Abstract
Two, simple, C5 compounds, dimethylally diphosphate and isopentenyl diphosphate, are the universal precursors of isoprenoids, a large family of natural products involved in numerous important biological processes. Two distinct biosynthetic pathways have evolved to supply these precursors. Humans use the mevalonate route whilst many species of bacteria including important pathogens, plant chloroplasts and apicomplexan parasites exploit the non-mevalonate pathway. The absence from humans, combined with genetic and chemical validation suggests that the non-mevalonate pathway holds the potential to support new drug discovery programmes targeting Gram-negative bacteria and the apicomplexan parasites responsible for causing serious human diseases, and also infections of veterinary importance. The non-mevalonate pathway relies on eight enzyme-catalyzed stages exploiting a range of cofactors and metal ions. A wealth of structural and mechanistic data, mainly derived from studies of bacterial enzymes, now exists for most components of the pathway and these will be described. Particular attention will be paid to how these data inform on the apicomplexan orthologues concentrating on the enzymes from Plasmodium spp.; these cause malaria, one the most important parasitic diseases in the world today.
Keywords: Antimicrobial drug discovery, isoprenoid precursor biosynthesis, malaria, structure-based inhibitor discovery, toxoplasmosis
The medical problem
The Apicomplexa are primitive, unicellular eukaryotes with a complex life cycle that involves a stage as a parasite in a diverse range of animals; for example lizards, birds or mammals. These protozoa share a common apical secretory apparatus that mediates movement and since it is involved in tissue or cell penetration is key to infection. Apicomplexan parasites are the causal agents for a range of important veterinary and human diseases [1]. For example, Eimeria tennella causes coccidiosis, a persistent and problematic disease in the poultry industry, while infection with Babesia species, Theileria annulata and T. parva curtails productivity in cattle herds. Cryptosporidium parvum is a water- and food-borne pathogen of livestock that also, like Babesia sp. infects humans. Toxoplasma gondii is a common parasite of cats that causes a mild disease in healthy humans but, like C. parvum, T. gondii becomes particularly problematic in immunocompromised individuals such as AIDS/HIV patients. The most important members of this family of parasites are the Plasmodium species, which are responsible for malaria, a disease that continues to exact a devastating toll on human populations in the tropics [2].
Vaccination would be an ideal method of dealing with these parasitic pathogens, and is especially appealing with respect to malaria. However, generous long term funding for malaria vaccine development has gained little reward. One reason is that Plasmodium spp. infect and shelter from the immune system within host cells and display antigenic variation, in their membrane-bound surface proteins [3]. This capability prolongs parasite circulation in the blood, increasing the likelihood of transmission and helps immune system evasion.
There are suitable drugs for some of these diseases but finances control what happens to infected animals or those at risk and it is often cheaper to cull than treat. In some parts of the world treatment is not even an option. With respect to human diseases then compounds such as pyrimethamine are used to treat toxoplasmosis and malaria, and the introduction of artemisinin a boon for control of malaria [4, 5]. However, issues that interfere with prevention and cure include the cost of goods and lack of healthcare infrastructure. The problem is compounded by the emergence of drug resistant forms of Plasmodium [5, 6], in part a consequence of inappropriate drug use. This means it is important to search out new treatments as one aspect of fighting malaria. In addition since approximately 10 % of emerging human diseases are due to parasites there is great encouragement to progress antiparasitic drug development [7].
The discovery of our current arsenal of antimalarial drugs owes much to colonialism and warfare. Now, advances in a number of scientific disciplines means that we can now couple access to extensive genomic information on the apicomplexan protists [e.g. 8; http://plasmodb.org/plasmo/], with improved understanding of their biology and of drug action. These data provide opportunities to apply medicinal chemistry approaches to modify known and successfully exploited chemical scaffolds to derive new drugs against already proven targets but in addition serve to map out potential new targets that might be exploited for structure based drug discovery [9]. It is the later area that is of interest here.
The ideal drug target in a protozoan parasite is one that provides a biological function required for survival or infectivity, that is unique to the parasite and absent from the mammalian host or sufficiently divergent that species selective inhibition is possible. For practical reasons the types of molecules that are ultimately sought must be stable, orally bioavailable, and cheap to manufacture. The molecules should have high affinity for one or more targets in the parasite, kill the pathogen quickly and should present little or no toxicity against humans. Oral bioavailability is a priority since it provides the practical benefit of using tablets in developing countries, and optimizes chances of treating parasites occupying an intracellular niche or which infect the central nervous system. The Lipinski guidelines addressing issues of bioavailability [10] are particularly relevant to antiparasite drug research. For protozoan parasites, drug research is influenced by the requirement for selectivity for one eukaryotic cell (the parasite) over another (the host) and in this respect the research is more similar to anticancer than antibacterial drug research.
Proven and new drug targets
Several areas of apicomplexan biology have already proven useful or attracted attention with respect to drug discovery. These include folate metabolism, digestion of hemoglobin, farnesylation and fatty acid biosynthesis and there are ongoing projects in those areas [11]. Of particular note was the recent discovery that the apicomplexa, with the exception of Cryptosporidium spp., possess a specialized non-photosynthetic plastid-like organelle called the apicoplast [12-15]. The organelle, which is essential for parasite survival, has a small circular genome that encodes a few genes. The majority of the apicoplast proteome is however nuclear encoded and an N-terminal leader sequence targets proteins into the organelle. The metabolic processes that occur in the apicoplast are primarily prokaryotic and typical of plastids; they include type II fatty acid and heme biosynthesis. These processes are distinct or absent from those found in mammals suggesting potential for therapeutic intervention. Indeed some drugs that are actually used to treat parasitic infections, for example ciprofloxacin, doxycycline and clindamycin act by inhibiting the apicoplast-located DNA gyrase and the translation machinery [4, 16, 17]. Also localized to the apicoplast is isoprenoid precursor biosynthesis [18, 19].
The simple C5 compounds isopentenyl pyrophosphate (IPP) and dimethylallyl pyrophosphate (DMAPP) are the universal precursors of isoprenoids, a large family of natural products that include geranyl, farnesyl, dolichols, sterols, ubiquinone, the prenyl groups used to modify proteins and isopentenylated tRNA’s [20, 21]. The diversity of isoprenoids support numerous important biological functions including respiration and photosynthesis, hormone-based signaling, the regulation of transcription and the post-translational processes that control lipid and glycoprotein biosynthesis, protein cleavage and degradation, meiosis and apoptosis. In addition, isoprenoids are structural components of membranes [20].
The mevalonate route to IPP/DMAPP
For many years it was assumed that the mevalonate (MVA) pathway, named after one of the metabolites used in the pathway, was the sole route to IPP and DMAPP. This pathway supplies the IPP and DMAPP in most eukaryotes (all mammals), archaea, a few eubacteria, the cytosol and mitochondria of plants, fungi and in the eukaryotic parasites Trypanosoma and Leishmania [22, 23]. The first stage is the production of acetoacetyl-CoA from acetyl-CoA in a reaction catalyzed by thiolase. Acetoacetyl-CoA is converted to 3-hydroxy-3-methylglutaryl-CoA (HMG-CoA) by HMG-CoA synthase and HMG-CoA reductase then converts the CoA derivative to (R)-MVA. Next, (R)-MVA is phosphorylated to (R)-MVA 5-diphosphate by firstly mevalonate kinase (MVK) then phosphomevalonate kinase (PMK). Decarboxylation by mevalonate diphosphate decarboxylase (MDD) produces IPP some of which is isomerized to DMAPP by an IPP isomerase. The MVA pathway is directly relevant to human health and inhibition of HMG-CoA reductase by statins controls sterol production and lowers blood pressure, and helps in the treatment of cardiovascular disease and inflammatory processes [24].
The non-mevalonate route to IPP/DMAPP
A second biosynthetic route to IPP and DMAPP has more recently been elucidated [18, 19, 25]. This pathway is called the non-mevalonate route or alternatively named after two intermediates, the 1-deoxy-D-xylulose-5-phosphate (DOXP) or 2C-methyl-D-erythritol-4-phosphate (MEP) pathway. A finding that sparked interest in the non-mevalonate route was the observation that the antibacterial drug fosmidomycin displayed activity against P. falciparum [26]. This pathway occurs in plant chloroplasts, algae, cyanobacteria, eubacteria, and apicomplexan parasites [16]. Characterization of this pathway is one of the best examples of modern proteomics research, driven by combination of genomic data and biological chemistry. Researchers have exploited Nuclear Magnetic Resonance (NMR) methodology to track and map out substrates and products, enzyme-assisted synthesis to acquire reagents necessary to characterise pathway components and crystallography to provide structural detail that complements detailed enzymatic studies [19]. The non-mevalonate pathway in bacteria consists of eight reactions catalyzed by nine enzymes. Each stage will be detailed with a description of what has been gleaned about enzyme structure, mechanism and reactivity. Studies designed to discover inhibitors of these enzymes are in their infancy but some early results are available and will be mentioned.
