Abstract
The key first step in evaluating pathogen levels in suspected contaminated water is concentration. Concentration methods tend to be specific for a particular pathogen group, for example US Environmental Protection Agency Method 1623 for Giardia and Cryptosporidium1, which means multiple methods are required if the sampling program is targeting more than one pathogen group. Another drawback of current methods is the equipment can be complicated and expensive, for example the VIRADEL method with the 1MDS cartridge filter for concentrating viruses2. In this article we describe how to construct glass wool filters for concentrating waterborne pathogens. After filter elution, the concentrate is amenable to a second concentration step, such as centrifugation, followed by pathogen detection and enumeration by cultural or molecular methods. The filters have several advantages. Construction is easy and the filters can be built to any size for meeting specific sampling requirements. The filter parts are inexpensive, making it possible to collect a large number of samples without severely impacting a project budget. Large sample volumes (100s to 1,000s L) can be concentrated depending on the rate of clogging from sample turbidity. The filters are highly portable and with minimal equipment, such as a pump and flow meter, they can be implemented in the field for sampling finished drinking water, surface water, groundwater, and agricultural runoff. Lastly, glass wool filtration is effective for concentrating a variety of pathogen types so only one method is necessary. Here we report on filter effectiveness in concentrating waterborne human enterovirus, Salmonella enterica, Cryptosporidium parvum, and avian influenza virus.
Keywords: Immunology, Issue 61, avian influenza virus, environmental sampling, Cryptosporidium, pathogen concentration, Salmonella, water, waterborne disease, waterborne pathogens
Protocol
1. Preparing the Glass Wool
Before and after making each batch of filters, sterilize the work area with 10% bleach solution.
Put on gloves and gown. Sterilize a bucket by autoclaving at 121°C and 15 psi for at least 20 minutes. Place the glass wool in the sterile bucket.
Saturate the glass wool with reverse osmosis water and let soak for 15 minutes.
Drain the reverse osmosis water from the bucket.
Saturate the glass wool with 1 M HCl and let soak for 15 minutes.
Drain the 1 M HCl from the bucket.
Rinse the glass wool with reverse osmosis water.
Mix thoroughly.
Check the pH using pH paper and repeat the reverse osmosis water rinse until a neutral pH is achieved.
Pour off the rinse water.
Saturate the glass wool with 1 M NaOH and let soak for 15 minutes.
Drain the 1 M NaOH from the bucket.
Repeat the reverse osmosis rinse until a neutral pH is achieved.
Pour off the rinse water.
Instead of a bucket, a glass wool washer can be constructed, which is similar in design to a glass pipette washer (Figure 1).
Cover the glass wool completely with sterile Phosphate Buffered Saline (PBS) adjusted to pH 6.8.
Use prepared glass wool immediately or store at 4°C. It can be stored for up to two weeks. Before using, make sure the pH is neutral as it will rise over time. If the pH is not neutral, re-rinse with sterile Phosphate Buffered Saline (pH 6.8).
2. Assembling the Glass Wool Filter
Drill a 11/16 inch hole into the PVC caps and tap threads so the male adapter nylon fittings can be screwed into the caps. This step is necessary only for the first assembly. Thereafter, the caps can be used for years. Apply Teflon tape to the nylon fittings threads and screw to the caps.
Pack the PVC pipe with small pieces of glass wool. Use a metal plunger, like a car engine valve, to pack tight. Packing does not require great force. Pack tight enough so that the glass wool stays in place and channels do not form. However, do not pack so tightly water cannot flow through the filter. When packed appropriately, a flow rate of 4 to 5 liters per minute should be attainable when the filter is attached to water taps with pressures between 40-60 psi. For the size of PVC pipe specified in this protocol, approximately 85 grams washed and packed glass wool is used per pipe. Tare the empty PVC pipe on a top-loading balance and pack with washed glass wool until the pipe mass increases 85 grams.
Insert polypropylene mesh into the PVC caps with male adapter nylon fittings attached.
Apply Teflon tape to the threaded portions of the PVC pipe.
Screw on PVC caps to the PVC pipe and label one filter end the inflow and the other end the outflow. This is important later for the elution step. Which end is labeled inflow does not matter.
