Abstract
The repeated and well-understood cellular architecture of the cerebellum make it an ideal model system for exploring brain topography. Underlying its relatively uniform cytoarchitecture is a complex array of parasagittal domains of gene and protein expression. The molecular compartmentalization of the cerebellum is mirrored by the anatomical and functional organization of afferent fibers. To fully appreciate the complexity of cerebellar organization we previously refined a wholemount staining approach for high throughput analysis of patterning defects in the mouse cerebellum. This protocol describes in detail the reagents, tools, and practical steps that are useful to successfully reveal protein expression patterns in the adult mouse cerebellum by using wholemount immunostaining. The steps highlighted here demonstrate the utility of this method using the expression of zebrinII/aldolaseC as an example of how the fine topography of the brain can be revealed in its native three-dimensional conformation. Also described are adaptations to the protocol that allow for the visualization of protein expression in afferent projections and large cerebella for comparative studies of molecular topography. To illustrate these applications, data from afferent staining of the rat cerebellum are included.
Keywords: Neuroscience, Issue 62, Gene expression, antibodies, compartmentalization, brain topography, circuitry, neuroanatomy
Protocol
1. Animal Perfusion and Cerebellum Dissection
Depending on the protein, perfusion may be essential for successful staining1,2. Transcardiac perfusion is an invasive, non-survival procedure that requires the proper use of anesthetics. Correct training, institutional approval, and IACUC approval are all necessary before attempting the procedure. It is always a good idea to consult the institution's veterinarians to get help in identifying experimental requirements and acquiring the correct training. Before the procedure, the animal should be deeply anesthetized and not responsive to stimuli such as foot or tail pinch. Using scissors make a cut in the abdominal skin and muscle wall and then continue to open the rib cage by cutting just adjacent to the sternum on each side. In addition, the experimenter can create a larger internal working space by cutting the fascia surrounding the diaphragm. By lifting the abdominal and thoracic wall superiorly, the heart will now be exposed. Make a small cut in the right atrium to allow blood to flow out and immediately insert the perfusion needle into the left ventricle. A basic hydrostatic pump is preferred to deliver the perfusion solutions (see Table 1 for details). First, flush the blood out with 0.1M phosphate buffered saline (PBS, see Table 3) until the fluid runs clear. Then, turn off the hydrostatic pump, quickly switch the solution delivery tube to a container of 4% paraformaldehyde (PFA, see Table 3) diluted in PBS and then turn the pump back on to deliver the fixative. The use of a pump is not necessary: with practice, two syringes filled with the PBS and 4% PFA can be used to slowly deliver the solutions.
It is critical to carefully dissect the brain out of the skull without introducing any lesions to the tissue. Even small lacerations to the brain during dissection will gradually expand into visible crevices that trap antibodies and cause artifactual staining during the processing of the tissue (e.g. red asterisks in Fig 2). For cerebellar dissections, it is typical to cut the skin down the middle of the head and then gently tease the flaps apart to expose the skull. Cut the skull from anterior to posterior along the dorsal midline, and also laterally along both sides of the skull. Do not cut beyond the bregma as this runs the risk of nicking the cerebellum. Using forceps gently lift the front of the skull away from the brain being careful not to tear away brain tissue that is attached to the meninges. Sever the cranial nerves on the ventral side of the brain.
Separation of the cerebellum from the rest of the brain is important for two reasons. First, it allows for exploration of expression patterns in the anterior lobules, which are hidden from view by the colliculi in situ. The second is that separating the tissue into smaller, isolated regions allows for more complete fixation, which can facilitate the staining of some proteins. Throughout this final dissection step, care should be taken to not touch the cerebellum with the tips of the forceps. Insert the tips of a pair of forceps into the middle of the superior and inferior colliculi. With a second set of forceps slowly tease away the inferior colliculi from the cerebellum. Then, lay the cerebellum so that its anterior side is facing down and the brainstem is pointing upwards. Insert the forceps into the fourth ventricle between the brainstem and lobules IX/X of the cerebellum. Cut the peduncles by pinching with the forceps on both sides and then gently lift the cerebellum away from the brainstem. Once the cerebellum is isolated, post-fix it by immersion in 4% PFA at 4 °C for a minimum of 24-48 hours. The cerebellum can be stored in 4% PFA for an extended period of time. The meninges should be removed either before or after staining for better visibility of the expression patterns. If the experimenter chooses to remove the meninges before staining there is a strong possibility that the cerebellum will be nicked during the process. Peeling away the meninges at the end of the staining procedure is much simpler because they become easier to locate after they acquire weak background staining and become frail post-processing.
2. Processing the Tissue for Wholemount Staining
Because the wholemount staining approach takes longer than immunohistochemical staining of tissue sections, it is helpful to plan the timeline for each of the experiments as shown in the example calendar provided (Table 2). Before starting, there are several key things to keep in mind. 1) The microtubes containing the tissue should be rotated on a nutator at all times, except during the freeze/thaw process. 2) Several solutions must be made fresh for each experiment (See Table 3 for solution recipes). 3) Throughout the protocol, when changing solutions, gently pour out the spent solution rather than removing the cerebellum with forceps, to avoid touching the cerebellum. Then, after mixing the new solution in another container, use a pipette to gently add the fresh solution to the tube.
2.1 Day 1:
Fix the cerebellum in Dent's fix3 at room temperature for 6-8 hours rocking gently on the nutator. If methanol is destructive to your antigen, consider replacing steps 2.1a through 2.3a with an antigen retrieval method. The heat and buffers used for antigen retrieval will help prepare the tissue for antibody penetration.
Bleach the cerebellum in Dent's Bleach3 overnight at 4 °C.
2.2 Day 2:
Dehydrate the cerebellum in two rounds of 100% MeOH at room temperature for 30 min each.
Then, to enhance the penetration of the solutions, subject the tissue to a series of freeze/thaw cycles. Carefully place the tube in a container with dry ice and freeze for 30 min. Then, remove the tube and leave it at room temperature on the bench for 15 minute. The freeze/thaw step should be performed four times (at least).
After the last thawing of the tissue, place the tube into a -80 °C freezer overnight.
The tissue can be stored for prolonged periods of time at -80 °C either immediately before or after the freeze/thaw steps. Otherwise, the protocol should be continued.
2.3 Day 3:
Rehydrate the cerebellum by washing it in 50% MeOH/50% PBS, 15% MeOH/85% PBS, and 100% PBS for 60-90 minutes each at room temperature.
Next, for adult mouse cerebella, subject the tissue to enzymatic digestion with 10μg/ml Proteinase K in PBS for 2-3 minutes. No digestion step is necessary for cerebella of young animals (P5 or younger in mouse). Longer digestion is suggested for larger brains. For instance, digestion could be as high as 5-6 minutes for primate brains4.
After digestion, immediately wash the cerebellum in PBS three times for 10 minutes each at room temperature to remove the Proteinase K.
Block the tissue in PMT (see Table 3) overnight at 4 °C. Milk powder is often the first choice when selecting a blocking agent for protein work because it is inexpensive, effectively lowers background staining, and typically does not interfere with antibody binding or access to the antigen. Animal serums can also be used, although the experimenter should be aware of the added cost and must first test the serum quality for cross-reactive immunoglobulins and ability to produce an optimal signal to noise ratio.
2.4 Day 4-5:
Incubate the tissue in primary antibodies diluted in PMT supplemented with 5% DMSO for 48 hours at 4 °C (larger cerebella will require increased incubation times).
2.5 Day 6:
Wash the cerebellum 2-3 times for 2-3 hours each in PMT at 4 °C to remove unbound primary antibodies.
Then, incubate the tissue in secondary antibodies in a solution containing PMT with 5% DMSO at 4 °C overnight (larger cerebella may require increased incubation times).
2.6 Day 7:
Wash the cerebellum 2-3 times for 2-3 hours each in PMT at 4 °C.
Give the tissue a final wash with PBT (see Table 3) for 1-2 hours. This step removes excess milk and enhances the clarity of the staining.
Finally, incubate the tissue in DAB (3,3'-diaminobenzidine) solution until the optimal staining intensity is reached. The brown reaction product should be clearly visible within ~10 minutes. It is best to use DAB in the dark as light causes the chromogen to precipitate, which can increase background staining.
2.7
When the optimal staining intensity is reached, stop the reaction by placing the cerebellum in PBS with 0.04% sodium azide. The tissue may be stored long-term in this solution. Sodium azide is a potent inhibitor of bacterial growth but should be avoided until this step as it also inhibits horseradish peroxidase.
2.8 Optional amplification procedure:
follow normal protocol but these steps should be performed in place of steps 2.5b-2.7 above.
Incubate the tissue overnight at 4 °C in biotin-conjugated secondary antibodies in PBST (see Table 3) with 5% DMSO.
Rinse the tissue for 2-3 hours each in 3-4 changes of PBST at room temperature.
Incubate the cerebellum overnight at 4 °C in ABC complex solution in PBST.
Rinse the tissue for 2-3 hours each at room temperature in 3-4 changes of PBS.
Incubate the tissue in freshly-prepared DAB solution, as described above.
When the staining is optimal, stop the reaction by washing the cerebellum in PBS with 0.04% sodium azide.
3. Imaging the Stained Tissue
Wholemount images can be captured by using, for instance, a Leica DFC3000 FX camera mounted on a Leica MZ16 FA stereomicroscope running Leica Application Suite FX software. If desired, deconvolution microscopy can be used to acquire multiple consecutive images in different focal planes, with each image having only a single lobule in crisp focus. Then, the images can be compressed into a single stack and software rendered to remove distortion. The final composite image will illustrate cerebellar surface expression patterns with all visible patterning in clear focus.
Wholemount stained tissue should be imaged while immersed in PBS (or another medium that will give an optimal refractive index). In order to easily position the wholemount for imaging, make a 1% agar gel in a plastic Petri dish. Cut small holes into the gel. The holes will allow the cerebellum to be wedged into the desired orientation.
Raw data are imported into Adobe Photoshop CS4 and adjusted for contrast and brightness levels.
Animals
All animal studies were carried out under an approved IACUC animal protocol according to the institutional guidelines at Albert Einstein College of Medicine. Male and female outbred Swiss Webster (Taconic, Albany, NY) mice were maintained in our colony and used for all studies. Euthanized adult rats were kindly provided by Dr. Bryen Jordan (Albert Einstein College of Medicine). All animals were at least one month old.
4. Representative Results
The cerebellum is compartmentalized by molecular expression into four transverse zones: the anterior zone (AZ: ~lobules I-V), the central zone (CZ: ~lobules VI-VII), the posterior zone (PZ: ~lobules VIII-dorsal IX) and the nodular zone (NZ: ~lobules IX ventral and X)5. Each zone contains a unique array of parasagittal stripes1,2,5,6 (Fig. 1). ZebrinII expression in Purkinje cells reveals stripes in the AZ and PZ (Fig. 2) and uniform expression in the CZ and NZ (Fig. 2). The parasagittal organization of Purkinje cells is mirrored by the terminal field topography of afferent fibers. Cocaine- and amphetamine-regulated transcript (CART) peptide is expressed in subsets of climbing fibers (Fig. 3a) that project to stripes of Purkinje cell dendrites in the molecular layer of the cerebellar cortex7 (Fig. 3b). With the appropriate modifications, such as amplification7, the wholemount protocol allows for the visualization of olivocerebellar patterns without the need for a laborious, time consuming reconstructions from staining tissue sections (Fig. 3b).
 Figure 1.
A. ZebrinII/aldolaseC expression reveals sagittal bands in transverse sections of the cerebellum. Scale bar = 500 μm. B. ZebrinII/aldolaseC is expressed exclusively in Purkinje cell somata and dendrites. Scale bar = 150μm (gl = granular layer; pcl = Purkinje cell layer; ml=molecular layer).
Figure 1.
A. ZebrinII/aldolaseC expression reveals sagittal bands in transverse sections of the cerebellum. Scale bar = 500 μm. B. ZebrinII/aldolaseC is expressed exclusively in Purkinje cell somata and dendrites. Scale bar = 150μm (gl = granular layer; pcl = Purkinje cell layer; ml=molecular layer).
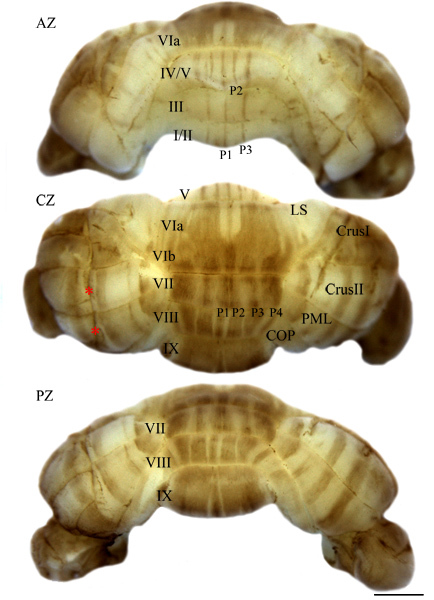 Figure 2. A wholemount cerebellum stained for ZebrinII/aldolaseC showing Purkinje cell stripes in images of the anterior (AZ), central (CZ), and posterior (PZ) zones of the cerebellum. