanines in this study are indicated by arrowheads. (D) Interaction of the Spartan UBZ4 domain with ubiquitin. In vitro GST pull-down assays were performed using indicated recombinant proteins produced in E. coli. MBP-Ub was examined by western blotting against MBP, and GST-fusion proteins in the precipitates were visualized by Coomassie staining. (E) Interaction of full-length Spartan with ubiquitin. In vitro GST pull-down assays were performed as in (D), except that GST fusion proteins were mixed with 293T cell lysates expressing indicated Spartan proteins. Spartan proteins were examined by western blotting against HA tag, and GST-fusion proteins in the precipitates were visualized by Coomassie staining. (F) Interaction of the Spartan C-terminal fragment with PCNA. In vitro GST pull-down assays were performed using the indicated recombinant proteins produced in E. coli. PCNA was examined by western blotting, and GST-fusion proteins in the precipitates were visualized by Coomassie staining.
Spartan interacts with ubiquitin and PCNA
Using in vitro GST pull-down assays, we first confirmed that the Spartan UBZ4 domain interacts with ubiquitin (Fig. 1D). As has been shown for other UBZ4 domains,20 alanine substitution of the highly conserved Asp473 (D473A) (Fig. 1B) abolished the ubiquitin interaction. Furthermore, full-length Spartan interacted with ubiquitin in GST pull-down assays, and again the D473A mutation disrupted the interaction (Fig. 1E). These results indicate that Spartan interacts with ubiquitin through its UBZ4 domain.
The conserved PIP box in Spartan is also functional. GST pull-down assays using recombinant proteins revealed that the C-terminal portion of Spartan (Spartan-C) containing the PIP box interacted with PCNA in vitro (Fig. 1F). Importantly, this interaction was abolished when two aromatic residues in the PIP box were mutated to alanines (YFAA). These results suggest that Spartan interacts with PCNA via the PIP box.
Spartan localizes to sites of stalled replication
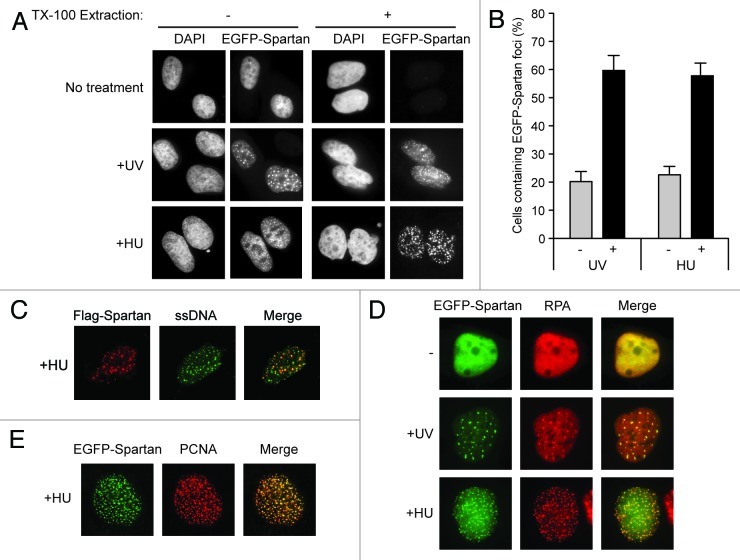
Considering that previously reported UBZ4-containing proteins localize to damaged DNA,20-22 we tested whether Spartan changes its localization in response to DNA damage. We expressed EGFP-tagged Spartan and monitored its localization after treatment with UV. In an unperturbed condition, EGFP-Spartan was evenly distributed in nuclei (Fig. 2A). In contrast, after treatment with UV, EGFP-Spartan localized to nuclear speckles (Fig. 2A and B). Because cell cycle profiles are largely unchanged 3 h after UV exposure (Fig. S1A), it is unlikely that the focus formation of Spartan is due to enrichment of UV-exposed cells in a particular phase of the cell cycle. These EGFP-Spartan foci were resistant to TX-100 extraction, a procedure that eliminates cytoplasmic and nucleoplasmic proteins (Fig. 2A), suggesting that EGFP-Spartan was loaded onto chromatin in response to DNA damage. Because Spartan formed foci after treatment with hydroxyurea (HU), which stalls replication forks via dNTP depletion (Fig. 2A and B), it appears that Spartan foci are primarily formed in response to stalled DNA replication. Consistent with this notion, HU-induced foci of EGFP-Spartan colocalized with single-strand DNA (ssDNA) (Fig. 2C). Moreover, UV- or HU-induced EGFP-Spartan foci colocalized with RPA foci, which mark sites of stalled replication (Fig. 2D). In agreement with its ability to interact with PCNA, Spartan foci colocalized with that of PCNA in response to HU treatment (Fig. 2E). Collectively, these data indicate that Spartan localizes to the sites of stalled replication.