Push 60 mL of sterile Phosphate Buffered Saline (pH = 6.8) into the filter using a catheter tipped syringe. Excess will come out opposite end.
Wrap ends tightly with Parafilm to avoid leaking. Filters can be stored up to 30 days at 4°C.
3. Sampling
Sterilize tubing used for sampling by recirculating or immersing items for 30 minutes in 0.525% NaClO (i.e., a 10% solution of standard household bleach). Follow this by draining the hypochlorite solution and neutralize with 0.05% sodium thiosulfate, made by adding 25 mL of a 2% stock anhydrous sodium thiosulfate solution to one liter of reverse osmosis water.
For sample water with a pH greater than 7.5, adjust the pH to between 6.5 and 7.0 with HCl. HCl concentration can range from 0.25 M to 1 M. Four liters 0.5 M HCl is generally sufficient for two 800 liter samples, depending on the water's ambient pH and buffering capacity. Inject HCl during sampling using a peristaltic pump or Venturi (Figure 2). Adjust the pump speed or Venturi opening to achieve the target pH. Measure the pH at the sample line outlet using a field pH meter.
Use a prefilter if the glass wool filter clogs from water with high turbidity. (Specifications for the prefilter and its housing are in the Materials list online.) Because pathogens can be attached to particles trapped by the prefilter it must be eluted along with the glass wool filter (see below).
Adjust the flow rate to between 2 and 4 liters/minute. Typical sample volumes are between 200 and 1,500 liters.
Disconnect the glass wool filter and store in a plastic sterile bag at 4°C for up to 48 hours.
4. Elution
Affix the glass wool filter to a ring stand with the sample inflow end pointing downward into a polypropylene bottle. Elute opposite of the direction of sample flow.
Push 80 mL of sterile 3% beef extract in 0.05 M glycine with a pH of 9.5 into the filter.
Wait 15 minutes.
Push another 80 mL of sterile 3% beef extract in 0.05 M glycine with a pH of 9.5 into the filter.
Push air thorough the filter until foam comes out of the filter inlet.
Adjust the eluate pH to between 7.0 and 7.5 with 1 M HCl.
Store eluate at 4°C for up to 24 hours or at -20°C for longer periods of time.
Elute the prefilter if one is used. Unscrew the top of the housing cartridge and pour off all residual water inside the housing while holding the prefilter in place with sterile gloved hands.
Remove the prefilter from the housing cartridge and slip it into a 15" x 6" zip-lock bag.
Pour 200 mL of sterile 3% beef extract in 0.05 M glycine with a pH of 9.5 into the bag and seal securely with the zip-lock.
Invert the bagged prefilter several times to ensure the entire surface comes in contact with the beef extract.
Wait 15 minutes, occasionally inverting and massaging the bagged prefilter.
Open the zip-lock. Grip the bag around the prefilter tightly to squeeze out as much beef extract as possible and pull the prefilter out with sterile gloved hands,.
Pour the beef extract eluate into a polypropylene bottle.
Adjust the eluate pH to between 7.0 and 7.5 with 1M HCl.
Store eluate at 4°C for up to 24 hours or at -20°C for longer periods of time. The prefilter eluate can be separately analyzed for pathogens or combined with the glass wool filter eluate.
