Abstract
p53 plays an important role in mitotic checkpoint, but what its role is remains enigmatic. Aurora A is a Ser/Thr kinase involved in correcting progression of mitosis. Here, we show that p53 is a negative regulator for Aurora A. We found that p53 deficiency leads to Aurora A elevation. Ectopic expression of p53 or DNA damage-induced expression of p53 can suppress the expression of Aurora A. Mechanistic studies show that p53 is a negative regulator for Aurora A expression through both transcriptional and posttranslational regulation. p53 knockdown in cancer cells reduces the level of p21, which, in turn, increases the activity of CDK2 followed by induction of Rb1 hyperphosphorylation and its dissociation with transcriptional factor E2F3. E2F3 can bind to Aurora A gene promoter, potentiating Aurora A gene expression and p53 deficiency, enhancing the binding of E2F3 on Aurora A promoter. Also, p53 deficiency leads to decelerating Aurora A’s turnover rate, due to the fact that p53 deficiency causes the downregulation of Fbw7α, a component of E3 ligase of Aurora A. Consistently, p53 knockdown-mediated Aurora A elevation is mitigated when Fbw7α is ectopically expressed. Thus, p53-mediated Aurora A degradation requires Fbw7α expression. Significantly, inverse correlation between p53 and Aurora A elevation is translated into the deregulation of centrosome amplification. p53 knockdown leads to high percentages of cells with abnormal amplification of centrosome. These data suggest that p53 is an important negative regulator of Aurora A, and that loss of p53 in many types of cancer could lead to abnormal elevation of Aurora A and dysregulated mitosis, which provides a growth advantage for cancer cells.
Keywords: cell cycle, gammaH2AX, human non-small cell lung carcinoma, mTOR, metronomic chemotherapy, oncogenes, personalized cancer treatment
Introduction
The p53 protein plays a central role in the regulation of several biological signaling processes, including cell cycle, apoptosis, differentiation, autophagy, senescence, angiogenesis and metabolism.1 The loss of p53 function is found in around 50% of all human tumors (reviewed in ref. 2). Furthermore, the status of p53 strongly affects the sensitivity of tumor cells to chemotherapeutic and/or radio therapeutic agents.3 p53 is activated in response to DNA damage, hypoxia and abnormal expression of oncogenes.4,5 Upon activation, p53 is stabilized by dissociation from MDM2 and can trigger the transcription of a set of target genes (p21, GADD45, Puma or Bax) that arrest the cell cycle or induce cellular apoptosis (reviewed in ref. 6). In addition to transcriptional transactivation, p53 can transcriptionally suppress certain target genes such as c-met and c-myc via the recruitment of histone deacetylase7 or by directly competing with the TATA-box binding protein to interfere with target gene expression.8 Although p53 can suppress oncogene expression, the full spectrum of its targets is not well known.
Aurora kinase, a family of serine/threonine kinases,9 is evolutionarily conserved from yeast to C. elegans, Drosophila, Xenopus and mammals. Three types of Aurora kinases, namely Aurora A, -B and -C have been identified in humans so far. Human Aurora A is the human homolog of the yeast/Drosophila Ipl1/Aurora kinase family molecule and is regulated in a cell cycle-dependent manner. Aurora A mRNA and protein levels are elevated between the G2 and M phases, and decrease quickly between the M and G1 phases. Importantly, increased mRNA and protein levels of Aurora A have been observed in many human cancers, including lung, breast, ovarian, colon, liver, head and neck, prostate cancer and leukemia. Furthermore, the overexpression of Aurora A is oncogenic.9,10 Overexpression of Aurora A might be achieved by gene amplification, transcription activation or inhibition of protein degradation. With regards to transcriptional activation, several transcription factors, such as E2F3, E4TF1 and TRAP220/MED1, have been identified as positive regulators of Aurora A gene expression.11-13 As for posttranscriptional regulation, Fbw7, CHFR, TPX2, phosphatase 2A and AURKAIP1 are involved in regulating the protein stability of Aurora A.14-20 However, how the upstream signal/mediators that affect these transcriptional/posttranscriptional regulators contribute to the expression of Aurora A in cells is largely unclear. Identifying such factors may provide further insight into the mechanism behind highly dysregulated Aurora A expression in cancer.
Here, we demonstrate that both the p53-Rb-E2F3 axis and p53-Fbw7 axis regulate Aurora A’s expression level through transcriptional and post-transcriptional regulation, respectively. Significantly, p53 deficiency leads to the enhancement of Aurora A expression and activity, resulting in abnormal amplification of centrosomes. Together, this study provides important insights into how p53 deficiency or mutation may impact Aurora A’s role in mitosis in cancer cells.
Results
p53 is a negative regulator of Aurora A
To examine the functional relationship between p53 and Aurora A, p53-knockdown A549 cells were used. Western blot analysis showed that Aurora A markedly increased in two p53-knockdown A549 subclones when compared with parental and vector control cells (Fig. 1A). Additionally, this result was not limited to lung cancer cells; knockdown of p53 by siRNA in hepatocellular carcinoma HepG2 cells also induced an elevation of Aurora A (Fig. S1). Furthermore, p21CIP1/WAF1, a well-known p53 downstream target involved in CDK inhibition,21 exhibited decreased levels in p53-knockdown A549 cells (Fig. 1A). We then showed that the expression levels of Aurora A were increased in p53-siRNA-treated A549 cells in a dose-dependent manner (Fig. 1B). Administration of Pifithrin-α (PFT), an inhibitor of p53 transactivation, also induced Aurora A expression in A549 cells in a dose-dependent manner (Fig. 1C). We also delivered p53 in p53 knockdown A549 cells or p53-null H1299 cells by infecting cells with adenoviruses (Adv) carrying p53 gene. The results showed that Aurora A levels decreased in cells treated with Adv-WT p53. It should be noted, however, that Adv-p53 mutant (DNA binding mutant) in p53 knockdown cells (Fig. 1D) and p53-null H1299 cells (Fig. 1E) did not lower Aurora A levels, suggesting that only wt p53 is able to downregulate the expression of Aurora A.
Figure 1. p53 is a negative regulator of Aurora A. (A) The parental and p53-knockdown A549 stable clones were harvested and subjected to western blotting using the indicated antibodies. (B) A549 cells were transfecetd with various dosages of p53 siRNA for 48 h, the cells were then collected and subjected to western blotting using the indicated antibodies. (C) Various dosages of p53 inhibitor PFT were added into A549 cells, 48 h later, the cells were collected and subjected to western blot. (D) The adenovirus particles containing p53-wild type or p53-mut (DNA binding mutant) gene were infected into A549-p53shRNA stable cells. The cells were then harvested and subjected to western blotting using the indicated antibodies. (E) p53-null H1299 cells were infected with adenovirus particles described in (D). (F) A549-control shRNA, A549-p53 shRNA and H1299 cells were treated with indicated doses of cisplatin for 48 h, the cells were then collected, lysed and subjected to western blotting using the indicated antibodies.
The clinical anticancer drug cisplatin, a DNA-damaging agent that causes p53 elevation,22,23 can significantly induce the protein level of p53, resulting in the downregulation of Aurora A in a dose-dependent manner (Fig. 1F). This indicates that the negative correlation between Aurora A and p53 exists in response to DNA damage. Noticeably, p53 knockdown compromises such a p53-mediated downregulation of Aurora A in A549 cells (Fig. 1F). p53-null H1299 cells also did not demonstrate DNA damage-mediated Aurora A downregulation (Fig. 1F), reemphasizing the role of p53 in Aurora A regulation. Cell cycle analysis in the wild type or p53-knockdown A549 cells indicates that p53-knockdown leads to a longer S phase, as expected, but there was no significant difference among wild type and p53-knockdown A549 cells in terms of the G2/M phase. These data suggest that the increasing protein level of Aurora A under p53-deficent circumstances is not due to an increased number of cells in the G2/M phases (Fig. S2).
p53-E2F3 axis is involved in regulating Aurora A transcriptional expression
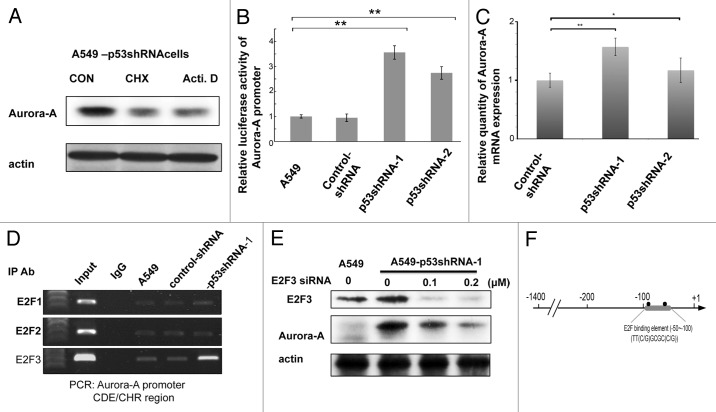
To understand the mechanism behind p53-mediated Aurora A downregulation, A549-p53shRNA cells were treated with either cycloheximide, a translational inhibitor or actinomycin D, a transcriptional inhibitor, respectively. As shown in Figure 2A, both cycloheximide and actinomycin D treatment can mitigate the expression level of Aurora A (Fig. 2A), suggesting that the elevation of Aurora A protein levels under p53 deficiency might be regulated via both posttranslational and transcriptional processes. Luciferase reporter gene assays indicate that Aurora A (+15~-1400 bp) promoters exhibited higher luciferase activity in p53-knockdown cells than parental cells (Fig. 2B). The qRT-PCR data confirmed that Aurora A mRNA in p53-knockdown cells was higher than that of A549 cells (Fig. 2C; Fig. S3). These results indicate that p53 is able to regulate Aurora A expression in a transcriptional manner. E2F3 is a member of cell cycle regulators and can directly bind to the CDE/CHR region of Aurora A promoters and induce Aurora A expression in the G2/M phase.11 The function of E2F3 is regulated by p53 via the p21-CDK-Rb1 pathway.24 To address whether E2F3 is involved in the upregulation of Aurora A in p53-knockdown cells, we performed a chromatin immunoprecipitation assay, analyzed the sequence of the Aurora A promoter, and found that the Aurora A promoter does contain an E2F binding site (Fig. 2D and F). Our results show that E2F3 exhibits a greater ability to bind to the Aurora A promoter region in p53-knockdown cells than parental cells (Fig. 2D), while the binding activity of E2F1 and E2F2 to CDE/CHR region of Aurora A promoter in wild type and p53-knockdown A549 cells remained the same (Fig. 2D). These results suggest that the binding affinity, to the promoter of Aurora A, of E2F3, but not E2F1 or E2F2, is increased in the presence of p53 deficiency. Further knockdown of E2F3 expression by siRNA in p53-knockdown cells decreased the expression of Aurora A in a dose-dependent manner (Fig. 2E), suggesting that E2F3 is directly involved in regulating the promoter of Aurora A via p53.
Figure 2. p53 affects the binding ability of E2F3 on Aurora A promoter region. (A) A549-p53shRNA cells were treated with actinomycin D (500 ng/ml) or cyclohexmide (50 μg/ml), respectively, for 9 h, the cells were then collected and subjected to western blot using the indicated antibodies. (B) Aurora A promoter (+15 to -1400 bps)-luciferase vector was cotransfected with internal control luciferase vector into wild type or p53-knockdown A549 cells for 48 h, the cells were then harvested and subjected to luciferase assay. (C) The expression levels of Aurora A of A549-shRNA, A549 p53-shRNA-1 and A549-p53shRNA-2 cells were determined by qPCR. (D) A549, A549-shRNA and A549-p53shRNA-1 cells were analyzed for chromatin immunoprecipitation (ChIP) using IgG, E2F1, E2F2, E2F3 antibody, respectively. The immunoprecipitated chromatin fragments were then amplified by PCR reaction using primer pairs that corresponded to E2F binding element (CDE/CHR) in Aurora A promoter region. (E) A549-p53shRNA-1 cells were transfected with various doses of E2F3 siRNA for 48 h followed by western blot using anti-E2F3 and Aurora A antibodies, respectively. (F) A diagram of the E2F3 binding region in Aurora A promoter.
p53 /p21/Rb1/E2F3 axis is critical in regulating Aurora A expression
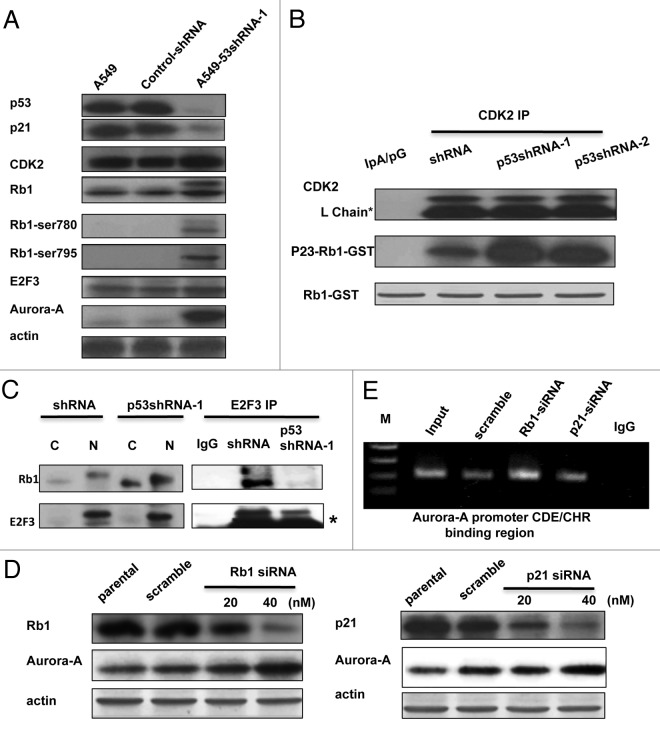
Since E2F3 is a downstream effector of the p21-CDK-Rb1 pathway, we analyzed the expression patterns of p21, Rb1 and E2F3 in wild type and p53-knockdown A549 cells. As expected, p21 is diminished in p53-knockdown cells when compared with both A549 cells. In this p53-knockdown context, Rb1 also exhibited stronger phosphorylation at Ser780 and Ser795, which are known indicators of CDK2/4 activation25 (Fig. 3A). These results suggest that p53 deficiency induces the loss of p21 expression, which leads to CDK2 activation and thus allows the phosphorylation of Rb1 (a CDK substrate).