Figure 2. Spartan localizes to sites of stalled replication. (A) Focus formation of EGFP-Spartan in response to DNA damage or replication stress. U2OS cells stably expressing EGFP-Spartan were treated as indicated. Cells were fixed 3 h after UV (20 J/m2) or 6 h after HU (10 mM) treatment. Where indicated, TX-100 extraction was performed to visualize EGFP-Spartan on chromain. (B) Quantitation of cells containing EGFP-Spartan Foci. Cells were treated as in (A), and percentage of cells containing more than ten EGFP-Spartan foci is presented as mean ± SD (n = 3). (C) Colocalization of EGFP-Spartan with ssDNA. Flag-Spartan was transiently expressed in U2OS cells prelabeled with BrdU for two doubling times. Cells were treated with 10 mM HU for 6 h and coimmunostained with anti-Flag and anti-BrdU in non-denaturing conditions. (D) Colocalization of EGFP-Spartan with RPA. U2OS cells stably expressing EGFP-Spartan were fixed 3 h after UV (20 J/m2) or 6 h after HU (10 mM) treatment and immunostained for RPA. (E) Colocalization of EGFP-Spartan with PCNA. U2OS cells stably expressing EGFP-Spartan were treated with 10 mM HU for 6 h and immunostained for PCNA.
Spartan focus formation is dependent on Rad18 and PCNA Lys164
The similarity in domain structure between Spartan and TLS polymerases prompted us to test whether Spartan focus formation is dependent on Rad18. EGFP-Spartan foci induced by UV or HU treatment were drastically diminished in Rad18−/− cells (Fig. 3A and B), indicating that Rad18 participates in recruitment of Spartan to sites of stalled replication.
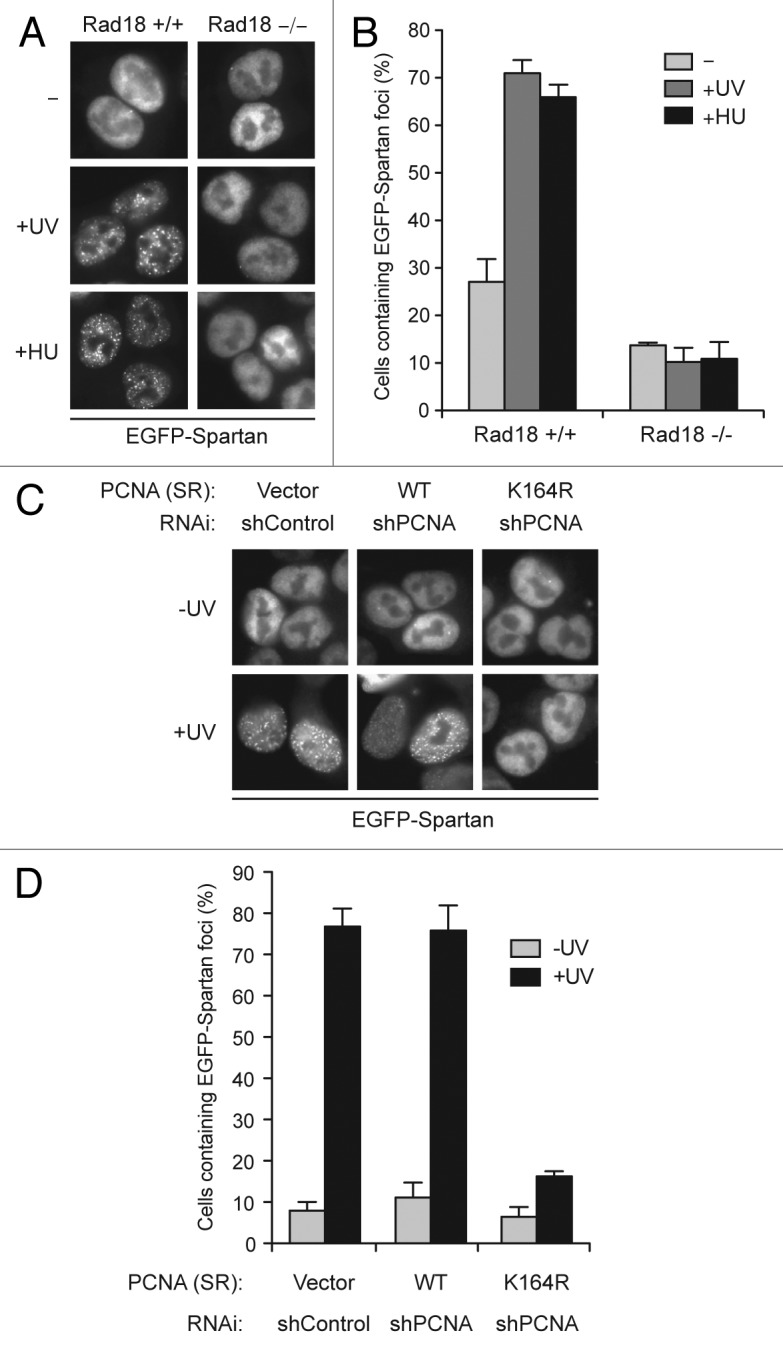
Figure 3. Focus formation of EGFP-Spartan is dependent on Rad18 and PCNA ubiquitination. (A) Focus formation of EGFP-Spartan in response to DNA damage. EGFP-Spartan was stably expressed in HCT116 cells (Rad18 +/+ and −/−) and visualized 3 h after UV (20 J/m2) or 6 h after HU (10 mM) treatment. (B) Quantitation of cells containing EGFP-Spartan foci. Cells were treated as in (A), and percentage of cells containing more than ten EGFP-Spartan foci is presented as mean ± SD (n = 3). (C) Focus formation of EGFP-Spartan in response to UV. EGFP-Spartan was stably expressed in HCT116 cells expressing wild-type or the K164R mutant PCNA and visualized 3 h after UV (20 J/m2) treatment. See also Figure S2. (D) Quantitation of cells containing EGFP-Spartan Foci. Cells were treated as in (C), and percentage of cells containing more than ten EGFP-Spartan foci is presented as mean ± SD (n = 3).
Rad18 monoubiquitinates PCNA at Lys164 to recruit downstream effectors in response to stalled replication. Therefore, we next explored the possibility that ubiquitinated PCNA is a signal for Spartan recruitment. To address this, we established cells that express the PCNA K164R mutant in which the ubiquitination site Lys164 was mutated to arginine. In these experiments, endogenous PCNA was depleted by RNAi and replaced with exogenously expressed wild-type or K164R PCNA (Fig. S2A). As expected, DNA damage-induced PCNA ubiquitination was abolished in the PCNA K164R cells, while it was intact in PCNA wild-type cells (Fig. S2B). By using these cells, we examined focus formation of EGFP-Spartan following UV-induced DNA damage. As shown in Figure 3C and D, cells expressing the PCNA K164R mutant formed far fewer EGFP-Spartan foci after UV exposure, providing evidence that PCNA ubiquitination participates in Spartan recruitment.
Spartan focus formation requires both the PIP box and the UBZ4 domain
Monoubiquitinated PCNA recruits TLS polymerases via interactions with two defined elements, PIP boxes and ubiquitin-binding domains, in TLS polymerases.1 Both interactions are necessary for TLS polymerases to form DNA damage-induced foci. Accordingly, we tested whether the PIP box and UBZ4 of Spartan are required for Spartan focus formation. As shown in Figure 4A and B, mutations in either the PIP box (YFAA) or the UBZ4 domain (D473A) greatly diminished focus formation of EGFP-Spartan following UV or HU treatment.
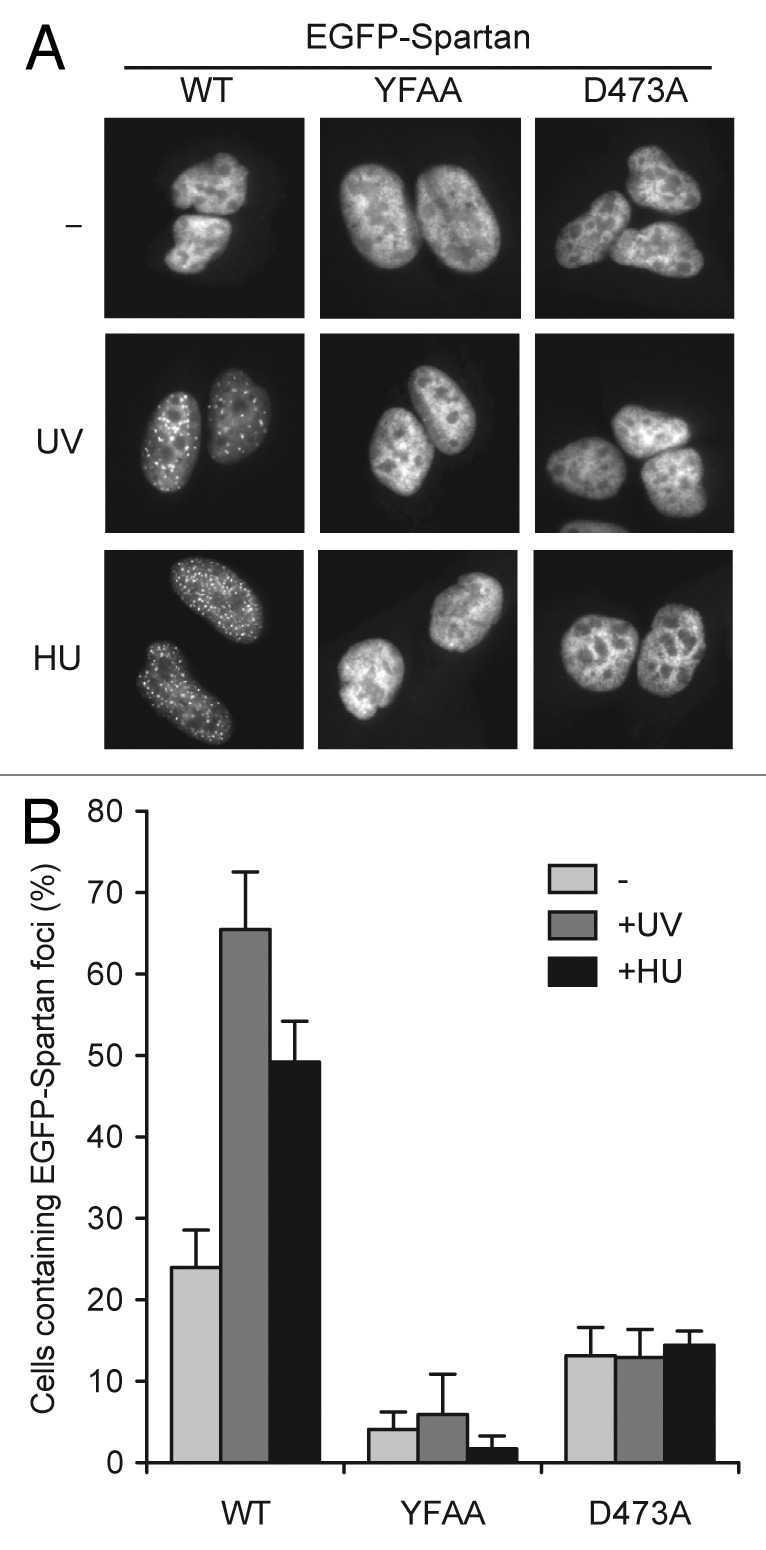
Figure 4. Focus formation of EGFP-Spartan is dependent on its PIP box and UBZ4 domain. (A) Focus formation of EGFP-Spartan in response to DNA damage. Wild-type or indicated mutants of Spartan were stably expressed as a fusion protein with EGFP in U2OS. Localization of EGFP-Spartan was visualized 3 h after UV (20 J/m2) or 6 h after HU (10 mM) treatment. (B) Quantitation of cells containing EGFP-Spartan Foci. Cells were treated as in (A), and percentage of cells containing more than ten EGFP-Spartan foci is presented as mean ± SD (n = 3).
Collectively, these experiments demonstrated that Spartan focus formation after DNA damage is not only regulated by Rad18 and PCNA ubiquitination, but requires the PIP box and UBZ4 domain of Spartan. These results confirm and extend the recent suggestion that Spartan is recruited to sites of DNA damage through its interaction with ubiquitinated PCNA.27
Spartan depletion increases damage-induced mutagenesis
Ubiquitinated PCNA regulates lesion bypass through TLS and TS pathways. TLS is potentially error-prone and is, therefore, responsible for some of damage-induced mutagenesis. To examine the impact of Spartan depletion on DNA damage-induced mutagenesis, we measured the frequency of UV-induced mutations during DNA replication using a shuttle vector system that scores mutations in a SupF tRNA gene.32 Knockdown of Spartan by RNAi was confirmed by western blotting (Fig. 5A). Endogenous Spartan proteins were detected as a doublet, of which the top band is likely a monoubiquitinated form (data not shown and ref. 27). As shown in Figure 5B, UV-induced mutagenesis was increased in Spartan-depleted cells. Sequencing of the mutated SupF genes confirmed that the mutation spectrum in the Spartan-depleted cells was consistent with UV-induced mutagenesis, in which the major mutation types are C:G to T:A transitions (Table 1). These results indicate that Spartan plays an important role in suppression of DNA damage-induced mutagenesis during replication.
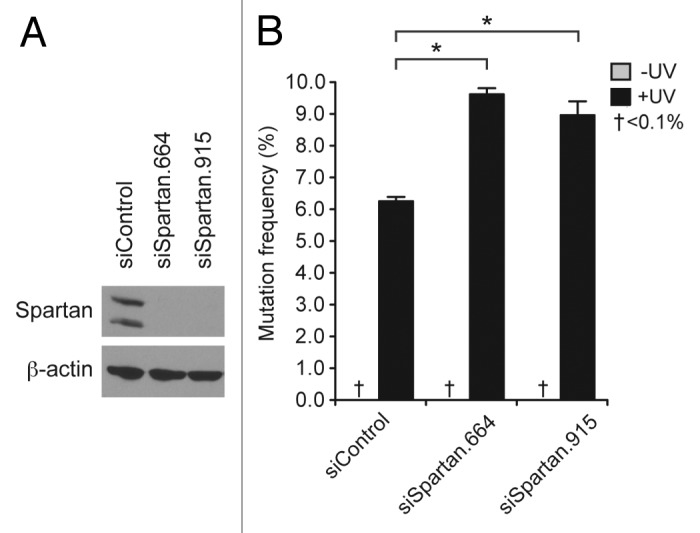
Figure 5. Spartan depletion increases UV-induced mutagenesis. (A) Knock down of Spartan by RNAi. 293T cells were transfected with the indicated siRNA oligos and Spartan levels were examined by western blotting. β-actin is shown as a loading control. (B) Increased mutagenesis in Spartan-depleted cells. UV-induced mutagenesis was measured using the SupF shuttle vector system in 293T. Cells were transfected with indicated siRNA oligos and then transfected with SupF plasmids after 24 h. Forty-eight hours later, plasmids were recovered and assayed for mutations in the SupF gene. Mutation frequencies are presented as percentage of mutant SupF genes. Experiments were performed in triplicate and results are shown as mean ± SD. Mutations were not detected in undamaged plasmids. (*, p < 0.0005, two-tailed Student’s t-test.)
Table 1. Mutation spectrum of SupF mutants (%).
| siControl (n = 57) | siSpartan.664 (n = 75) | |
|---|---|---|
|
Transisions |
|
|
| C:G to T:A |
77.2 |
76.0 |
| T:A to C:G |
3.5 |
4.0 |
|
Transversions |
|
|
| C:G to A:T |
8.8 |
9.3 |
| C:G to G:C |
1.8 |
1.4 |
| T:A to A:T |
8.8 |
4.0 |
| T:A to G:C | 0.0 | 5.3 |
Discussion
In this study, we demonstrate that Spartan is recruited to ubiquitinated PCNA via its PIP box and the UBZ4 domain (Figs. 1, 3 and 4) and that Spartan is important for prevention of damage-induced mutagenesis (Fig. 5). While this manuscript was in preparation, Zou’s group independently demonstrated that Spartan is recruited to sites of UV damage in a manner dependent on the PIP box, UBZ4 domain and Rad18.27 Our data confirm and extend this study. In particular, we demonstrated that Spartan focus formation is dependent not only on Rad18, but also lysine 164 of PCNA, the site for Rad18-mediated ubiquitination, demonstrating that PCNA ubiquitination is indeed crucial for Spartan recruitment. In addition, our study showed that Spartan forms foci in response to HU-induced stalled DNA replication (Fig. 2). We presented evidence that HU treatment induces colocalization of Spartan to ssDNA and RPA, the markers for sites of stalled DNA replication (Fig. 2C and D). Because Spartan colocalizes with RPA foci after UV treatment as well (Fig. 2D), it is likely that Spartan foci seen after UV exposure represent sites of stalled replication. Finally, our data demonstrated that Spartan-depleted cells exhibit increased mutation frequencies during replication of UV-induced DNA damage (Fig. 5), supporting the conclusion that Spartan plays a role at sites of stalled replication.
How does depletion of Spartan cause increased mutagenesis during replication of UV-damaged DNA? PCNA ubiquitination plays a central role in lesion bypass through the TLS or TS mechanisms. Because TLS is potentially error-prone, whereas TS is error-free, one possibility is that Spartan might regulate the choice between TLS and TS, favoring TS over TLS. However, given that TLS is the major mechanism for lesion bypass in mammalian systems,33 a more likely possibility is that Spartan depletion effects fidelity of TLS. Recent study from Zou’s group27 is in agreement with this possibility. The authors demonstrated that Spartan interacts with Rad18 and forms a feedforward loop to enhance PCNA ubiquitination. PCNA ubiquitination and focus formation of DNA polymerase η, a high fidelity TLS polymerase for UV-induced cyclobutane pyrimidine dimers, are diminished in Spartan-depleted cells.27 Thus, it is possible that reduced PCNA ubiquitination underlies the increased mutation frequency observed after Spartan depletion in our study. However, a recent report demonstrated that mutation frequencies during TLS across UV-induced DNA damage are unchanged or even reduced in cells lacking PCNA ubiquitination,34 suggesting that defects in PCNA ubiquitination by itself may not cause increased mutagenesis. Further studies of how Spartan depletion causes increased mutagenesis will be necessary to fully understand the role of Spartan in the DNA damage response.
Materials and Methods
Plasmids
Full-length Spartan cDNA was amplified by RT-PCR using LA Taq (Takara Bio) from MCF10A mRNAs, cloned into the transient mammalian expression vectors pEFF-N (for an N-terminal Flag tag) and pEF4H (for an N-terminal 4HA tag), and a lentiviral expression vector pLVX6-IRES-Neo (for an N-terminal EGFP tag). cDNAs encoding wild-type or the K164R mutant of PCNA were subcloned from pSPC (a gift from Larry Karnitz)35 and expressed from pLVX-IRES-puro. For production of recombinant proteins in E. coli, cDNAs encoding Spartan-C (amino acids 212–489) were cloned in pGexHis (an N-terminal GST-tag and a C-terminal His6 tag). A C-terminal fragment (amino acids 441–489) spanning the UBZ4 domain was expressed as a GST-fusion protein from pGEX-6P-2. cDNA for PCNA was cloned in pET28a (Novagen) for expression of His6/T7-tagged PCNA. Recombinant ubiquitin proteins were expressed from pGEX-6P-2 or pMAL-c2x (New England Biolabs). Site-directed mutagenesis was performed using pfu Turbo (Agilent Technologies), and all introduced mutations were confirmed by sequencing. shRNAs were expressed from lentiviral vector pSIH1-H1-Neo.
Cell culture and RNAi
Human osteosarcoma cell line U2OS and colon cancer cell line HCT116 were obtained from American Type Culture Collection (ATCC) and cultured in McCoy’s 5A medium supplemented with 10% FBS. HCT116 Rad18−/− has been reported previously.36 Human embryonic kidney cell line 293T (ATCC) was maintained in RPMI1640 containing 10% FBS. Recombinant lentiviruses were produced by cotransfection of viral and packaging plasmids in 293T cells. For virus infection, cells were incubated with supernatants containing viruses in the presence of 4 μg/ml polybrene (Sigma-Aldrich) and selected with 1 μg/ml puromycin (Sigma-Aldrich) or 400 μg/ml G418 (Sigma-Aldrich). For RNAi, cells were transfected with siRNA oligos using RNAiMAX (Invitrogen) for 48–72 h. For stable knockdown, cells were infected with lentiviruses expressing shRNAs. The target sequences of siRNAs and shRNAs are: siControl, CGUACGCGGAAUACUUCGA; siSpartan.664, GACCCUGUGUGCUGGGAUA; siSpartan.915, ACGAUGAGGUGGAUGAGUA; shControl, ACAAGAUGAAGAGCACCAA; shPCNA, GGAGGAAGCUGUUACCAUA.
Immunocytochemistry and microscopy
For immunostaining, cells cultured on coverslips were fixed with 4% paraformaldehyde for 10 min at room temperature (for anti-RPA32 staining) or with methanol/acetone mixture (3:1) for 10 min at -20°C (for anti-PCNA staining) and permeabilized in 0.2% TX-100 in PBS for 10 min. After blocking with PBS containing 3% BSA for 1 h, cells were incubated with primary antibodies anti-RPA32 (Santa Cruz Biotechnology, 1:200) or anti-PCNA (Santa Cruz Biotechnology, 1:200) at room temperature for 1 h. After washing, cells were incubated with Alexa fluor 594-conjugated goat anti-mouse IgG (Invitrogen) in a 1:2,000 dilution for 30 min. Cells were mounted with VectaShield containing 4',6-diamidino-2-phenylindole (Vector Laboratories) and viewed and photographed on a Carl Zeiss fluorescent microscope. Detection of ssDNA was performed as described elsewhere.37 Briefly, cells were prelabeled with 10 μM BrdU for two doubling times and immunostained as described above with rabbit anti-Flag (Sigma) and mouse anti-BrdU (Roche) antibodies in non-denaturing conditions. For visualization of EGFP-tagged proteins, cells were fixed with 4% paraformaldehyde and permeabilized. Where indicated, cells were extracted for 10 min with CSK buffer (10 mM Hepes-KOH pH7.4, 300 mM sucrose, 100 mM NaCl and 3 mM MgCl2) supplemented with 0.5% Triton X-100 before fixing. For quantitation of EGFP-Spartan focus formation, cells containing at least 10 foci were considered positive for focus formation, and at least 100 cells were counted in each sample.
Western blotting
Cells were lysed in NP-40 lysis buffer (50 mM TRIS-HCl pH7.4, 150 mM NaCl, 0.1% Nonidet P-40, 5 mM EDTA, 50 mM NaF, 1 mM Na3VO4, 10% Glycerol) supplemented with protease inhibitor mix (Sigma). For western blotting, 30 μg of proteins were separated by SDS-PAGE, transferred to nitrocellulose membrane and probed with antibodies. Mouse monoclonal anti-Spartan antibody (IgG2a, clone 11A) was produced at the Mayo Clinic Monoclonal Antibody Core using GST-Spartan-C as an antigen. Anti-PCNA and anti-HA antibodies were from Santa Cruz Biotechnology. Anti-β-actin and anti-MBP antibodies were purchased from Sigma-Aldrich and New England Biolabs, respectively.
Recombinant proteins and GST pull-down assays
All recombinant proteins were produced in E. coli BL21(DE3) and purified on glutathione beads (Sigma-Aldrich; for GST-tagged proteins), TALON metal affinity resins (Novagen; for His6-tagged proteins) or amylose resins (New England Biolabs; for MBP-tagged proteins). For in vitro GST pull-down assays, GST-fusion proteins (5 μg) were incubated with glutathione beads in GST binding buffer (20 mM TRIS-HCl (pH7.4), 150 mM NaCl, 1.0% Nonidet P-40, 25 μg/ml BSA 1 mM DTT) for 1 h at 4°C. Beads were then incubated for 2 h at 4°C with 1 μg of recombinant PCNA in GST binding buffer or cell lysates containing 1 mg proteins. Beads were washed five times with GST binding buffer and bound proteins were eluted by boiling in 2 × SDS-PAGE sample buffer. After SDS-PAGE, GST-proteins were visualized by Coomassie brilliant blue staining while coprecipitated proteins were detected by western blotting.
SupF mutation assays
Mutation frequencies were measured using the SupF shuttle vector system as described previously (Parris et al., 1994). 293T cells were transfected with siRNAs for 24 h and then with 2 μg pSP189 that were unirradiated or irradiated with 1,000 J/m2 UVC (XL-1000; Spectroline) in a 40 μl droplet. After 48 h, plasmids were recovered by a miniprep kit (Qiagen) and treated with DpnI for 4 h to eliminate unreplicated plasmids. Plasmid DNA was purified by phenol/chloroform extraction, concentrated by ethanol precipitation, and electroporated into E. coli MBM7070. Transformed E. coli cells were grown on LB agar plates containing 200 μM isopropyl β-D-1-thiogalactopyranoside (Sigma-Aldrich), 100 μg/ml Bluo-gal (Invitrogen), and 100 μg/ml ampicillin. Blue (wild-type SupF) and white (mutant SupF) colonies were counted and mutation frequencies were presented as a percentage of white colonies over total colonies. Experiments were performed in triplicate and typically 10,000 colonies were screened in each experiment. To verify mutations, SupF genes in white colonies were sequenced as described previously.32
Supplementary Material
Acknowledgments
We thank S. Kaufmann, L. Karnitz, D. Billadeau and Z. Lou for critical reading of the manuscript. This work was supported by American Cancer Society Research Scholar Grant (to Y.J.M.) and the Minnesota Partnership for Biotechnology and Medical Genomics (to Y.J.M.).
Glossary
Abbreviations:
- PIP
PCNA-interacting peptide
- UV
ultraviolet
- UBZ
ubiquitin-binding zinc-finger
- RNAi
RNA interference
- HU
hydroxyurea
- TLS
translesion synthesis
- TS
template switching
- ssDNA
single-strand DNA
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Supplemental Material
Supplemental material may be found here: www.landesbioscience.com/journals/cc/article/21694
Footnotes
Previously published online: www.landesbioscience.com/journals/cc/article/21694
References
- 1.Chang DJ, Cimprich KA. DNA damage tolerance: when it’s OK to make mistakes. Nat Chem Biol. 2009;5:82–90. doi: 10.1038/nchembio.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hoege C, Pfander B, Moldovan GL, Pyrowolakis G, Jentsch S. RAD6-dependent DNA repair is linked to modification of PCNA by ubiquitin and SUMO. Nature. 2002;419:135–41. doi: 10.1038/nature00991. [DOI] [PubMed] [Google Scholar]
- 3.Kannouche PL, Wing J, Lehmann AR. Interaction of human DNA polymerase eta with monoubiquitinated PCNA: a possible mechanism for the polymerase switch in response to DNA damage. Mol Cell. 2004;14:491–500. doi: 10.1016/S1097-2765(04)00259-X. [DOI] [PubMed] [Google Scholar]
- 4.Watanabe K, Tateishi S, Kawasuji M, Tsurimoto T, Inoue H, Yamaizumi M. Rad18 guides poleta to replication stalling sites through physical interaction and PCNA monoubiquitination. EMBO J. 2004;23:3886–96. doi: 10.1038/sj.emboj.7600383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bienko M, Green CM, Crosetto N, Rudolf F, Zapart G, Coull B, et al. Ubiquitin-binding domains in Y-family polymerases regulate translesion synthesis. Science. 2005;310:1821–4. doi: 10.1126/science.1120615. [DOI] [PubMed] [Google Scholar]
- 6.Kannouche PL, Wing J, Lehmann AR. Interaction of human DNA polymerase eta with monoubiquitinated PCNA: a possible mechanism for the polymerase switch in response to DNA damage. Mol Cell. 2004;14:491–500. doi: 10.1016/S1097-2765(04)00259-X. [DOI] [PubMed] [Google Scholar]
- 7.Guo C, Tang TS, Bienko M, Dikic I, Friedberg EC. Requirements for the interaction of mouse Polkappa with ubiquitin and its biological significance. J Biol Chem. 2008;283:4658–64. doi: 10.1074/jbc.M709275200. [DOI] [PubMed] [Google Scholar]
- 8.Plosky BS, Vidal AE, Fernández de Henestrosa AR, McLenigan MP, McDonald JP, Mead S, et al. Controlling the subcellular localization of DNA polymerases iota and eta via interactions with ubiquitin. EMBO J. 2006;25:2847–55. doi: 10.1038/sj.emboj.7601178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sale JE, Lehmann AR, Woodgate R. Y-family DNA polymerases and their role in tolerance of cellular DNA damage. Nat Rev Mol Cell Biol. 2012;13:141–52. doi: 10.1038/nrm3289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Motegi A, Liaw HJ, Lee KY, Roest HP, Maas A, Wu X, et al. Polyubiquitination of proliferating cell nuclear antigen by HLTF and SHPRH prevents genomic instability from stalled replication forks. Proc Natl Acad Sci USA. 2008;105:12411–6. doi: 10.1073/pnas.0805685105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Motegi A, Sood R, Moinova H, Markowitz SD, Liu PP, Myung K. Human SHPRH suppresses genomic instability through proliferating cell nuclear antigen polyubiquitination. J Cell Biol. 2006;175:703–8. doi: 10.1083/jcb.200606145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Unk I, Hajdú I, Fátyol K, Szakál B, Blastyák A, Bermudez V, et al. Human SHPRH is a ubiquitin ligase for Mms2-Ubc13-dependent polyubiquitylation of proliferating cell nuclear antigen. Proc Natl Acad Sci USA. 2006;103:18107–12. doi: 10.1073/pnas.0608595103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bergink S, Jentsch S. Principles of ubiquitin and SUMO modifications in DNA repair. Nature. 2009;458:461–7. doi: 10.1038/nature07963. [DOI] [PubMed] [Google Scholar]
- 14.Hofmann K. Ubiquitin-binding domains and their role in the DNA damage response. DNA Repair (Amst) 2009;8:544–56. doi: 10.1016/j.dnarep.2009.01.003. [DOI] [PubMed] [Google Scholar]
- 15.Kratz K, Schöpf B, Kaden S, Sendoel A, Eberhard R, Lademann C, et al. Deficiency of FANCD2-associated nuclease KIAA1018/FAN1 sensitizes cells to interstrand crosslinking agents. Cell. 2010;142:77–88. doi: 10.1016/j.cell.2010.06.022. [DOI] [PubMed] [Google Scholar]
- 16.Liu T, Ghosal G, Yuan J, Chen J, Huang J. FAN1 acts with FANCI-FANCD2 to promote DNA interstrand cross-link repair. Science. 2010;329:693–6. doi: 10.1126/science.1192656. [DOI] [PubMed] [Google Scholar]
- 17.MacKay C, Déclais AC, Lundin C, Agostinho A, Deans AJ, MacArtney TJ, et al. Identification of KIAA1018/FAN1, a DNA repair nuclease recruited to DNA damage by monoubiquitinated FANCD2. Cell. 2010;142:65–76. doi: 10.1016/j.cell.2010.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shereda RD, Machida Y, Machida YJ. Human KIAA1018/FAN1 localizes to stalled replication forks via its ubiquitin-binding domain. Cell Cycle. 2010;9:3977–83. doi: 10.4161/cc.9.19.13207. [DOI] [PubMed] [Google Scholar]
- 19.Smogorzewska A, Desetty R, Saito TT, Schlabach M, Lach FP, Sowa ME, et al. A genetic screen identifies FAN1, a Fanconi anemia-associated nuclease necessary for DNA interstrand crosslink repair. Mol Cell. 2010;39:36–47. doi: 10.1016/j.molcel.2010.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Crosetto N, Bienko M, Hibbert RG, Perica T, Ambrogio C, Kensche T, et al. Human Wrnip1 is localized in replication factories in a ubiquitin-binding zinc finger-dependent manner. J Biol Chem. 2008;283:35173–85. doi: 10.1074/jbc.M803219200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Guo C, Tang TS, Bienko M, Dikic I, Friedberg EC. Requirements for the interaction of mouse Polkappa with ubiquitin and its biological significance. J Biol Chem. 2008;283:4658–64. doi: 10.1074/jbc.M709275200. [DOI] [PubMed] [Google Scholar]
- 22.Huang J, Huen MS, Kim H, Leung CC, Glover JN, Yu X, et al. RAD18 transmits DNA damage signalling to elicit homologous recombination repair. Nat Cell Biol. 2009;11:592–603. doi: 10.1038/ncb1865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yan Z, Guo R, Paramasivam M, Shen W, Ling C, Fox D, 3rd, et al. A Ubiquitin-Binding Protein, FAAP20, Links RNF8-Mediated Ubiquitination to the Fanconi Anemia DNA Repair Network. Mol Cell. 2012;47:61–75. doi: 10.1016/j.molcel.2012.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Leung JW, Wang Y, Fong KW, Huen MS, Li L, Chen J. Fanconi anemia (FA) binding protein FAAP20 stabilizes FA complementation group A (FANCA) and participates in interstrand cross-link repair. Proc Natl Acad Sci USA. 2012;109:4491–6. doi: 10.1073/pnas.1118720109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ali AM, Pradhan A, Singh TR, Du C, Li J, Wahengbam K, et al. FAAP20: a novel ubiquitin-binding FA nuclear core-complex protein required for functional integrity of the FA-BRCA DNA repair pathway. Blood. 2012;119:3285–94. doi: 10.1182/blood-2011-10-385963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kim H, Yang K, Dejsuphong D, D’Andrea AD. Regulation of Rev1 by the Fanconi anemia core complex. Nat Struct Mol Biol. 2012;19:164–70. doi: 10.1038/nsmb.2222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Centore RC, Yazinski SA, Tse A, Zou L. Spartan/C1orf124, a reader of PCNA ubiquitylation and a regulator of UV-induced DNA damage response. Mol Cell. 2012;46:625–35. doi: 10.1016/j.molcel.2012.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ponting CP. Novel domains and orthologues of eukaryotic transcription elongation factors. Nucleic Acids Res. 2002;30:3643–52. doi: 10.1093/nar/gkf498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Moldovan GL, Pfander B, Jentsch S. PCNA, the maestro of the replication fork. Cell. 2007;129:665–79. doi: 10.1016/j.cell.2007.05.003. [DOI] [PubMed] [Google Scholar]
- 30.Colaiácovo MP, Stanfield GM, Reddy KC, Reinke V, Kim SK, Villeneuve AM. A targeted RNAi screen for genes involved in chromosome morphogenesis and nuclear organization in the Caenorhabditis elegans germline. Genetics. 2002;162:113–28. doi: 10.1093/genetics/162.1.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Maeda I, Kohara Y, Yamamoto M, Sugimoto A. Large-scale analysis of gene function in Caenorhabditis elegans by high-throughput RNAi. Curr Biol. 2001;11:171–6. doi: 10.1016/S0960-9822(01)00052-5. [DOI] [PubMed] [Google Scholar]
- 32.Parris CN, Levy DD, Jessee J, Seidman MM. Proximal and distal effects of sequence context on ultraviolet mutational hotspots in a shuttle vector replicated in xeroderma cells. J Mol Biol. 1994;236:491–502. doi: 10.1006/jmbi.1994.1160. [DOI] [PubMed] [Google Scholar]
- 33.Lehmann AR, Niimi A, Ogi T, Brown S, Sabbioneda S, Wing JF, et al. Translesion synthesis: Y-family polymerases and the polymerase switch. DNA Repair (Amst) 2007;6:891–9. doi: 10.1016/j.dnarep.2007.02.003. [DOI] [PubMed] [Google Scholar]
- 34.Hendel A, Krijger PH, Diamant N, Goren Z, Langerak P, Kim J, et al. PCNA ubiquitination is important, but not essential for translesion DNA synthesis in mammalian cells. PLoS Genet. 2011;7:e1002262. doi: 10.1371/journal.pgen.1002262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Geng L, Huntoon CJ, Karnitz LM. RAD18-mediated ubiquitination of PCNA activates the Fanconi anemia DNA repair network. J Cell Biol. 2010;191:249–57. doi: 10.1083/jcb.201005101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shiomi N, Mori M, Tsuji H, Imai T, Inoue H, Tateishi S, et al. Human RAD18 is involved in S phase-specific single-strand break repair without PCNA monoubiquitination. Nucleic Acids Res. 2007;35:e9. doi: 10.1093/nar/gkl979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Despras E, Daboussi F, Hyrien O, Marheineke K, Kannouche PL. ATR/Chk1 pathway is essential for resumption of DNA synthesis and cell survival in UV-irradiated XP variant cells. Hum Mol Genet. 2010;19:1690–701. doi: 10.1093/hmg/ddq046. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.