5. Representative Results
| Pathogen | Water Turbidity Level (NTU) a | Amount Seeded/L b,c,d | % Recovery ± 1 SD | Number Independent Trials |
| Enterovirus- Poliovirus Sabin III | 0.5 | 500 | 81% ± 11 | 7 |
| Enterovirus- Poliovirus Sabin III | 0.5 | 5000 | 67% ± 12 | 8 |
| Enterovirus- Poliovirus Sabin III | 215 | 500 | 59% ± 32 | 7 |
| Enterovirus- Poliovirus Sabin III | 215 | 5000 | 38% ± 22 | 6 |
| Enterovirus- Poliovirus Sabin III | 447 | 500 | 56% ± 18 | 8 |
| Enterovirus- Poliovirus Sabin III | 447 | 5000 | 63% ± 37 | 8 |
| Cryptosporidium parvum | 0.5 | 5 | 38% ± 14 | 7 |
| Cryptosporidium parvum | 0.5 | 50 | 53% ± 19 | 8 |
| Cryptosporidium parvum | 215 | 5 | 40% ± 16 | 7 |
| Cryptosporidium parvum | 215 | 50 | 30% ±6 | 6 |
| Cryptosporidium parvum | 447 | 5 | 33% ± 13 | 8 |
| Cryptosporidium parvum | 447 | 50 | 28% ± 11 | 8 |
| Salmonella enterica | 0.5 | 5 | 29% ± 24 | 7 |
| Salmonella enterica | 0.5 | 500 | 56% ± 16 | 8 |
| Salmonella enterica | 215 | 5 | 32% ± 24 | 7 |
| Salmonella enterica | 215 | 500 | 34% ±11 | 6 |
| Salmonella enterica | 447 | 5 | 34% ± 18 | 8 |
| Salmonella enterica | 447 | 500 | 31% ± 24 | 8 |
Nephelometric Turbidity Unit
Enteroviruses enumerated by qPCR as genomic copies/L
C. parvum enumerated by immunofluorescence as oocysts
S. enterica enumerated by culture as colony-forming-units
Table 1. Glass wool concentration with varying water turbidities and pathogen densities.
| Pathogen | Water Sample Location | Amount Seeded/La | % Recovery |
| Avian Influenza H5N2 | Sundi Lake, Anchorage Borough | 2500 | 42.9% |
| Avian Influenza H5N2 | Minto Flats, Fairbanks North Star Borough | 2500 | 36.7% |
| Avian Influenza H5N2 | Portage Valley, Anchorage Borough | 2500 | 7.8% |
| Avian Influenza H5N2 | Potter Marsh, Anchorage Borough | 2500 | 41.5% |
| Avian Influenza H5N2 | Willow Lake, Yukon-Koyukuk Borough | 2500 | 15.5% |
Measured by qPCR as genomic copies/L
Table 2. Glass wool concentration of avian influenza virus using water from five sites in Alaska.
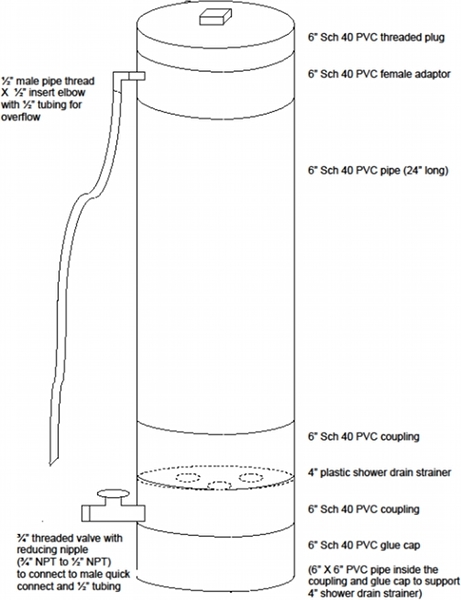 Figure 1. Diagram of glass wool washer. This can be used instead of a bucket, saving time from rinsing. The concept is similar to a glass pipette washer.
Figure 1. Diagram of glass wool washer. This can be used instead of a bucket, saving time from rinsing. The concept is similar to a glass pipette washer.
 Figure 2. Glass wool filtration with acid injection by peristaltic pump. Note the "T" connector where the pump tubing introduces acid into the sample line between the water tap and glass wool filter. pH adjustment is necessary only if the water sampled has a pH > 7.5.
Figure 2. Glass wool filtration with acid injection by peristaltic pump. Note the "T" connector where the pump tubing introduces acid into the sample line between the water tap and glass wool filter. pH adjustment is necessary only if the water sampled has a pH > 7.5.