Figure 3. The p21-cdk-Rb1-E2F3 pathway is involved in the upregulation of Aurora A. (A) The western blot of p21, Rb1, Rb1-Ser 780, Rb1-Ser795, E2F3, p53 and Aurora A of parental-, vehicle- and A549 p53-shRNA-1 cells were performed. (B) The endogenous CDK2 of A549-shRNA, p53-shRNA-1 and p53shRNA-2 cells were immunoprecipitated this was then followed by a kinase reaction using Rb1-GST as a substrate in the presence of γ-p32 [ATP]. Asterisk indicates IgG light chain. (C) Cytoplasmic and nuclear fractions were prepared from the A549-shRNA and A549-p53shRNA-1 cells and were then immunoprecipitated with anti-E2F3 antibody or with IgG alone. The immunoprecipitation complex was then immunoblotted with the indicated antibodies. Asterisk indicates IgG light chain. (D) A549-shRNA cells were transfected with different doses of Rb1 siRNA (left panel), p21 siRNA (right panel) or scrambled siRNA, respectively. The cell lysates were subjected to western blot using the indicated antibodies. (E) Similar transfection experiments described above were performed, and the cells were collected 60 h after transfection followed by chromatin-immunoprecipitation using anti E2F3 antibody. The CDE/CHR region of the Aurora A promoter was then amplified by PCR using the indicated primer set.
To determine this link, we analyzed the activity of CDK in wild type and p53-knockdown A549 cells. Endogenous CDK2 was immunoprecipitated for use in the kinase assays. Recombinant Rb1-C protein was used as a substrate (Fig. 3B). The results show that CDK2 is highly activated in p53-knockdown A549 cells when compared with knockdown control cells (Fig. 3B). Furthermore, the interaction between Rb1 and E2F3 is mitigated in p53-knockdown cells but not in parental cells (Fig. 3C). These results demonstrate that the p21-CDK-Rb1-E2F3 pathway, which is dysregulated under p53 deficiency, correlates with the increasing expression of Aurora A in p53-knockdown cells. To confirm the roles of Rb1 and p21 in the upregulation of Aurora A expression levels, we knocked down these two genes in A549 cells via siRNA transfection, and showed that the knockdown of either Rb1 or p21 increased Aurora A expression in a dose-dependent manner (Fig. 3D). Accordingly, the binding affinity of E2F3 onto the CDE/CHR region of the Aurora A promoter region also increases when Rb1 is knocked down (Fig. 3E). Taken together, we can conclude that p53 deficiency results in the dysregulation of the p21-CDK-pRb1-E2F3 signaling axis, contributing to the upregulation of Aurora A expression levels through promoter regulation.
Fbw7 has a critical role in p53-mediated Aurora A downregulation
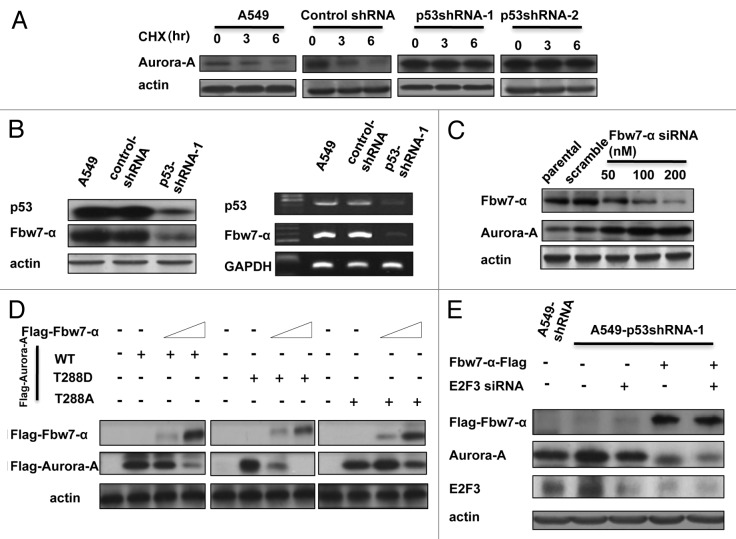
Although Aurora A’s expression level is stringently controlled at both the transcriptional and translational level as the cell cycle progresses,26,27 a detailed mechanistic understanding of this regulation remains elusive. To determine the posttranscriptional regulation of Aurora A, we employed cycloheximide, an inhibitor of de novo protein synthesis, for the following studies. The turnover rate of endogenous Aurora A was compromised in p53 knockdown cells in a cycloheximide chase assay (Fig. 4A). Interestingly, the level of Fbw7α, an E3 ligase of Aurora A and a downstream target gene of p53, was lower with respect to both the protein and mRNA level when p53 was knocked down (Fig. 4B). To determine the role of Fbw7α in regulating Aurora A, the siRNA of Fbw7α was transfected into cells at various dosages. The results showed that knockdown of Fbw7α increases the steady-state level of Aurora A in a dose-dependent manner (Fig. 4C). Accordingly, overexpression of exogenous Fbw7α into p53-knockdown or 293T cells decreases the steady-state level of the Aurora A protein in a dose-dependent manner (Fig. S4). These results indicate that Fbw7α is indeed involved in the downregulation of Aurora A in cells. We scanned the sequence of Fbw7α and found that there is a predicted phosphodegron14,28(T-P-P-X-T/S) on Aurora A at threonine 288 (T288LCGT). To characterize the phosphorylation role of T288 in Fbw7α-mediated Aurora A degradation, we constructed T288A and T288D Aurora A and assayed the impact of Fbw7α on these constructs. As expected, the expression level of wt Aurora A was degraded by Fbw7α, while the Aurora A-T288A was resistant to Fbw7α–mediated degradation. Importantly, the Aurora A T288D, which mimics phosphorylation, exhibited faster degradation in the presence of Fbw7α when compared with wt Aurora A (Fig. 4D). These results point to Aurora A T-288 as a potential phosphodegron for Fbw7α. To determine whether E2F3 knockdown and Fbw7α overexpression play a synergistic role in the downregulation of Aurora A, E2F3 siRNA and Fbw7α-Flag were transfected into p53-knockdown cells, both individually and simultaneously. The results showed that ectopic expression of Fbw7α with simultaneous knockdown of E2F3 can attenuate the steady-state expression level of Aurora A more efficiently (Fig. 4E).
Figure 4. Fbw-7a acts as a regulator in p53-mediated downregulation of Aurora A. (A) A549 cells were transfected with indicated shRNA-vector or p53-knockdown RNA and treated with cyclohexamide (CHX) for the indicated hours; the cells were then lysed and subjected to western blot using anti-Aurora A antibody. (B) The proteins or total RNAs of indicated cells were extracted, and subjected to western blot, or RT-PCR analysis was performed to examine the protein and RNA levels of Fbw7-α. (C) Various doses of Fbw7-α siRNA were transfected into A549 cells, 48 h later, the cells were collected and subjected to western blot using anti-Fbw7 or Aurora A antibody, respectively. (D) Indicated Aurora A constructs were transfected into 293T cells, followed by transfection with various dosages of Fbw7-α. The cell lysates were subjected to western blot using anti-Flag antibody. (E) The indicated plasmids or siRNA were transfected into A549-p53shRNA-1 cells. The cell lysates were subjected to western blot using anti-Flag, E2F3 and Aurora A antibodies, respectively.
p53-Aurora A axis controls centrosome amplification
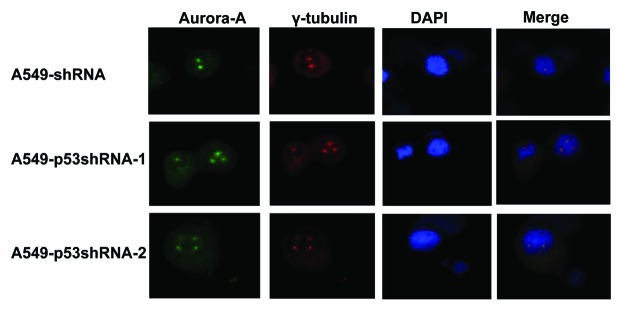
We have shown that knockdown of p53 inversely increases the Aurora A protein level via both transcriptional and posttranslational manners in cancer cells. Given that overexpression of Aurora A also induces the formation of multiple centrosomes,29 we looked into whether or not p53-mediated Aurora A expression has any impact on centrosome formation. Our results showed that knockdown of p53 induced the amplification of multiple centrosomes (Fig. 5 and Table 1). The centrosome abnormality during the M phase was assessed, and the results indicated that high percentages of p53-deficient cells have centrosome abnormalities (Table 1). The formation of multiple centrosomes might be due to Aurora A overexpression resulting from p53 deficiency.
Figure 5. The knockdown of p53 induces the amplification of multiple centrosomes in cells. A549-shRNA, A549-p53shRNA-1 and A549-p53shRNA-2 cells were immunostained with indicated antibodies for fluorescent microscopy. Cell nuclei were stained with DAPI.
Table1. The centrosome abnormality in A549 cells treated with p53 shRNA.
| A549 cells | Centrosome abnormality at M phase |
|---|---|
|
Control ShRNA |
1.5% ± 0.2% |
|
p53 ShRNA-1 |
32.6% ± 3.2% |
| p53 ShRNA-2 | 21.1% ± 2.2% |
p53-Aurora A regulation is recapitulated in primary cancer sample analysis
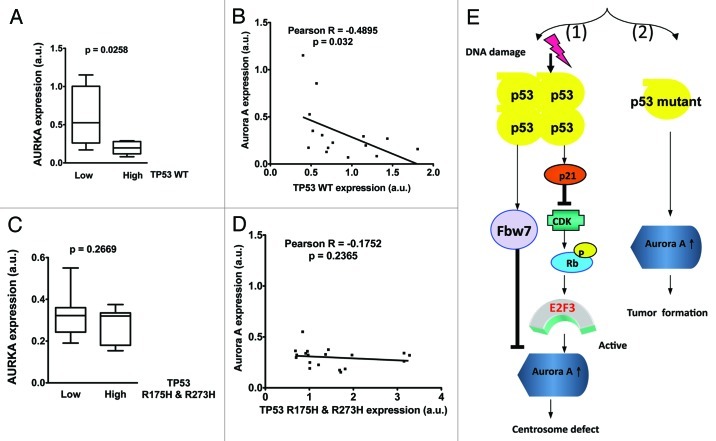
To investigate the relevance of p53-mediated Aurora A regulation during cancer development, we evaluated primary specimens of ovarian cancer, since it’s a disease with a high rate of p53 mutation. p53 status was sequenced and Aurora A gene expression level quantitated in these primary cancer samples. Importantly, Aurora A expression was found to be suppressed in ovarian cancer patients with high level of wt TP53 expression. Aurora A gene expression in primary ovarian cancer samples negatively correlates with wt p53 expression level at the time of diagnosis (Fig. 6A and B). Further analyses showed that Aurora A gene expression is not affected by mutated p53 (R175H and 273H) (Fig. 6C and D). Together, our studies indicate that p53 status correlates with Aurora A expression in cancer; and that p53 may function as a tumor suppressor through negative regulation of Aurora A during tumorigenesis.
Figure 6. wt p53 expression level inversely correlates with Aurora A expression in human ovarian cancer samples. (A) Aurora A expression is suppressed in ovarian cancer patients with high levels of TP53. Gene expression profiles of p53 and Aurora A were retrieved from The Cancer Genome Atlas’ ovarian cancer patients microarray data sets and analyzed with Nexus Expression 2.0 (BioDiscovery). Unpaired Student’s t-test was done using Prism 5.0d. (B) Aurora A expression is inversely correlated with wt TP53 level in ovarian cancer patients. Pearson Correlation Test was done using Prism 5.0d (C) Aurora A expression levels are not affected by mutated p53 in ovarian cancer patients. Gene expression profiles were retrieved from The Cancer Genome Atlas’ ovarian cancer patient microarray data sets and analyzed with Nexus Expression 2.0 (BioDiscovery). TP53 mutations R175H and R273H were determined by sequencing analysis. Unpaired Student t-test was done using Prism 5.0d. (D) There is no inverse correlation between Aurora A expression and TP53 mutants R175H and R273H levels in ovarian cancer patients. Pearson Correlation Test was done using Prism 5.0d. (E) Conclusion model.
Disscusion
In mammalian cells, mitotic kinase Aurora A regulates various cellular targets, including stress-responsive transcription factors and the p53 tumor suppressor.30 Aurora A can phosphorylate p53 and cause p53 degradation, thereby inhibiting its transcriptional activity.30 However, it is not clear how p53 regulates Aurora A. Here, our results indicate that p53 is a negative regulator of Aurora A, suggesting there is feedback or reciprocal regulation between Aurora A and p53.
Both p53 and Aurora A are diagnostic and prognostic markers in many cancers and serve as promising therapeutic targets. Aurora A is overexpressed and/or activated in many types of cancers including NSCLC,9,31 while the p53 gene is frequently mutated or lost.32,33 Aurora A phosphorylates p53 and promotes p53 degradation.30 Additionally, Aurora A also blocks p53 functions by directly phosphorylating p53 and inhibiting its DNA binding affinity, indicating that Aurora A has a negative regulatory impact on p53 functions. Another mitotic kinase, Aurora B, also seems to have a negative impact on p53 during interphase and mitosis.34 Interestingly, our studies suggest that Aurora A and p53 may reciprocally regulate one another. Especially since, we show that even though p53, originally deemed an Aurora A substrate, is negatively regulated by Aurora A in terms of stability, it can regulate Aurora A gene and protein expression. Significantly, cisplatin, a chemotherapeutic drug, can induce the activation of p53 via a DNA-damage pathway in various cancer cells22,23,35-37 and elevate p53 protein levels, thereby reducing the expression of Aurora A (Fig. 1). Cancer sample studies also indicate that expression levels of wt p53 but not mutant p53 suppress the expression of Aurora A during tumorigenesis (Fig. 6). Although it has been reported that positive p53 expression correlates with low expression of Aurora A in breast cancer, the underlying mechanism behind this observation is not clear, and gene status of p53 has yet to be characterized.38 Furthermore, in mouse tumor studies, Aurora A is highly amplified in the lymphomas of p53+/− mice, while Aurora A is frequently deleted in p53−/− mice.38 These observations reemphasize the possible mutual regulation between Aurora A and p53, and reaffirm the need for a detailed mechanism to be characterized. Our studies provide a mechanistic explanation that defines the p53→Aurora A regulatory direction.
Our mechanistic studies indicate that p53 negatively regulates Aurora A expression level via both transcriptional and posttranslational manners. First, in the transcriptional regulation, p53 deficiency causes a reduction in the expression of p21 and induces the activation of CDK2. This is followed by the phosphorylation of Rb1, which, in turn, dissociates with E2F3. The released E2F3 binds to the promoter region of the Aurora A promoter and eventually induces the transcription of Aurora A. Surprisingly, out of all the E2F family members, only E2F3 is involved in this process. Such specificity indicates a delicate controlling process. Although the E2F site is known to be present in Aurora A promoter,11 our studies provide insight into the link between p53 and E2F3 in the regulation of Aurora A transcription and fill an important knowledge gap. It is important to point out that that Aurora A also phosphorylates recombinant Rb1 in vitro (data not shown), which may further enhance the dissociation of E2F3 from Rb1. This, in turn, will lead to more Aurora A gene expression. This suggests that Aurora A is able to form a feedback loop that induces its own expression via the Rb1-E2F3 pathway. It is important to mention that, Aurora B, as a member of the Aurora family, does not regulate mitosis alone31 and can also phosphorylate Rb1 at serine 780.39 This suggests that Aurora B may also contribute to Aurora A gene expression. Second, p53 upregulates Fbw7α, an F-box protein component of Aurora A’s E3 ligase to facilitate p53-mediated Aurora A degradation.14,20 Importantly, we show that Fbw7α regulates Aurora A through a phosphodegron on Aurora A at threonine 288 (T288LCGT). It is important to point out that the phosphorylation of Aurora A-T288 is considered as a sign of Aurora A activation during the mitotic phase.40 We went on to characterize Aurora A-T288A as resistant to Fbw7α-mediated degradation, while the Aurora A T288D exhibited faster degradation in the presence of Fbw7α (Fig. 4) . This demonstrated for the first time that an Fbw7α target degoron is characterized on Aurora A. The important kinase responsible for this phosphorylation is known to be Aurora A, although other potential kinases may be involved.
Previous reports demonstrate that the overexpression of Aurora A causes the formation of multiple centrosomes29 and, subsequently, unequal separation of chromosome. Our results also show that knockdown of p53 induces the formation of multiple centromes (Fig. 5). Aurora A is involved in centrosome regulation.29 Conceivably, p53 deficiency leads to the elevation of Aurora A levels, thereby increasing centrosome amplification. As a genomic guardian, it is important for p53 to be capable of such action in order to control Aurora A-regulated centrosome amplification. Otherwise, p53 deficiency would allow for high levels of Aurora A expression, thereby resulting in the formation of polyploidy. It is possible that Aurora A is regulated by p53 to regulate the progression through mitosis. When p53 is mutated or lost in cancers, p53 cannot control the Aurora A level properly, which causes mitotic defects allowing the polyploidy that is commonly observed in cancers. It is important to point out that overexpressing Aurora A in p53−/− MEF leads to a drastic increase in centorsome numbers when compared with p53wt MEF,29 suggesting that p53 status is critical in determining Aurora A-mediated amplification of centrosomes. Based on our biochemical studies, we propose the following model: DNA damage-mediated stabilization of p53 leads to the expression of p21 and subsequent inhibition of CDK-mediated Rb phosphorylation, impairing the E2F3-mediated transcriptional activation of Aurora A. Also p53 can induce Fbw7α to downregulate Aurora A. In both scenarios, p53 can regulate Aurora A and thus maintain centrosome numbers [Fig. 6E, (1)]. On the other hand, p53 mutation leads to attenuation of inhibiting Aurora A gene expression, facilitating tumor formation [Fig. 6E, (2)]. Our ovarian cancer analysis data provides firm support for this second notion. Therefore, the impact of p53 on Aurora A gene expression plays a significant role in tumorigenesis.
Aurora A and p53 regulate each other reciprocally, suggesting that the equilibrium between p53 and Aurora A has to be maintained. Imbalance caused by p53 deficiency or Aurora A ovexpression is manifested in cancer diseases. Our data fills an important gap of knowledge regarding a direction that had not been completely defined before, i.e., p53’s role in controlling the level of Aurora A and maintaining the correct maturation of the centrosome.
Material and Methods
Cell culture and reagents
The human lung cancer cell line, A549, p53-knockdown A549, p53-null H1299 hepatocellular carcinoma cell lines and HepG2, were maintained in RPMI 1640 medium (Gibco/BRL) supplemented with 10% heat inactivated fetal bovine serum (Hyclone), 2 mM glutamine and antibiotics (100 unit/ml penicillin and 100 mg/ml streptomycin) at 37°C in a humidified atmosphere of 5% CO2. The Adeno-p53-WT or -DN virus (unable to bind DNA) was kindly provided by Dr. Shyue, Song-Kun. The PFT, doxorubicin, cisplatin, cyclohexamide and actinomycin D were obtained from either Sigma or Merck.
Preparation of cell extracts and immunoblot analysis
To prepare proteins for immunoblotting, untreated or various compound-treated cells were lysed in protein lysis buffer (RIPA, 50 mM Tris–HCl, pH 7.4, 150 mM NaCl, 1 mM EDTA, 1 mM EGTA, 0.5 mM dithiothreitol, 1% NP-40, 0.1% deoxycholate, 10 mg/ml aprotinin, 10 μg/ml soybean trypsin inhibitor, 10 μg/ml leupeptin, 0.5 mM phenylmethylsulfonyl fluoride), and the protein concentration was determined using the Bradford method. Equal amounts of sample lysates were subjected to sodium dodecyl-PAGE (SDS–PAGE) and electrophoretically transferred onto a PVDF membrane (Millipore). The membrane was blocked with 5% nonfat milk in TBST buffer (20 mM Tris–HCl, pH 7.4, 150 mM NaCl, 0.1% Tween-20) and incubated overnight at 4°C with specific primary antibodies. Subsequently, the membrane was washed with TBST buffer and incubated with the appropriate secondary antibody (horseradish, peroxidase-conjugated goat, anti-mouse or anti-rabbit IgG). Signal detection was performed using enhanced chemiluminescence kits (Amersham; ECL kit).
Chromatin immunoprecipitation assay
This procedure was performed according to previous repost.41 Briefly, 1 × 107 A549-shRNA or A549-p53shRNA cells were crosslinked by 1% formaldehyde for 15 min at room temperature. This was followed by quenching with glycine. After centrifugation the cell pellet was rinsed with an ice-cold PBS/AEBSF solution and then suspended in ChIP lysis buffer (Tris-SDS) followed by subjecting to sonication. The sheared chromatin was then cleared by centrifugation at 4°C (10 min at 10,621 × g) and re-suspended in immunoprecipitation buffer. The protein A/G beads were then added into the immunoprecipitation mixture and rotated for 1 h at 4°C. After the spin down, the supernatant was transferred to a new eppendorff and followed by addition of 2 μg of anti-E2F1, -2 or -3 antibody, respectively. (Santa Cruz) and incubated over night at 4°C on a rotator. The immune complexes were washed two times with 1 ml ChIP immunoprcipitation buffer, two times with Low salt wash buffer, two times with high salt and once with 1 × TE buffer (10 mM Tris base, 1 mM EDTA). All washing steps were performed at 4°C. The IP-beads compex were then added with 150 μl SDS elution buffer (1% SDS, 0.1 M NaHCO3) and incubated in a shaking water bath for O/N at 67°C to reverse the crosslink. The supernatants were then purified by DNA-clean UP kit (promega) following the manufacturer's instructions. The elute DNA then underwent PCR, using indicated primer to amplify the CDE/CHR region in Aurora A promoter.
Quantitative polymerase chain reactions
Total cellular RNA was extracted by RNA-BeeTM RNA isolation kit (TEL-TEST) following the manufacturer’s instructions. One microgram of total RNA was reverse-transcribed using Advantage RT for PCR Kit (Clontech) at 42°C for 1 h as described in the manufacturer’s protocol. The qPCR primers for Aurora A were forward (847): TCTTCCAGGAGGACCACTCTCT and reverse (917): TGCATCCGACCTTCAATCATT. For each combination of primers, the kinetics of PCR amplification was studied. The number of cycles corresponding to plateau was determined and PCR was performed at an exponential range. PCR products were then separated through a 1% agarose gel and visualized by ethidium bromide staining under UV irradiation. The mRNA levels were also determined by real-time PCR with an ABI PRISM 7900 Sequence Detector system according to the manufacturer’s instructions. β-actin was used as endogenous control. The PCR reaction mixture contained the SYBR PCR master mix (Applied Biosystems), cDNA and the primers. Relative gene expression level (the amount of target, normalized to endogenous control gene) was calculated using the comparative Ct method formula E-DDCt.
Cytosolic and nuclear fractionation
Briefly, the A549-shRNA or p53shRNA cells were scraped with cold PBS and collected by centrifugation at 2,000 × R.P.M for 5 min followed by resuspending in hypotonic buffer A (10 mM Hepes-KOH, 10 mM NaCl, 1 mM KH2PO4, 5 mM NaHCO3, 1 mM CaCal2 and 0.5 mM MgCl2). The cells were then homogenized with 10 strokes in a Dounce homogenizer after 5 min incubation on ice, and repeated once. Cells were then spun at 2,000 × R.P.M. for 15 min. The supernatant was collected as cytosol fractions. The pellet from the low-speed centrifugation was washed twice and resuspended in a nuclear isolation buffer (10 mM Tris, pH 7.5/300 mM sucrose/0.1% Nonidet P-40). This was followed by stroking with homogenizer and incubation on ice for 20 min. The mixture was then centrifuged at 14,000 R.P.M. for 30 min; the supernatant was then collected as nuclear fractions.
Immunofluorescence
Cells were fixed with 2% paraformaldehyde for 20 min prior to incubation with 0.1% Triton X-100 for 30 min and then blocked with 1% BSA for 30 min. Cells were probed with an anti-p65 antibody (Santa Cruz) overnight at 4°C, followed by incubation with a FITC-conjugated goat anti-mouse/rabbit IgG antibody (Sigma, diluted 1:200) for 1 h at 37°C, washing with PBS three times and staining with propidium iodide for 15 min. The expression and location of target proteins were observed with a laser scanning confocal microscope.
Immunoprecipitation and kinase assay
Experimental cells were lysed and immunoprecipitated with the indicated antibody. The immunoprecipitated complexes were then subjected to SDS-PAGE and western blotting with the indicated antibody. For CDK2 activity assays, protein lysates were subjected to immunoprecipitation in immunoprecipitation buffer at 4°C overnight in the presence of anti-Aurora A antibody and protein A agarose beads. Phosphorylation of GST-Rb is measured by incubating the beads with 40 μL “hot” kinase solution (containing γ-32p-[ATP]) for 30 min at 30°C. Samples were analyzed by 13% SDS-PAGE and the gels were dried and subjected to autoradiography.
Transfection procedure and luciferase reporter assay
The related siRNAs of Aurora A were transfected into A549-p53shRNA, H1299 or 293T cells by Lipofectamine 2000 (Invitrogen) for 16 h. The transfection solution was replaced with fresh culture medium and then used in the following experiments. The working concentration of each siRNA was based on the manufacturer’s recommendation (Santa Cruz). Transfection of the plasmid was performed in a similar manner. The concentration of each WT-, KR- or T228D Aurora A plasmid was 1 μg. For the reporter assay, 2 μg p-NF-κB-LUC (4X) and 0.2 μg b-galactosidase internal control (pTRElacZ) were cotransfected into 293T or A549-p53shRNA cells using Lipofectamine 2000 (Invitrogen). After 12 h, the transfection medium was replaced with free culture medium. After the desired experiments were conducted, the luciferase activity of each experimental set of cells was determined and normalized to the β-galactosidase activity using the dual luciferase assay system according to the manufacturer’s protocol (Tropsix) using a luminometer (Minilumate LB 9506).
Statistical analysis
Figures were generated from at least three independent experiments with similar patterns. All data are presented as means ± SD of nine replicates from three separate experiments. Statistical differences were evaluated using Student’s t-test (* denotes p < 0.05, ** denotes p < 0.01 and *** denotes p < 0.001; these were considered significant) or by the calculation and grouped using the SAS program.
Supplementary Material
Acknowledgments
This work was supported in part by grants from the National Institutes of Health (NIH) (R01CA089266) and Susan G. Komen Breast Cancer Foundation (KG081048). We also appreciate the support from National Science Council of Taiwan (NSC99–3112-B-075A-001 to SLH) and VGH grant (TCVGH-1017305C to SLH). We thank Stephen Skerl for editing.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Supplemental Material
Supplemental material may be found here: www.landesbioscience.com/journals/cc/article/ 21732
Footnotes
Previously published online: www.landesbioscience.com/journals/cc/article/21732
References
- 1.Toledo F, Wahl GM. Regulating the p53 pathway: in vitro hypotheses, in vivo veritas. Nat Rev Cancer. 2006;6:909–23. doi: 10.1038/nrc2012. [DOI] [PubMed] [Google Scholar]
- 2.Soussi T, Béroud C. Assessing TP53 status in human tumours to evaluate clinical outcome. Nat Rev Cancer. 2001;1:233–40. doi: 10.1038/35106009. [DOI] [PubMed] [Google Scholar]
- 3.Brown JM, Attardi LD. The role of apoptosis in cancer development and treatment response. Nat Rev Cancer. 2005;5:231–7. doi: 10.1038/nrc1560. [DOI] [PubMed] [Google Scholar]
- 4.Koumenis C, Alarcon R, Hammond E, Sutphin P, Hoffman W, Murphy M, et al. Regulation of p53 by hypoxia: dissociation of transcriptional repression and apoptosis from p53-dependent transactivation. Mol Cell Biol. 2001;21:1297–310. doi: 10.1128/MCB.21.4.1297-1310.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lakin ND, Jackson SP. Regulation of p53 in response to DNA damage. Oncogene. 1999;18:7644–55. doi: 10.1038/sj.onc.1203015. [DOI] [PubMed] [Google Scholar]
- 6.Vousden KH, Prives C. Blinded by the light: the growing complexity of p53. Cell. 2009;137:413–31. doi: 10.1016/j.cell.2009.04.037. [DOI] [PubMed] [Google Scholar]
- 7.Barlési F, Tchouhadjian C, Doddoli C, Villani P, Greillier L, Kleisbauer JP, et al. Gefitinib (ZD1839, Iressa) in non-small-cell lung cancer: a review of clinical trials from a daily practice perspective. Fundam Clin Pharmacol. 2005;19:385–93. doi: 10.1111/j.1472-8206.2005.00323.x. [DOI] [PubMed] [Google Scholar]
- 8.Laptenko O, Prives C. Transcriptional regulation by p53: one protein, many possibilities. Cell Death Differ. 2006;13:951–61. doi: 10.1038/sj.cdd.4401916. [DOI] [PubMed] [Google Scholar]
- 9.Giet R, Petretti C, Prigent C. Aurora kinases, aneuploidy and cancer, a coincidence or a real link? Trends Cell Biol. 2005;15:241–50. doi: 10.1016/j.tcb.2005.03.004. [DOI] [PubMed] [Google Scholar]
- 10.Agnese V, Bazan V, Fiorentino FP, Fanale D, Badalamenti G, Colucci G, et al. The role of Aurora-A inhibitors in cancer therapy. Ann Oncol. 2007;18(Suppl 6):vi47–52. doi: 10.1093/annonc/mdm224. [DOI] [PubMed] [Google Scholar]
- 11.He L, Yang H, Ma Y, Pledger WJ, Cress WD, Cheng JQ. Identification of Aurora-A as a direct target of E2F3 during G2/M cell cycle progression. J Biol Chem. 2008;283:31012–20. doi: 10.1074/jbc.M803547200. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 12.Tanaka M, Ueda A, Kanamori H, Ideguchi H, Yang J, Kitajima S, et al. Cell-cycle-dependent regulation of human aurora A transcription is mediated by periodic repression of E4TF1. J Biol Chem. 2002;277:10719–26. doi: 10.1074/jbc.M108252200. [DOI] [PubMed] [Google Scholar]
- 13.Udayakumar TS, Belakavadi M, Choi KH, Pandey PK, Fondell JD. Regulation of Aurora-A kinase gene expression via GABP recruitment of TRAP220/MED1. J Biol Chem. 2006;281:14691–9. doi: 10.1074/jbc.M600163200. [DOI] [PubMed] [Google Scholar]
- 14.Mao JH, Perez-Losada J, Wu D, Delrosario R, Tsunematsu R, Nakayama KI, et al. Fbxw7/Cdc4 is a p53-dependent, haploinsufficient tumour suppressor gene. Nature. 2004;432:775–9. doi: 10.1038/nature03155. [DOI] [PubMed] [Google Scholar]
- 15.Kufer TA, Silljé HH, Körner R, Gruss OJ, Meraldi P, Nigg EA. Human TPX2 is required for targeting Aurora-A kinase to the spindle. J Cell Biol. 2002;158:617–23. doi: 10.1083/jcb.200204155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Eyers PA, Erikson E, Chen LG, Maller JL. A novel mechanism for activation of the protein kinase Aurora A. Curr Biol. 2003;13:691–7. doi: 10.1016/S0960-9822(03)00166-0. [DOI] [PubMed] [Google Scholar]
- 17.Lim SK, Gopalan G. Aurora-A kinase interacting protein 1 (AURKAIP1) promotes Aurora-A degradation through an alternative ubiquitin-independent pathway. Biochem J. 2007;403:119–27. doi: 10.1042/BJ20061272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Portier N, Audhya A, Maddox PS, Green RA, Dammermann A, Desai A, et al. A microtubule-independent role for centrosomes and aurora a in nuclear envelope breakdown. Dev Cell. 2007;12:515–29. doi: 10.1016/j.devcel.2007.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yu X, Minter-Dykhouse K, Malureanu L, Zhao WM, Zhang D, Merkle CJ, et al. Chfr is required for tumor suppression and Aurora A regulation. Nat Genet. 2005;37:401–6. doi: 10.1038/ng1538. [DOI] [PubMed] [Google Scholar]
- 20.Fujii Y, Yada M, Nishiyama M, Kamura T, Takahashi H, Tsunematsu R, et al. Fbxw7 contributes to tumor suppression by targeting multiple proteins for ubiquitin-dependent degradation. Cancer Sci. 2006;97:729–36. doi: 10.1111/j.1349-7006.2006.00239.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gottifredi V, Karni-Schmidt O, Shieh SS, Prives C. p53 down-regulates CHK1 through p21 and the retinoblastoma protein. Mol Cell Biol. 2001;21:1066–76. doi: 10.1128/MCB.21.4.1066-1076.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yeh PY, Chuang SE, Yeh KH, Song YC, Chang LLY, Cheng AL. Phosphorylation of p53 on Thr55 by ERK2 is necessary for doxorubicin-induced p53 activation and cell death. Oncogene. 2004;23:3580–8. doi: 10.1038/sj.onc.1207426. [DOI] [PubMed] [Google Scholar]
- 23.Yang C, Kaushal V, Haun RS, Seth R, Shah SV, Kaushal GP. Transcriptional activation of caspase-6 and -7 genes by cisplatin-induced p53 and its functional significance in cisplatin nephrotoxicity. Cell Death Differ. 2008;15:530–44. doi: 10.1038/sj.cdd.4402287. [DOI] [PubMed] [Google Scholar]
- 24.Dyson N. The regulation of E2F by pRB-family proteins. Genes Dev. 1998;12:2245–62. doi: 10.1101/gad.12.15.2245. [DOI] [PubMed] [Google Scholar]
- 25.Cooper CS, Nicholson AG, Foster C, Dodson A, Edwards S, Fletcher A, et al. Nuclear overexpression of the E2F3 transcription factor in human lung cancer. Lung Cancer. 2006;54:155–62. doi: 10.1016/j.lungcan.2006.07.005. [DOI] [PubMed] [Google Scholar]
- 26.Adams RR, Carmena M, Earnshaw WC. Chromosomal passengers and the (aurora) ABCs of mitosis. Trends Cell Biol. 2001;11:49–54. doi: 10.1016/S0962-8924(00)01880-8. [DOI] [PubMed] [Google Scholar]
- 27.Carmena M, Ruchaud S, Earnshaw WC. Making the Auroras glow: regulation of Aurora A and B kinase function by interacting proteins. Curr Opin Cell Biol. 2009;21:796–805. doi: 10.1016/j.ceb.2009.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ye X, Nalepa G, Welcker M, Kessler BM, Spooner E, Qin J, et al. Recognition of phosphodegron motifs in human cyclin E by the SCF(Fbw7) ubiquitin ligase. J Biol Chem. 2004;279:50110–9. doi: 10.1074/jbc.M409226200. [DOI] [PubMed] [Google Scholar]
- 29.Meraldi P, Honda R, Nigg EA. Aurora-A overexpression reveals tetraploidization as a major route to centrosome amplification in p53-/- cells. EMBO J. 2002;21:483–92. doi: 10.1093/emboj/21.4.483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Katayama H, Sasai K, Kawai H, Yuan ZM, Bondaruk J, Suzuki F, et al. Phosphorylation by aurora kinase A induces Mdm2-mediated destabilization and inhibition of p53. Nat Genet. 2004;36:55–62. doi: 10.1038/ng1279. [DOI] [PubMed] [Google Scholar]
- 31.Vader G, Lens SM. The Aurora kinase family in cell division and cancer. Biochim Biophys Acta. 2008;1786:60–72. doi: 10.1016/j.bbcan.2008.07.003. [DOI] [PubMed] [Google Scholar]
- 32.Feng Z, Hu W, Rajagopal G, Levine AJ. The tumor suppressor p53: cancer and aging. Cell Cycle. 2008;7:842–7. doi: 10.4161/cc.7.7.5657. [DOI] [PubMed] [Google Scholar]
- 33.Wang W, El-Deiry WS. Restoration of p53 to limit tumor growth. Curr Opin Oncol. 2008;20:90–6. doi: 10.1097/CCO.0b013e3282f31d6f. [DOI] [PubMed] [Google Scholar]
- 34.Gully CP, Velazquez-Torres G, Shin JH, Fuentes-Mattei E, Wang E, Carlock C, et al. Aurora B kinase phosphorylates and instigates degradation of p53. Proc Natl Acad Sci USA. 2012;109:E1513–22. doi: 10.1073/pnas.1110287109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Huang H, Huang SY, Chen TT, Chen JC, Chiou CL, Huang TM. Cisplatin restores p53 function and enhances the radiosensitivity in HPV16 E6 containing SiHa cells. J Cell Biochem. 2004;91:756–65. doi: 10.1002/jcb.10769. [DOI] [PubMed] [Google Scholar]
- 36.Sedletska Y, Giraud-Panis MJ, Malinge JM. Cisplatin is a DNA-damaging antitumour compound triggering multifactorial biochemical responses in cancer cells: importance of apoptotic pathways. Curr Med Chem Anticancer Agents. 2005;5:251–65. doi: 10.2174/1568011053765967. [DOI] [PubMed] [Google Scholar]
- 37.Zhou M, Gu L, Li F, Zhu Y, Woods WG, Findley HW. DNA damage induces a novel p53-survivin signaling pathway regulating cell cycle and apoptosis in acute lymphoblastic leukemia cells. J Pharmacol Exp Ther. 2002;303:124–31. doi: 10.1124/jpet.102.037192. [DOI] [PubMed] [Google Scholar]
- 38.Mao JH, Wu D, Perez-Losada J, Jiang T, Li Q, Neve RM, et al. Crosstalk between Aurora-A and p53: frequent deletion or downregulation of Aurora-A in tumors from p53 null mice. Cancer Cell. 2007;11:161–73. doi: 10.1016/j.ccr.2006.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nair JS, Ho AL, Tse AN, Coward J, Cheema H, Ambrosini G, et al. Aurora B kinase regulates the postmitotic endoreduplication checkpoint via phosphorylation of the retinoblastoma protein at serine 780. Mol Biol Cell. 2009;20:2218–28. doi: 10.1091/mbc.E08-08-0885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Crane R, Gadea B, Littlepage L, Wu H, Ruderman JV. Aurora A, meiosis and mitosis. Biol Cell. 2004;96:215–29. doi: 10.1016/j.biolcel.2003.09.008. [DOI] [PubMed] [Google Scholar]
- 41.Cheng CC, Hsueh CM, Liang KW, Ting CT, Wen CL, Hsu SL. Role of JNK and c-Jun signaling pathway in regulation of human serum paraoxonase 1 gene transcription by berberine in human HepG2 cells. Eur J Pharmacol. 2011;650:519–25. doi: 10.1016/j.ejphar.2010.10.051. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.