The non-mevalonate pathway enzymes are present in all intraerythrocytic stages of P. falciparum [27] and information about these enzymes will be given since this is both the most important apicomplexan parasite and can be taken as representative of the family. However, the apicomplexan enzymes are in general poorly characterized, in part due to difficulties in obtaining recombinant expression of soluble enzymes from Plasmodium spp. or indeed other apicomplexans. Most progress has been made studying the enzymes derived from bacterial sources with information derived from amino acid sequence and structure-based comparisons informing about the apicomplexan orthologues. Such comparisons will form part of the discussion.
Target validation
The non-mevalonate pathway is absent from humans so this should help in the search for compounds that kill parasites but which have little effect on human biological processes. The non-mevalonate enzymes constitute metabolic chokepoints i.e. they uniquely consume substrates or generate specific products and their function is not compensated for by another enzyme [28]. Genes encoding the component enzymes of the non-mevalonate pathway have been proven essential for survival in bacteria [29] and in one case that will be described, in P. falciparum itself [30]. Genetic approaches to characterize essentiality of a gene should be complemented by chemical validation, i.e. the use of specific small molecule inhibitors that elicit a biological effect. Two pieces of data suggest chemical validation of isoprenoid precursor biosynthesis as a drug target. First, fosmidomycin, a potent inhibitor of the enzyme 1-deoxy-D-xylulose-5-phosphate reductoisomerase, the second enzyme in the non-mevalonate pathway has been used in the clinic as an antibacterial and displays antimalarial properties [26]. Further details of this system will be described. Secondly, inhibitors of protein farnesylation, a process dependent on the supply of DMAP/IPP, also display anti-parasitic properties [31] highlighting the importance of isoprenoid metabolism to parasite survival.
Having put the non-mevalonate biosynthetic route to isoprenoid precursors into context, the different stages of the pathway will now be described.
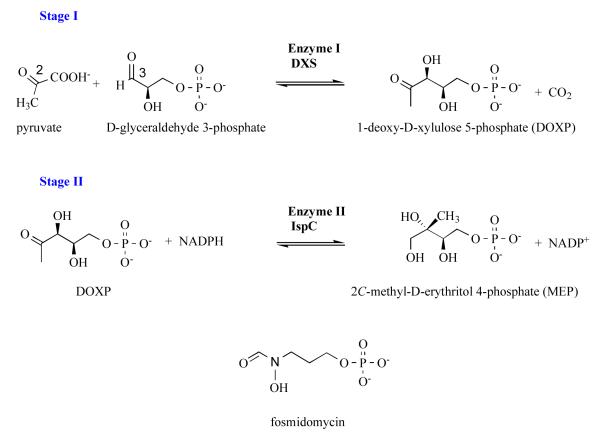
Stage I: 1-deoxy-D-xylulose 5-phosphate synthase (DXS, EC 2.2.1.7)
The pathway starts with production of DOXP following the acyloin condensation of pyruvate and glyceraldehyde 3-phosphate, at C2 and C3 respectively (Figure 1), by 1-deoxy-D-xylulose 5-phosphate synthase (DXS) in a thiamine-dependent manner. Structures of Escherichia coli and Deinococcus radiodurans DXS have been determined [32]. The DXS subunit is formed by three domains similar to the equivalent domains found in thiamine-dependent enzymes although the relative position of the domains varies. The active site of the dimeric DXS is created around the cofactor thiamine diphosphate which is buried within a single subunit, placing the thiazolium group at the base of a polar cavity. The key residues that activate the cofactor have been recognised. An active site histidine is likely involved in proton transfer during catalysis and an arginine that contributes to glyceraldehyde 3-phosphate binding have been noted. There are 25 amino acids that contribute to the formation of the bacterial DXS active site by virtue of binding cofactor, creating the polar substrate binding site and providing the functional groups to assist catalysis [32]. Of, these 22 are strictly conserved in the P. falciparum DXS sequence (gene id: MAL13P1.186) and the three differences are conservative; a methionine is substituted for a valine, isoleucine for leucine and serine for threonine (data not shown). This indicates a high level of conservation between the bacterial and apicomplexan DXS in and around the active site. The P. falciparum enzyme is predicted to be much larger consisting of a subunit of approximate mass 140 kDa compared to the bacterial orthologues with typical mass of around 68 kDa per subunit. The P. falciparum DXS carries the apicoplast targetting signal peptide and also shows, in common with many other Plasmodium sequences, several asparagine rich segments.
Figure 1.
The non-mevalonate pathway; stages I and II and the structure of fosmidomycin, a known inhibitor of IspC.
Stage II: 1-deoxy-D-xylulose-5-phosphate reductoisomerase (IspC, EC 1.1.1.267)
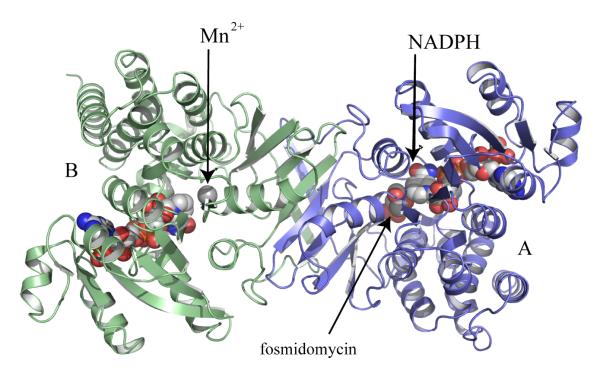
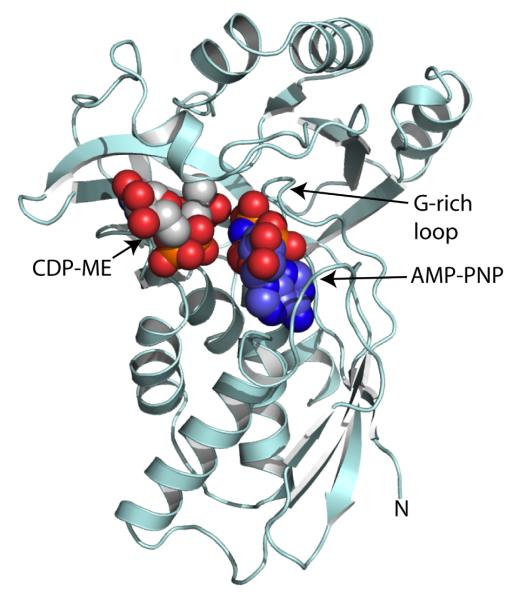
The enzyme that catalyses stage II has been pivotal to analysis of this pathway with the discovery that it is a target for fosmidomycin [26]. IspC catalyzes the conversion of DOXP to MEP (Figure 1). IspC is a homodimer (Figure 2) with a subunit of three domains, an N-terminal cofactor binding domain, a central domain carrying many of the active site residues together with a flexible segment that acts as a lid or gate for the catalytic center, and a C-terminal α-helical domain. Studies of E. coli IspC with Mn2+, NADPH, fosmidomycin and other inhibitors in conjunction with additional structures, e.g of the enzymes from Mycobacterium tuberculosis and Zymomonas mobilis have advanced our understanding of structure and reactivity [33-40].
Figure 2.
Ribbon diagram of M. tuberculosis IspC [40]. Subunits are colored pale blue and green respectively. The cofactor, NADPH, divalent cation and inhibitor fosmidomycin, which is present in one active site, are depicted as van der Waals spheres. Molecular graphics were prepared using PyMol [73].
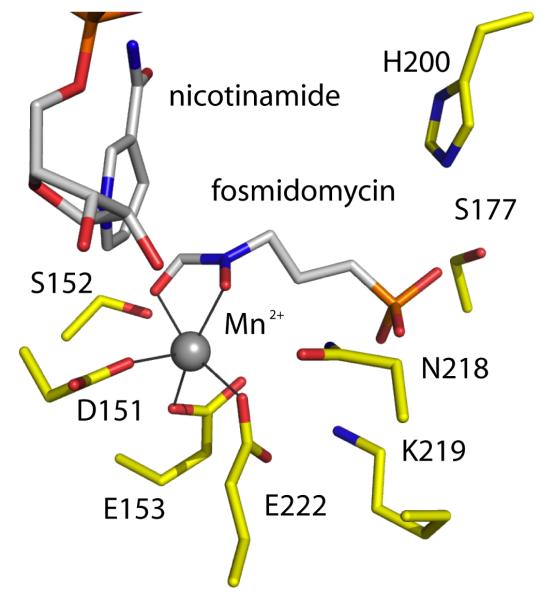
IspC is a class B dehydrogenase using the NADPH proS hydride during catalysis. The mechanism is ordered sequential with cofactor binding first followed by the substrate DOXP. A divalent cation binds, orients and polarizes the substrate and reaction intermediate. Two mechanisms have been suggested for the initial isomerization that converts DOXP to a methylerythrose intermediate, an α-ketol rearrangement or a retro-adol/aldol reaction. The intermediate is then reduced by the hydride from NADPH. Fosmidomycin contains a hydroxamate moiety (Figure 1) and inhibits by coordinating the metal ion to mimic the substrate. The phosphate group provides a tether for the inhibitor (or substrate) at the catalytic center (Figure 3). Genetic analysis has shown that in P. falciparum this reductoisomerase activity is required for intraerythrocytic development of the parasite [30]. This observation is crucial and correlates with the antiparasitic properties of fosmidomycin. Crystallization of a truncated P. falciparum IspC (gene id PF14_0641), in which the apicoplast targeting peptide has been omitted, has recently been reported [41] and the structural results are eagerly awaited. The Plasmodium and bacterial orthologues share almost 40 % sequence identity with, as observed for DXS, very high conservation of residues in and around the active site. Consider the eight amino acid residues of the M. tuberculosis IspC shown to bind the inhibitor fosmidomycin and the divalent cation required for function (Figure 3). All eight residues are strictly conserved in the E. coli and P. falciparum enzymes and this conservation of key residues extends to those involved in binding the NADPH cofactor (not shown).
Figure 3.
The fosmidomycin binding site in M. tuberculosis IspC. Atoms are colored; C of IspC yellow, of the cofactor and inhibitor grey; P orange, O red, N blue. The five-coordinate manganese (II) is depicted as a grey sphere with coordinating groups linked by a dark line.
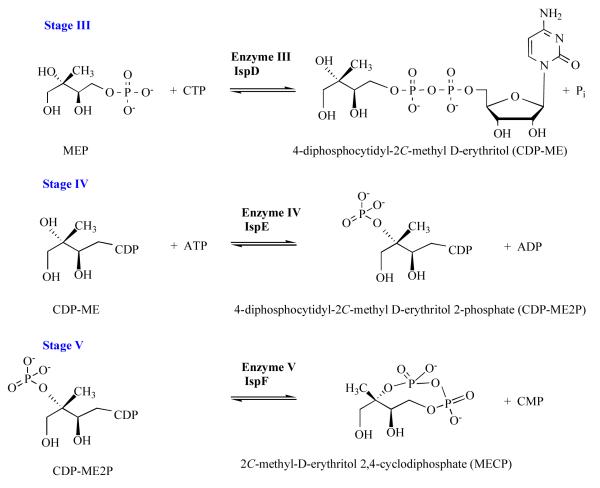
Stage III: 4-diphosphocytidyl-2C-methyl-D-erythritol cytidylyltransferase (IspD, EC 2.7.7.60)
The central stages of the pathway, III-V, utilize cytidine-containing substrates (Figure 4). In stage III the cytidylyltransferase IspD catalyses the reaction of MEP with CTP to produce 4-diphosphocytidyl-2C-methyl-D-erythritol (CDP-ME) and pyrophosphate. A comprehensive study of the dimeric IspD and dissection of the E. coli enzyme structure-mechanism relationship has been carried out exploiting the powerful combination of informative structures and steady state kinetic analyses [42, 43].
Figure 4.
The non-mevalonate pathway; stages III to V.
The enzyme subunit displays similarities to the nucleotide-binding Rossmann fold, primarily a single domain α/β structure constructed around a seven-stranded twisted β-sheet, into which is inserted an extended “β-arm”. Two β-arms associate by hydrogen bonding, salt bridge interactions and van der Waals interactions to create the dimer interface where the active site is located. Seven polypeptide segments, six from one subunit and one from the partner contribute to the active site. The study of a plant IspD, the Arabidopsis thaliana enzyme, identified alterations in the alignment of subunits with respect to each other and highlighted the role that such adjustments might contribute to enzyme activity [44]. An ordered sequential mechanism applies with CTP, as an ion pair with Mg2+, binding first followed by MEP. A highly basic active site binds and the four phosphates present on the substrates. Two lysine residues (Lys27 and Lys213 in E. coli IspD) are critical to stabilize a pentavalent transition state that results following an in-line nucleophilic attack by the MEP phosphate on the CTP α-phosphate. A hydrogen bond network serves to position MEP in support of catalysis. Additional structures of E. coli [45], Thermatoga maritima, Neisseria gonorrhea IspD and the bifunctional IspDF of Camphylobacter jejuni are published [46] or deposited in the Protein Data Bank (PDB).
The P. falciparum enzyme (gene id: PFA0340w) is predicted to have a mass of 88.2 kDa, very much bigger than the typical IspD enzymes, which are around 25-28 kDa. The parasite protein contains a large number of insertions compared to the bacterial orthologues in addition to the apicoplast N-terminal targeting sequence, and this complicates comparisons. Nevertheless, amino acid alignments suggest that the basic residues in the IspD active site, in particular the E. coli IspD Lys27/Lys213 combination are strictly conserved.
Stage IV: 4-diphosphocytidyl-2C-methyl-D-erythritol kinase (IspE, EC 2.7.1.148)
The ATP-dependent IspE is a member of the GHMP kinase superfamily so called after the founding members namely Galacto, Homoserine, Mevalonate and Phosphomevalonate kinases [47]. A dead-end ternary complex of E. coli IspE:CDP-ME and the non-hydrolysable ATP derivative adenosine 5′-(β,γ-imino) triphosphate (AMP-PNP) has been instrumental in delineating the features important for substrate recognition and enzyme mechanism [47]. In addition structures of another crystal form of that orthologue, together with structures of the enzyme from Thermus thermophilus and Aquifex aeolicus are published [48-50].
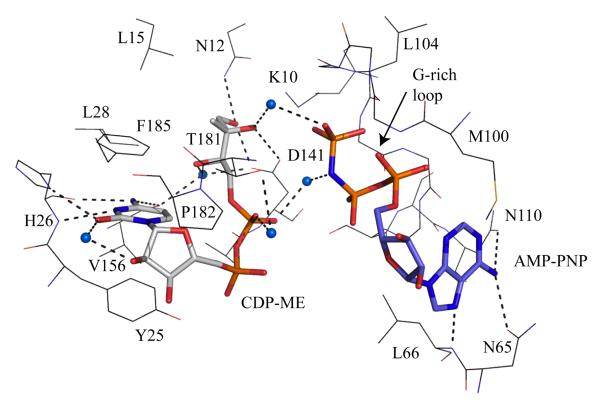
The enzyme subunit displays the characteristic GHMP kinase α/β fold (Figure 5), arranged into a cofactor and a substrate-binding domain. Structural comparisons with other GHMP kinases indicate that the core of each domain is highly conserved with differences primarily restricted to the substrate-binding and purine-binding pockets. It is intriguing that the IspE fold resembles that of three enzymes in the mevalonate pathway, namely MVK, PMK and MDD. The catalytic center is located in a deep polar cavity near the interface of the cofactor and substrate-binding domains. Here, in E. coli IspE, a lysine-aspartate pair (Lys10 and Asp141) form hydrogen bonds with substrate and polarize the hydroxyl group O2M (Figure 6) to facilitate proton abstraction with Asp141 acting as a general base to generate a nucleophile. The cofactor γ-phosphate is positioned to accept a nucleophilic attack from CDP-ME O2M likely with an associative in-line mechanism to form a pentacovalent intermediate, and subsequently production of 4-diphosphocytidyl-2C-methyl-D-erythritol-2-phosphate (CDP-ME2P) and ADP. It is noteworthy that the cofactor purine forms hydrogen-bonding interactions that stabilize the uncommon syn orientation of the base with respect to the glycosidic bond. The vast majority of ATP-dependent enzymes bind the purine in anti conformation and this suggests opportunities for inhibition of IspE at the ATP binding as well as at the substrate-binding site. Assay development has progressed [51, 52] such that screening of compound libraries can be carried out and the first novel inhibitors of this enzyme published [53, 54].
Figure 5.
Ribbon diagram of E. coli IspE [47]. The cofactor and substrate are depicted as van der Waals spheres colored with C of ATP blue, C of substrate grey, O red, N blue, P orange.
Figure 6.
The active site of E. coli IspE [47]. The IspE residues are shown as lines, the substrate and AMP-PNP as sticks using the same color scheme employed in Figure 6. Water molecules are blue spheres. Dashed lines represent potential hydrogen bonds.
The P. falciparum enzyme (gene id: PFE0150c) has a predicted mass of 63 kDa, much greater then the bacterial orthologues, which are around 31 kDa. The key residues involved in forming the E. coli IspE active site are shown in Figure 6 [47] and sequence comparisons indicate a high level of conservation with the parasite sequence. Two residues important for catalysis namely Lys10 and Asp141 are conserved, as are the Asn65/Asn110 combination that binds the syn adenine. In addition the G-rich loop that binds the ATP phosphate tail and Tyr25, which stacks on the cytidine moiety of substrate are strictly conserved. In E. coli IspE, His26 froms hydrogen bonds from main chain and side chain to the cytidine and in the P. falciparum sequence this is changed to an asparagine, a conservative difference.
Stage V: 2C-methyl-D-erythritol-2, 4-cyclodiphosphate synthase (IspF, EC 4.6.1.12)
IspF is a small trimeric enzyme and crystal structures of orthologues from E. coli, Haemophilus influenzae, Shewanella oneidensis, T. thermophilus, Burkholderia pseudomallei and the bifunctional IspDF of C. jejuni have been determined with a number of ligand complexes reported [e.g. 55-60] or deposited in the PDB.
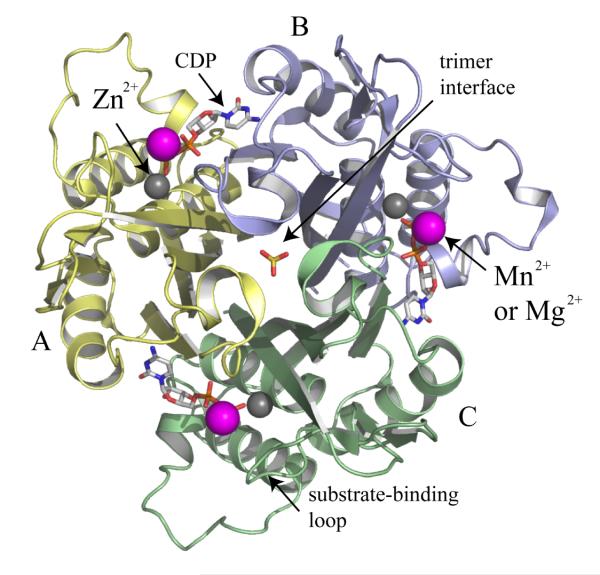
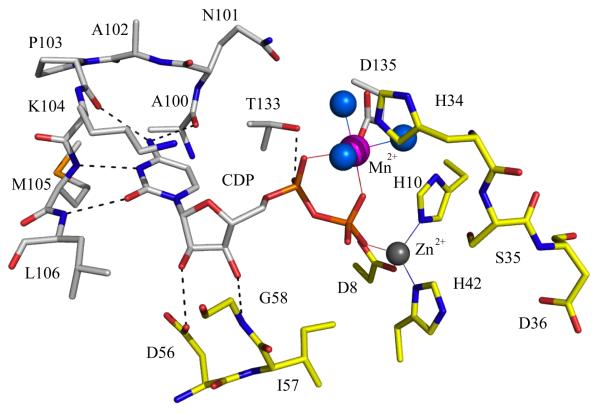
The IspF subunit is a small single α/β domain with a four-stranded mixed β-sheet on one side of the molecule and three α-helices on the other. Three subunits form a compact bell-shaped assembly approximately 45 Å by 60 Å in the axial and equatorial dimensions respectively (Figure 7). Each active site is constructed by contributions from two adjacent subunits to provide the residues, which recognize, bind and orient the nucleotide substrate in the active site. Here, Zn2+ displays tetrahedral coordination to a pair of histidines, an aspartate and either a water molecule or, if substrate is present, a phosphate (Figure 8). A second divalent cation (Mn2+ or Mg2+) with octahedral coordination is positioned between α and β-phosphates [55].
Figure 7.
Ribbon diagram of the trimeric E. coli IspF [55]. The subunits are colored pale yellow, blue and green. CDP is shown as a stick model colored C grey, O red, N blue, P orange and the metal ions Mn2+ and Zn2+ depicted as magenta and grey spheres respectively. A phosphate group occupies the entrence to the trimer interface.
Figure 8.
The active site of E. coli IspF [55]. A similar color scheme as in Figure 7 is used and enzyme residues and CDP are depicted as sticks. Water molecules are blue spheres, potential hydrogen bonds are depicted as dashed lines and metal ion coordination is shown by thin continuous lines. The segment comprising His34, Ser35, Asp36 is involved in binding the ME2P part of the substrate CDP-MEP, and is strictly conserved in all IspF sequences.
The enzyme requires both divalent cations for catalysis, one of which (Zn2+), is always present in the active site, the other presumably brought in with the substrate. Both metal ions contribute to the precise alignment of α and β-phosphates and as suitable Lewis acids to polarize the phosphate groups. An associative in-line mechanism is most likely with, as the first step, nucleophilic attack by the terminal phosphate of CDP-ME2P on the β-phosphate. This generates a pentacoordinate transition state that is also stabilized by coordination with the metal ions, and this collapses to release CMP and 2C-methyl-D-erythritol-2, 4-cyclodiphosphate (MECP).
The P. falciparum enzyme (gene id: PFB0420w) has a predicted mass of 27 kDa whilst E. coli IspF has a mass of 17 kDa. A truncated version of the P. falciparum enzyme, in which the apicoplast targeting sequence was removed, has been studied after refolding of the inclusion bodies produced in E. coli expression system [61]. The structure of IspF from P. vivax has been deposited in the PDB by a structural genomics consortium (PDB code 3B6N). Although the quality of this structure leaves something to be desired it highlights that, as predicted, bacterial IspF provides a suitable template for the enzyme from protozoan sources [55]. The coordination of the metal ions and recognition of substrate in E. coli IspF involves some 17 residues (Figure 8) of which 15 are strictly conserved in the P. falciparum sequence [50]. The two differences occur at the cytidine-binding site where Met105 and Leu106 are changed to an isoleucine and a serine in P. falciparum IspF. Since the interaction with substrate involves hydrogen bond donation from the main chain amides (Figure 8) then the nature of the side chain is not critical.
Inhibitors of E. coli IspF have been identified by virtual screening and molecular modeling methods [60]. These inhibitors bind copying the interactions formed between the cytidine moiety of substrate and the enzyme but these molecules do not yet display the strength of binding required to support a program of optimization and it is likely that further screening to find better starting points may be required. That the enzyme is dependent on a Zn2+ ion offers scope to exploit metal chelating groups to gain affinity.
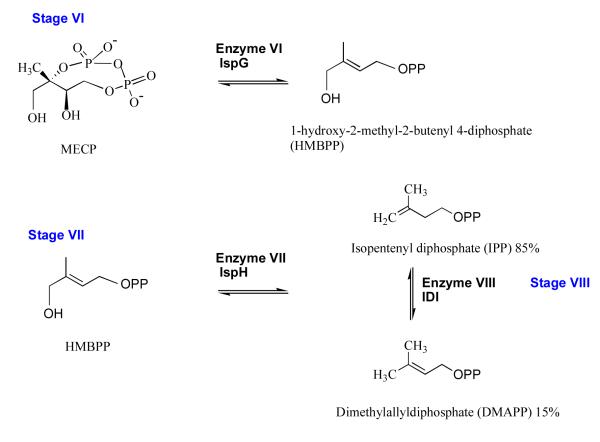
Stage VI: 1-hydroxy-2-methyl-2-(E)-butenyl-4-diphosphate synthase (IspG, EC 1.17.4.3)
The cyclodiphosphate MECP undergoes a two-electron reduction and elimination to form 1-hydroxy-2-methyl-2-butenyl-4-diphosphate (HMBPP) in a reaction catalyzed by IspG (formerly known as GcpE, Figure 9). The putative assignment of the gene encoding this protein in P. falciparum (gene id: PF10_0221), and other apicomplexans, has been made but there is no characterization of the enzyme from parasites and no crystal structure available for any IspG orthologue. The enzyme possesses a 4Fe-4S cluster [62, 63] and database mining of the PDB using the amino acid sequence of the P. falciparum orthologue identifies a short 70-residue segment of sulfite reductase (code 1AOP), which displays 33% identity. Interestingly, this segment is placed between the heme cofactor and 4Fe-4S cluster used by sulfite reductase [64].
Figure 9.
The non-mevalonate pathway; stages VI to VIII.
Stage VII: 1-hydroxy-2-methyl-2-(E)-butenyl-4-diphosphate reductase (IspH, EC 1.17.1.2)
IspH, previously known as lytB, catalyzes production of IPP (85%) and some DMAPP (15%) from the pool of HMBPP (Figure 9). Like IspG, IspH contains a 4Fe-4S cluster and is oxygen sensitive. Under anaerobic conditions, catalysis is supported by a combination of NADPH, flavodoxin reductase and flavodoxin supplying reducing equivalents. Recent efforts have resulted in structures of IspH from E. coli [65] and A. aeolicus [66]; complex structures in particular that have helped to delineate aspects of specificity and mechanism. The enzyme forms a clover-like structure consisting of three domains of similar structure with the active site at the core of the protein. Here the iron-sulfur cluster, which provides reducing capacity, is held in a polar cavity. A proposed mechanism involves water-assisted catalysis with a proton relay system, and with the substrate itself providing a proton via the β-phosphate. The gene encoding the putative IspH of P. falciparum has been identified, (gene id: PFA0225w) and again, as seen in every other case the predicted mass (63 kDa) significantly exceeds that of the bacterial orthologues (E. coli IspH mass 35 kDa). The amino acid sequence identity with the E. coli and A. aeolicus proteins is around 32 %. Of note are eight key residues, in addition to the three iron coordinating cysteines, involved in binding the substrate and providing the functional groups to support catalysis [65] and these are all strictly conserved in the P. falciparum sequence (data not shown). The need to maintain anaerobic conditions to assay IspH could complicate the use of this protein as a target for high throughput screening.
Stage VIII: Isopentenyl diphosphate isomerase (IDI, EC 5.3.3.2)
Isomerization or 1,3-allylic rearrangement of the IPP C=C bond to produce DMAPP creates an electrophile that supports alkylation reactions downstream in the biosynthetic routes that branch off from the non-mevalonate route and provision of chemical diversity (Figure 9). Two, distinct IPP isomerases, IDI-I and -II maintain a supply of DMAPP. IDI-I is dependent on divalent cations for activity and structures of E. coli and human enzymes have been determined [67]. IDI-I is a small, compact globular α/β-protein with a divalent cation (Mn2+ or Mg2+) helping to create a polar active site where three important residues (Cys67, Glu116 and Tyr104 in E. coli IDI-I [68]) participate in protonation and deprotonation of the isoprenoid unit. IDI-II adopts the triosephosphate isomerase-type α/β8 fold, and oligomerizes to a cage-like octamer. This enzyme, which is restricted to the archaea and some eubacteria, requires a metal ion, flavin mononucleotide and, under aerobic conditions NADPH although their contributions to catalysis remains unclear. The plant A. thaliana possesses two isoforms of IDI-I but searches of the P. falciparum and T. gondii genomic data using those sequences and also bacterial sequences have not revealed any IDI orthologues. The gene may have been lost from the Apicomplexa and the isomerase activity lacking or a completely different enzyme catalyzes the reaction. It is tempting to speculate that the mixed pool of IPP/DMAPP resulting from IspH activity may suffice for parasite needs.
How is the non-mevalonate pathway regulated?
There are different mechanisms to regulate a biosynthetic pathway. This can depend on the levels of products of reactions catalyzed within the pathway or on the presence of effector molecules. IspF binds phosphate, IPP or DMAPP, geranyl pyrophosphate and farnesyl pyrophosphate in a well-conserved hydrophobic core created by trimer formation [69]. These ligands, with the exception of phosphate, are synthesized by enzymes two, three or four steps downstream of IspF [70]. This may be coincidence but raises the possibility of feedback regulation of isoprenoid biosynthesis with IspF as a point of control although there are no kinetic data to support this idea.
Covalent modification, for example phosphorylation of HMG-CoA reductase in the MVA pathway [22], can regulate enzyme activity and influence a pathway or activity can be regulated at the level of gene expression. In Gram-positive bacteria the genes encoding enzymes of the MVA pathway form operons and are likely regulated at the level of transcription. The genes encoding the non-mevalonate pathway enzymes are dispersed throughout individual genomes with no evidence of a transcriptional regulator to coordinate expression. However, ispD and ispF are transcriptionally coupled or in a few organisms fused, encoding a bifunctional enzyme. IspDF is therefore unusual since, unlike most bifunctional enzymes which catalyze consecutive reactions, it catalyses non-consecutive steps. The crystal structure of the hexameric C. jejuni IspDF has been determined and associations of bifunctional and monofunctional enzymes (IspD-E-F) have been observed [46, 71]. A complex comprising three IspD and IspE dimers together with two IspF trimers creates an assembly localizing 18 catalytic centers. With a shorter distance over which diffusion, of product from one active site to become substrate in another, takes place this could support efficient catalysis of three reactions at the core of isoprenoid precursor biosynthesis. There is however, no evidence for enhanced catalytic rates on complex formation or any proof of substrate channeling [71].
The compartmentation of enzymes in organs or organelles can influence pathways and we note that with respect to plants the MVA pathway occurs in the cytosol and mitochondria, whilst the non-mevalonate pathway is compartmentalized into the chloroplasts. In the Apicomplexa the predicted proteins are all nuclear encoded but carry an apicoplast-targeting signal peptide. The inference is that the relevant enzymes are transported into and are functional within the apicoplast suggesting that compartmentation makes a contribution to the pathway activity.
There may not be a single mode of control of the non-mevalonate pathway and distinct mechanisms might be employed by different organisms. However, an understanding of how the non-mevalonate pathway is regulated, if indeed it is, might assist identifying where therapeutic attack would provide most benefit.
Concluding remarks
In theory, the enzymes of the non-mevalonate pathway present attractive new targets for the development of broad-spectrum antimicrobial drugs and can be considered to be chemically and genetically validated for drug discovery. They are enticing with respect to targeting important diseases that result from infection with apicomplexan parasites in particular malaria. However, one complicating issue with respect to the apicomplexans is that the enzymes reside in the apicoplast. To reach the enzyme targets small molecules would have to be able to enter the parasite and then cross the organelle membranes. This may prove difficult. Most information on the constituent enzymes has been derived from studies on bacterial orthologues and there have been difficulties in getting samples of some parasite enzymes for study. In any case, sequence-structure comparisons indicate that the key residues involved in substrate or cofactor binding and that contribute to catalysis are strictly conserved across bacterial and apicomplexan species and therefore the bacterial orthologues represent suitable templates to be exploited in screening and structure-based approaches to drug development. A structure-based approach to inhibitor discovery is highly complementary to high-throughput screening of compound libraries. For such screening it is critical to have an appropriate assay and suitable screening formats for non-mevalonate pathway enzymes are reported [51, 52]. This last factor is critical because although the indicators are promising (the biology suggests these are good targets) these enzymes should only be considered potential targets for drug discovery and their suitability in this regard should be reconsidered after we know the outcome of large scale, real and virtual, compound library screening. It is this type of data that, together with structural studies, can inform on the druggability of these targets (i.e. what does the chemistry of the active site indicate?). Ultimately it is the properties of the small molecule inhibitors that will dictate progress or lack of it in drug discovery targeting the non-mevalonate pathway.
Acknowledgements
The author is funded by The Wellcome Trust [grants WT082596, WT083481], the Biotechnology and Biological Sciences Research Council (Structural Proteomics of Rational Targets) and the European Union [FP7 Health-223461].
Abbreviations
- AIDS/HIV
acquired immune deficiency syndrome / human immunodeficiency virus
- AMP-PNP
adenosine 5′-(β,γ-imino)triphosphate
- CDP-ME
4-diphosphocytidyl-2C-methyl-D-erythritol
- CDP-ME2P
4-diphosphocytidyl-2C-methyl-D-erythritol-2-phosphate
- DMAPP
dimethylallyl diphosphate
- DOXP
1-deoxy-D-xylulose 5-phosphate; 3-hydroxy-3-methylglutaryl-CoA
- DXS
1-deoxy-D-xylulose 5-phosphate synthase
- GHMP
galactose kinase, homoserine kinase, mevalonate kinase, phosphomevalonate kinase
- HMBPP
1-hydroxy-2-methyl-2-butenyl 4-diphosphate
- HMG-CoA
3-hydroxy-3-methylglutaryl-CoA
- IDI
isopentenyl diphosphate isomerase
- IPP
isopentenyl diphosphate
- MDD
mevalonate 5-diphosphate decarboxylase
- MECP
2C-methyl-D-erythritol-2,4-cyclodiphosphate
- MEP
2C-methyl-D-erythritol-4-phosphate
- MVA
mevalonate
- MVK
mevalonate kinase
- NMR
Nuclear Magnetic Resonance
- PMK
phosphomevalonate kinase
- PDB
Protein Data Bank
Footnotes
Proteins are designated as DXS, IspC-D-E-F-G-H, IDI-I, and II as defined in the text.
References
- [1].Levine ND. Progress in taxonomy of the apicomplexan protozoa. J. Protozol. 1988;35:518–520. doi: 10.1111/j.1550-7408.1988.tb04141.x. [DOI] [PubMed] [Google Scholar]
- [2].Snow RW, Guerra CA, Noor AM, Myint HY, Hay HI. The global distribution of clinical episodes of Plasmodium falciparum malaria. Nature. 2005;434:214–217. doi: 10.1038/nature03342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Kyes SA, Kraemer SM, Smith JD. Antigenic variation in Plasmodium falciparum: gene organization and regulation of the var multigene family. Eukaryot. Cell. 2007;6:1511–1520. doi: 10.1128/EC.00173-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Lalloo DG, Shingadia D, Pasvol G, Chiodini PL, Whitty CJ, Beeching NJ, Hill DR, Warrell DA, Bannister BA. UK malaria treatment guidelines. J. Infect. 2007;54:111–121. doi: 10.1016/j.jinf.2006.12.003. [DOI] [PubMed] [Google Scholar]
- [5].Na-Bangchang K, Karbwang J. Current status of malaria chemotherapy and the role of pharmacology in antimalarial drug research and development. Fund. Clin. Pharm. 2009;23:387–409. doi: 10.1111/j.1472-8206.2009.00709.x. [DOI] [PubMed] [Google Scholar]
- [6].Dondorp AM, Yeung S, White L, Nguon C, Day NP, Socheat D, von Seidlein L. Artemisinin resistance: current status and scenarios for containment. Nat. Rev. Microbiol. 2010;8:272–280. doi: 10.1038/nrmicro2331. [DOI] [PubMed] [Google Scholar]
- [7].Jones KE, Patel NG, Levy MA, Storeygard A, Balk D, Gittleman JL, Daszak P. Global trends in emerging infectious diseases. Nature. 2008;451:990–993. doi: 10.1038/nature06536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Aurrecoechea C, Heiges M, Wang H, Wang Z, Fischer S, Rhodes P, Miller J, Kraemer E, Stoeckert CJ, Jr, Roos DS, Kissinger JC. ApiDB: integrated resources for the apicomplexan bioinformatics resource center. Nucleic Acids Res. 2007;35(Database issue):D427–30. doi: 10.1093/nar/gkl880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Hunter WN. Structure-based ligand design and the promise held for antiprotozoan drug discovery. J. Biol. Chem. 2009;284:11749–11753. doi: 10.1074/jbc.R800072200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Lipinski CA. Drug-like properties and the causes of poor solubility and poor permeability. J. Pharmacol. Toxicol. Methods. 2000;44:235–249. doi: 10.1016/s1056-8719(00)00107-6. [DOI] [PubMed] [Google Scholar]
- [11].Olliaro P, Wells TN. The global portfolio of new antimalarial medicines under development. Clin. Pharmacol. Ther. 2009;85:584–595. doi: 10.1038/clpt.2009.51. [DOI] [PubMed] [Google Scholar]
- [12].Roos DS, Crawford MJ, Donald RGK, Fraunholz M, Harb OS, He CY, Kissinger JC, Shaw MK, Striepen B. Mining the Plasmodium genome database to define organellar function: what does the apicoplast do? Phil. Trans. R. Soc. Lond. B. 2002;357:35–46. doi: 10.1098/rstb.2001.1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Foth BJ, McFadden GI. The apicoplast: a plastid in Plasmodium falciparum and other Apicomplexan parasites. Int. Rev. Cytol. 2003;224:57–110. doi: 10.1016/s0074-7696(05)24003-2. [DOI] [PubMed] [Google Scholar]
- [14].Wiesner J, Reichenberg A, Heinrich S, Schlitzer M, Jomaa H. The plastid-like organelle of apicomplexan parasites as drug target. Curr Pharm Des. 2008;14:855–871. doi: 10.2174/138161208784041105. [DOI] [PubMed] [Google Scholar]
- [15].Lim L, McFadden GI. The evolution, metabolism and functions of the apicoplast. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2010;365:749–763. doi: 10.1098/rstb.2009.0273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Moreno SN, Li ZH. Anti-infectives targeting the isoprenoid pathway of Toxoplasma gondii. Expert Opin. Ther. Targets. 2008;12:253–263. doi: 10.1517/14728222.12.3.253. [DOI] [PubMed] [Google Scholar]
- [17].Lell B, Kremsner PG. Clindamycin as an antimalarial drug: review of clinical trials. Antimicrob. Agents Chemother. 2002;46:2315–2320. doi: 10.1128/AAC.46.8.2315-2320.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Hunter WN. The non-mevalonate pathway of isoprenoid precursor biosynthesis. J. Biol Chem. 2007;282:21573–21577. doi: 10.1074/jbc.R700005200. [DOI] [PubMed] [Google Scholar]
- [19].Eisenreich W, Bacher A, Arigoni D, Rohdich F. Biosynthesis of isoprenoids via the non-mevalonate pathway. Cell Mol. Life Sci. 2004;61:1401–1426. doi: 10.1007/s00018-004-3381-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Rohmer M. Isoprenoids including carotenoids and steroids. In: Barton D, Nakanishi K, editors. Comprehensive Natural Products Chemistry. Vol. 2. Elsevier Science; Amsterdam: 1999. pp. 45–67. Vol. 2, pp 45-67. [Google Scholar]
- [21].Sacchettini JC, Poulter CD. Creating isoprenoid diversity. Science. 1997;277:1788–1789. doi: 10.1126/science.277.5333.1788. [DOI] [PubMed] [Google Scholar]
- [22].Goldstein JL, Brown MS. Regulation of the mevalonate pathway. Nature. 1990;343:425–430. doi: 10.1038/343425a0. [DOI] [PubMed] [Google Scholar]
- [23].Byres E, Alphey MS, Smith TK, Hunter WN. Crystal structures of Trypanosoma brucei and Staphylococcus aureus mevalonate diphosphate decarboxylase inform on the determinants of specificity and reactivity. J. Mol. Biol. 2007;371:540–553. doi: 10.1016/j.jmb.2007.05.094. [DOI] [PubMed] [Google Scholar]
- [24].Liao JK, Laufs U. Pleiotropic effects of statins. Annual Review of Pharmacology and Toxicology. 2005;45:89–118. doi: 10.1146/annurev.pharmtox.45.120403.095748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Rohmer M. From molecular fossils of bacterial hopanoids to the formation of isoprene units: discovery and elucidation of the methylerythritol phosphate pathway. Lipids. 2008;43:1095–1107. doi: 10.1007/s11745-008-3261-7. [DOI] [PubMed] [Google Scholar]
- [26].Jomaa H, Weisner J, Sanderbrand S, Altinicicek B, Weidemeyer C, Hintz M, Turbachova I, Eberl M, Zeidler J, Lichtenthaler HK, Soldati D, Beck E. Inhibitors of the nonmevalonate pathway of isoprenoid biosynthesis as antimalarial drugs. Science. 1999;285:1573–1576. doi: 10.1126/science.285.5433.1573. [DOI] [PubMed] [Google Scholar]
- [27].Cassera MB, Gozzo FC, D’Alexandri FL, Merino EF, del Portillo HA, Peres VJ, Almeida IC, Eberlin MN, Wunderlich G, Wiesner J, Jomaa H, Kimura EA, Katzin AM. The methylerythritol phosphate pathway is functionally active in all intraerythrocytic stages of Plasmodium falciparum. J. Biol. Chem. 2004;279:51749–51759. doi: 10.1074/jbc.M408360200. [DOI] [PubMed] [Google Scholar]
- [28].Ye I, Hanekamp T, Tsoka S, Karp PD, Altman RB. Computational analysis of Plasmodium falciparum metabolism: organizing genomic information to facilitate drug discovery. Genome Research. 2004;14:917–924. doi: 10.1101/gr.2050304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Essential genes of E. coli were extracted. from, http://www.shigen.nig.ac.jp/ecoli/pec/index.jsp, with information collated from a large number of references.
- [30].Odom AR, Van Voorhis WC. Functional genetic analysis of the Plasmodium falciparum deoxyxylulose 5-phosphate reductoisomerase gene. Mol. Biochem Parasitol. 2010;170:108–111. doi: 10.1016/j.molbiopara.2009.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Eastman RT, Buckner FS, Yokoyama K, Gelb MH, Van Voorhis WC. Lipid posttranslational modifications. Fighting parasitic disease by blocking protein farnesylation. J. Lipid Res. 2006;47:233–240. doi: 10.1194/jlr.R500016-JLR200. [DOI] [PubMed] [Google Scholar]
- [32].Xiang S, Usunow G, Lange G, Busch M, Tong L. Crystal structure of 1-deoxy-D-xylulose 5-phosphate synthase, a crucial enzyme for isoprenoids biosynthesis. J. Biol. Chem. 2007;282:2676–2682. doi: 10.1074/jbc.M610235200. [DOI] [PubMed] [Google Scholar]
- [33].Yajima S, Nonaka T, Kuzuyama T, Seto H, Ohsawa K. Crystal structure of 1-deoxy-D-xylulose 5-phosphate reductoisomerase complexed with cofactors: implications of a flexible loop movement upon substrate binding. J. Biochem. (Tokyo) 2002;131:313–317. doi: 10.1093/oxfordjournals.jbchem.a003105. [DOI] [PubMed] [Google Scholar]
- [34].Reuter K, Sanderbrand S, Jomaa H, Wiesner J, Steinbrecher I, Beck E, Hintz M, Klebe G, Stubbs MT. Crystal structure of 1-deoxy-D-xylulose-5-phosphate reductoisomerase, a crucial enzyme in the non-mevalonate pathway of isoprenoid biosynthesis. J. Biol. Chem. 2002;277:5378–5384. doi: 10.1074/jbc.M109500200. [DOI] [PubMed] [Google Scholar]
- [35].Steinbacher S, Kaiser J, Eisenreich W, Huber R, Bacher A, Rohdich F. Structural basis of fosmidomycin action revealed by the complex with 2-C-methyl-D-erythritol 4-phosphate synthase (IspC). Implications for the catalytic mechanism and anti-malaria drug development. J. Biol. Chem. 2003;278:18401–18407. doi: 10.1074/jbc.M300993200. [DOI] [PubMed] [Google Scholar]
- [36].MacSweeney A, Lange R, Fernandes RP, Schulz H, Dale GE, Douangamath A, Proteau PJ, Oefner C. The crystal structure of E. coli 1-deoxy-D-xylulose-5-phosphate reductoisomerase in a ternary complex with the antimalarial compound fosmidomycin and NADPH reveals a tight-binding closed enzyme conformation. J. Mol. Biol. 2005;345:115–127. doi: 10.1016/j.jmb.2004.10.030. [DOI] [PubMed] [Google Scholar]
- [37].Yajima S, Hara K, Sanders JM, Yin F, Ohsawa K, Wiesner J, Jomaa H, Oldfield E. Crystallographic structures of two bisphosphonate:1-deoxyxylulose-5-phosphate reductoisomerase complexes. J. Am. Chem. Soc. 2004;126:10824–10825. doi: 10.1021/ja040126m. [DOI] [PubMed] [Google Scholar]
- [38].Ricagno S, Grolle S, Bringer-Meyer S, Sahm H, Lindqvist Y, Schneider G. Crystal structure of 1-deoxy-D-xylulose-5-phosphate reductoisomerase from Zymomonas mobilis at 1.9-Å resolution. Biochem. Biophys. Acta. 2004;1698:37–44. doi: 10.1016/j.bbapap.2003.10.006. [DOI] [PubMed] [Google Scholar]
- [39].Henriksson LM, Bjorkelid C, Mowbray SL, Unge T. The 1.9 Å resolution structure of Mycobacterium tuberculosis 1-deoxy-D-xylulose 5-phosphate reductoisomerase, a potential drug target. Acta Crystallogr. D Biol. Crystallogr. 2006;62:807–813. doi: 10.1107/S0907444906019196. [DOI] [PubMed] [Google Scholar]
- [40].Henriksson LM, Unge T, Carlsson J, Åqvist J, Mowbray SL, Jones TA. Structures of Mycobacterium tuberculosis 1-deoxy-D-xylulose-5-phosphate reductoisomerase provide new insights into catalysis. J. Biol. Chem. 2007;282:19905–19916. doi: 10.1074/jbc.M701935200. [DOI] [PubMed] [Google Scholar]
- [41].Umeda T, Tanaka N, Kusakabe Y, Nakanishi M, Kitade Y, Nakamura KT. Crystallization and preliminary X-ray crystallographic study of 1-deoxy-D-xylulose 5-phosphate reductoisomerase from Plasmodium falciparum. Acta Crystallogr. Sect F Struct. Biol. Cryst. Commun. 2010;66:330–332. doi: 10.1107/S1744309110001739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Richard SB, Bowman ME, Kwiatkowski W, Kang I, Chow C, Lillo AM, Cane DE, Noel JP. Structure of 4-diphosphocytidyl-2-C-methylerythritol synthetase involved in mevalonate-independent isoprenoid biosynthesis. Nature Struct. Biol. 2001;8:641–648. doi: 10.1038/89691. [DOI] [PubMed] [Google Scholar]
- [43].Richard SB, Lillo AM, Tetzlaff CN, Bowman ME, Noel JP, Cane DE. Kinetic analysis of Escherichia coli 2-C-methyl-D-erythritol-4-phosphate cytidyltransferase, wild type and mutants, reveals roles of active site amino acids. Biochemistry. 2004;43:12189–12197. doi: 10.1021/bi0487241. [DOI] [PubMed] [Google Scholar]
- [44].Gabrielsen M, Kaiser J, Rohdich F, Eisenreich W, Laupitz R, Bacher A, Bond CS, Hunter WN. The crystal structure of a plant 2C-methyl-D-erythritol 4-phosphate cytidylyltransferase exhibits a distinct quaternary structure compared to bacterial homologues and a possible role in feedback regulation for cytidine monophosphate. FEBS J. 2006;273:1065–1073. doi: 10.1111/j.1742-4658.2006.05133.x. [DOI] [PubMed] [Google Scholar]
- [45].Kemp LE, Bond CS, Hunter WN. Structure of a tetragonal crystal form of Escherichia coli 2-C-methyl-D-erythritol 4-phosphate cytidylyltransferase. Acta Crystallogr. D Biol. Crystallogr. 2003;59:607–610. doi: 10.1107/s090744490202365x. [DOI] [PubMed] [Google Scholar]
- [46].Gabrielsen M, Bond CS, Hallyburton I, Hecht S, Bacher A, Eisenreich W, Rohdich F, Hunter WN. Hexameric assembly of the bifunctional methylerythritol 2,4-cyclodiphosphate synthase and protein-protein associations in the deoxy-xylulose-dependent pathway of isoprenoid precursor biosynthesis. J. Biol. Chem. 2004;279:52753–52761. doi: 10.1074/jbc.M408895200. [DOI] [PubMed] [Google Scholar]
- [47].Miallau L, Alphey MS, Kemp LE, Leonard GA, McSweeney SM, Hecht S, Bacher A, Eisenreich W, Rohdich F, Hunter WN. Biosynthesis of isoprenoids: crystal structure of 4-diphosphocytidyl-2C-methyl-D-erythritol kinase. Proc. Natl. Acad. Sci. USA. 2003;100:9173–9178. doi: 10.1073/pnas.1533425100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Wada T, Kuzuyama T, Satoh S, Kuramitsu S, Yokoyama S, Unzai S, Tame JR, Park SY. Crystal structure of 4-(cytidine 5′-diphospho)-2-C-methyl-D-erythritol kinase, an enzyme in the non-mevalonate pathway of isoprenoid synthesis. J. Biol. Chem. 2003;278:30022–30027. doi: 10.1074/jbc.M304339200. [DOI] [PubMed] [Google Scholar]
- [49].Kalinowska-Tłuścik J, Miallau L, Gabrielsen M, Leonard GA, McSweeney SM, Hunter WN. A triclinic crystal form of Escherichia coli 4-diphosphocytidyl-2C-methyl-D-erythritol kinase and reassessment of the quaternary structure. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 2010;66:237–41. doi: 10.1107/S1744309109054591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Sgraja T, Alphey MS, Ghilagaber S, Marquez R, Robertson MN, Hemmings JL, Lauw S, Rohdich F, Bacher A, Eisenreich W, Illarionova V, Hunter WN. Characterization of Aquifex aeolicus 4-diphosphocytidyl-2C-methyl-d-erythritol kinase–ligand recognition in a template for antimicrobial drug discovery. FEBS. J. 2008;275:2779–2794. doi: 10.1111/j.1742-4658.2008.06418.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Bernal C, Mendez E, Terencio J, Boronat A, Imperial SA. Spectrophotometric assay for the determination of 4-diphosphocytidyl-2-C-methyl-D-erythritol kinase activity. Anal. Biochem. 2005;340:245–251. doi: 10.1016/j.ab.2005.01.055. [DOI] [PubMed] [Google Scholar]