Glass wool filters are effective in concentrating pathogens from water with a wide range of turbidity levels and pathogen densities (Table 1). To test this, 20 liters dechlorinated tap water was mixed with dried silt loam soil (0, 1.27, or 2.75 g/L) to achieve the desired level of turbidity and then seeded with pathogens at various densities. The water was passed through a glass wool filter, the concentrated pathogens in the eluate were enumerated, and this value was the numerator in the percent recovery calculation. The quantity of pathogens seeded into the water, that is, the denominator of the percent recovery calculation, was determined by first seeding the pathogens into a negative eluate then enumerating the pathogens. The negative eluate was prepared by passing an unseeded 20 liter sample through a filter and eluting. Quantifying the seeded pathogens in a negative eluate avoids differences in pathogen enumeration that could result from matrix differences created by the glass wool filter. The importance of this step when quantifying pathogens by qPCR is discussed in Lambertini et al.3. A 20 liter negative control sample was concentrated via glass wool filtration to determine there were no native pathogens present that could confound the percent recovery calculation. A 10 μm nominal pore size prefilter was used when the turbidity level was ≥ 215 NTU.
Poliovirus was quantified by real-time qPCR using the secondary concentration and nucleic acid extraction procedures and primers and probe described in Lambertini et al.3. Cryptosporidium parvum was quantified in the final concentrated sample volume (FCSV) created by the secondary concentration procedure for poliovirus. Oocysts were visualized by immunofluorescence (MeriFluor Cryptosporidium & Giardia Detection Kit, Meridian Life Science, Inc., Cincinnati, OH). Salmonella enterica was quantified in the FCSV by plating onto XLD agar (Remel, Lenexa, KS) and counting colony-forming-units.
Glass wool filters are effective in concentrating avian influenza viruses (Table 2). Low pathogenicity avian influenza virus (H5N2) was seeded into water from several locations in Alaska and the percent recovery calculated as described above. Secondary concentration and nucleic acid procedures were performed as for poliovirus; the virus was quantified by qPCR using the primers and probe described in Spackman et al.4
Discussion
Glass wool filters have been used by several research teams3,5,6 to concentrate human enteric viruses from a variety of water sources such as finished drinking water7, groundwater8,9, surface water10, sea water11, wastewater12, and agricultural runoff13. Here we report the filters are also effective in concentrating avian influenza virus as well as the bacterial and protozoan pathogens Salmonella enterica (serovar Typhimurium) and Cryptosporidium parvum, respectively. Deboosere et al. also recently reported glass wool concentration of avian influenza virus14.
The filters are advantageous in that they are inexpensive, highly portable, usable in a wide range of water matrices, and effective for concentrating many types of waterborne pathogens. They can be constructed to any size, depending on research needs. After disinfection, filter housings are reusable.
Glass wool filters, however, do have limitations. As with any virus concentration method that relies on electropositively charged media for virus adsorption (e.g., 1MDS filter, CUNO Inc., Meriden, CT), filter effectiveness depends on ambient water pH. In our laboratory we have selected pH 7.5 as the cut-off, above which the water pH is adjusted downward by continuously pumping 0.25 M HCl into the filter input line during sampling. Higher pH waters can be sampled without pH adjustment, but at the cost of filter effectiveness3. Another limitation is shelf-life. We have shown for filters stored at 4°C for six weeks that pathogen concentration effectiveness does not decline (unpublished data). Nonetheless, longer storage times have not been tested so, conservatively, to ensure data quality, we do not use filters older than 30 days. Usually, filters are made as needed. Another potential roadblock in some countries is obtaining the glass wool from the French source specified in earlier papers. Recently, we demonstrated standard unfaced fiberglass insulation is equally effective, and this is readily available from hardware or home improvement stores (see Materials list online).
For all experiments, it is important to run two sets of controls, an equipment blank to ensure glass wool filters were not contaminated during construction and a recovery control to ensure the filters work as designed. These controls are necessary for any waterborne pathogen concentration method.
Using a glass wool filter can be as simple as attaching it to a faucet and turning on the tap or as complicated as sampling a sediment-laden river in a remote location, requiring pumps, pH adjustment, and a prefilter to prevent clogging. For our research group, the largest benefit of using glass wool filters is the capability to collect and analyze thousands of water samples for human and livestock pathogens, yielding data on pathogen abundance and distribution in the environment that would not have been as feasible with more costly, complicated methods13,15.
Disclosures
No conflicts of interests declared.
Acknowledgments
We thank William T. Eckert for narrating the video. Development of the glass wool protocol was part of the Wisconsin Water And Health Trial for Enteric Risks (WAHTER Study), funded by US EPA STAR Grant R831630. Alaska samples were collected by A. Reeves, A. Ramey, and B. Meixell with financial support from USGS. Any use of trade, product, or firm names is for descriptive purposes only and does not imply endorsement by the U.S. Government.
References
- US Environmental Protection Agency. Method 1623: Cryptosporidium and Giardia in Water by Filtration/IMS/FA. EPA 815-R-05-002. 2012.
- Cashdollar JL, Dahling DR. Evaluation of a method to re-use electropositive cartridge filters for concentrating viruses from tap and river water. J. Virol. Methods. 2006;132:13–17. doi: 10.1016/j.jviromet.2005.08.016. [DOI] [PubMed] [Google Scholar]
- Lambertini E. Concentration of enteroviruses, adenoviruses, and noroviruses from drinking water by use of glass wool filters. Appl. Environ. Microbiol. 2008;74:2990–2996. doi: 10.1128/AEM.02246-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spackman E. Development of a real-time reverse transcription PCR assay for Type A influenza virus and the avian H5 and H7 hemagglutinin subtypes. J. Clin. Microbiol. 2002;40:3256–3260. doi: 10.1128/JCM.40.9.3256-3260.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Environment Agency. Report No. NC/99/40. West Midlands, United Kingdom: Environment Agency, National Groundwater and Contaminated Land Centre; 2000. Optimisation of a new method for detection of viruses in groundwater. [Google Scholar]
- Vilaginés P, Sarrette B, Husson G, Vilaginés R. Glass wool for virus concentration at ambient water pH level. Water Sci. Technol. 1993;27:299–306. [Google Scholar]
- Vivier JC, Ehlers MM, Grabow WO. Detection of enteroviruses in treated drinking water. Water Res. 2004;38:2699–2705. doi: 10.1016/S0043-1354(01)00433-X. [DOI] [PubMed] [Google Scholar]
- Powell KL. In: Enteric virus detection in groundwater using a glass wool trap. In: Groundwater: Past Achievements and Future Challenges. Sililo O, editor. Balkema, Rotterdam: 2000. pp. 813–816. [Google Scholar]
- Hunt RJ, Borchardt MA, Richards KD, Spencer SK. Assessment of sewer source contamination of drinking water wells using tracers and human enteric viruses. Environ. Sci. Technol. 2010;44:7956–7963. doi: 10.1021/es100698m. [DOI] [PubMed] [Google Scholar]
- van Heerden J, Ehlers MM, Heim A, Grabow WO. Prevalence, quantification and typing of adenoviruses detected in river and treated drinking water in South Africa. J. Appl. Microbiol. 2005;99:234–242. doi: 10.1111/j.1365-2672.2005.02617.x. [DOI] [PubMed] [Google Scholar]
- Vilaginés P. Round robin investigation of glass wool method for poliovirus recovery from drinking water and sea water. Water Sci. Technol. 1997;35:445–449. [Google Scholar]
- Gantzer C, Senouci S, Maul A, Levi Y, Schwartzbrod L. Enterovirus genomes in wastewater: concentration on glass wool and glass powder and detection by RT-PCR. J. Virol. Methods. 1997;65:265–271. doi: 10.1016/s0166-0934(97)02193-9. [DOI] [PubMed] [Google Scholar]
- Borchardt MA, Jokela WE, Spencer SK, editors. Pathogen losses in surface water runoff from dairy manure applied to corn fields. American Society for Microbiology General Meeting; New Orleans, LA. 2011. [Google Scholar]
- Deboosere N. Development and validation of a concentration method for the detection of influenza A viruses from large volumes of surface water. Appl. Environ. Microbiol. 2011;77:3802–3808. doi: 10.1128/AEM.02484-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambertini E. Virus contamination from operation and maintenance practices in small drinking water distribution systems. J. Water Health. 2011;9:799–812. doi: 10.2166/wh.2011.018. [DOI] [PubMed] [Google Scholar]