- [52].Illarionova V, Kaiser J, Ostrozenkova E, Bacher A, Fischer M, Eisenreich W, Rohdich F. Nonmevalonate terpene biosynthesis enzymes as antiinfective drug targets: substrate synthesis and high-throughput screening methods. J. Org. Chem. 2006;71:8824–8834. doi: 10.1021/jo061466o. [DOI] [PubMed] [Google Scholar]
- [53].Hirsch AK, Alphey MS, Lauw S, Seet M, Barandun L, Eisenreich W, Rohdich F, Hunter WN, Bacher A, Diederich F. Inhibitors of the kinase IspE: structure-activity relationships and co-crystal structure analysis. Org. Biomol. Chem. 2008;6:2719–2730. doi: 10.1039/b804375b. [DOI] [PubMed] [Google Scholar]
- [54].Crane CM, Hirsch AK, Alphey MS, Sgraja T, Lauw S, Illarionova V, Rohdich F, Eisenreich W, Hunter WN, Bacher A, Diederich F. Synthesis and characterization of cytidine derivatives that inhibit the kinase IspE of the non-mevalonate pathway for isoprenoid biosynthesis. ChemMedChem. 2008;3:91–101. doi: 10.1002/cmdc.200700208. [DOI] [PubMed] [Google Scholar]
- [55].Kemp LE, Bond CS, Hunter WN. Structure of 2C-methyl-D-erythritol 2,4-cyclodiphosphate synthase: an essential enzyme for isoprenoid biosynthesis and target for antimicrobial drug development. Proc. Natl. Acad. Sci. USA. 2002;99:6591–6596. doi: 10.1073/pnas.102679799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Steinbacher S, Kaiser J, Wungsintaweekul J, Hecht S, Eisenreich W, Gerhardt S, Bacher A, Rohdich F. Structure of 2C-methyl-D-erythritol-2,4-cyclodiphosphate synthase involved in mevalonate-independent biosynthesis of isoprenoids. J. Mol. Biol. 2002;316:79–88. doi: 10.1006/jmbi.2001.5341. [DOI] [PubMed] [Google Scholar]
- [57].Richard SB, Ferrer JL, Bowman ME, Lillo AM, Tetzlaff CN, Cane DE, Noel JP. Structure and mechanism of 2C-methyl-D-erythritol 2,4-cyclodiphosphate synthase. An enzyme in the mevalonate-independent isoprenoid biosynthetic pathway. J. Biol. Chem. 2002;277:8667–8672. doi: 10.1074/jbc.C100739200. [DOI] [PubMed] [Google Scholar]
- [58].Kishida H, Wada T, Unzai S, Kuzuyama T, Takagi M, Terada T, Shirouzu M, Yokoyama S, Tame JR, Park SY. Structure and catalytic mechanism of 2C-methyl-D-erythritol 2,4-cyclodiphosphate (MECDP) synthase, an enzyme in the non-mevalonate pathway of isoprenoid synthesis. Acta Crystallogr. D Biol. Crystallogr. 2003;59:23–31. doi: 10.1107/s0907444902017705. [DOI] [PubMed] [Google Scholar]
- [59].Ni S, Robinson H, Marsing GC, Bussiere DE, Kennedy MA. Structure of 2C-methyl-D-erythritol-2,4-cyclodiphosphate synthase from Shewanella oneidensis at 1.6 Å: identification of farnesyl pyrophosphate trapped in a hydrophobic cavity. Acta Crystallogr. D Biol. Crystallogr. 2004;60:1949–1957. doi: 10.1107/S0907444904021791. [DOI] [PubMed] [Google Scholar]
- [60].Ramsden NL, Buetow L, Dawson A, Kemp LA, Ulaganathan V, Brenk R, Klebe G, Hunter WN. A structure-based approach to ligand discovery for 2C-methyl-D-erythritol-2,4-cyclodiphosphate synthase: a target for antimicrobial therapy. J. Med. Chem. 2009;52:2531–2542. doi: 10.1021/jm801475n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Rohdich F, Eisenreich W, Wungsintaweekul J, Hecht S, Schuhr CA, Bacher A. Biosynthesis of terpenoids. 2C-Methyl-D-erythritol 2,4-cyclodiphosphate synthase (IspF) from Plasmodium falciparum. Eur. J. Biochem. 2001;268:3190–3197. doi: 10.1046/j.1432-1327.2001.02204.x. [DOI] [PubMed] [Google Scholar]
- [62].Xiao Y, Zahariou G, Sanakis Y, Liu P. IspG enzyme activity in the deoxyxylulose phosphate pathway: roles of the iron-sulfur cluster. Biochemistry. 2009;48:10483–10485. doi: 10.1021/bi901519q. [DOI] [PubMed] [Google Scholar]
- [63].Zepeck F, Gråwert T, Kaiser J, Schramek N, Eisenreich W, Bacher A, Rohdich F. Biosynthesis of isoprenoids. purification and properties of IspG protein from Escherichia coli. J. Org. Chem. 2005;70:9168–9174. doi: 10.1021/jo0510787. [DOI] [PubMed] [Google Scholar]
- [64].Crane BR, Siegel LM, Getzoff ED. Structures of the siroheme- and Fe4S4-containing active center of sulfite reductase in different states of oxidation: heme activation via reduction-gated exogenous ligand exchange. Biochemistry. 1997;36:12101–12119. doi: 10.1021/bi971065q. [DOI] [PubMed] [Google Scholar]
- [65].Gråwert T, Span I, Eisenreich W, Rohdich F, Eppinger J, Bacher A, Groll M. Probing the reaction mechanism of IspH protein by x-ray structure analysis. Proc. Natl. Acad. Sci. U S A. 2010;107:1077–1081. doi: 10.1073/pnas.0913045107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Wang W, Wang K, Liu YL, No JH, Li J, Nilges MJ, Oldfield E. Bioorganometallic mechanism of action, and inhibition, of IspH. Proc. Nat. Acad. Sci. U S A. 2010;107:4522–4527. doi: 10.1073/pnas.0911087107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Rohdich F, Bacher A, Eisenreich W. Perspectives in anti-infective drug design. The late steps in the biosynthesis of the universal terpenoid precursors, isopentenyl diphosphate and dimethylallyl diphosphate. Bioorg. Chem. 2004;32:292–308. doi: 10.1016/j.bioorg.2004.05.012. [DOI] [PubMed] [Google Scholar]
- [68].de Ruyck J, Rothman SC, Poulter CD, Wouters J. Catalytic mechanism of Escherichia coli isopentenyl diphosphate isomerase involves Cys-67, Glu-116, and Tyr-104 as suggested by crystal structures of complexes with transition state analogues and irreversible inhibitors. Biochem. Biophys. Res. Commun. 2005;338:1515–1518. doi: 10.1074/jbc.M212823200. [DOI] [PubMed] [Google Scholar]
- [69].Kemp LE, Alphey MS, Bond CS, Ferguson MA, Hecht S, Bacher A, Eisenreich W, Rohdich F, Hunter WN. The identification of isoprenoids that bind in the intersubunit cavity of Escherichia coli 2C-methyl-D-erythritol-2,4-cyclodiphosphate synthase by complementary biophysical methods. Acta Crystallogr. D Biol. Crystallogr. 2005;61:45–52. doi: 10.1107/S0907444904025971. [DOI] [PubMed] [Google Scholar]
- [70].Kellogg BA, Poulter CD. Chain elongation in the isoprenoid biosynthetic pathway. Curr. Opin. Chem. Biol. 1997;1:570–578. doi: 10.1016/s1367-5931(97)80054-3. [DOI] [PubMed] [Google Scholar]
- [71].Gabrielsen M, Rohdich F, Eisenreich W, Grawert T, Hecht S, Bacher A, Hunter WN. Biosynthesis of isoprenoids: a bifunctional IspDF enzyme from Campylobacter jejuni. Eur. J. Biochem. 2004;271:3028–3035. doi: 10.1111/j.1432-1033.2004.04234.x. [DOI] [PubMed] [Google Scholar]
- [72].Lherbet C, Pojer F, Richard SB, Noel JP, Poulter CD. Absence of substrate channeling between active sites in the Agrobacterium tumefaciens IspDF and IspE enzymes of the methyl erythritol phosphate pathway. Biochemistry. 2006;45:3548–3553. doi: 10.1021/bi0520075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].DeLano WL. The PyMOL Molecular Graphics System. 2002 http://www.pymol.org.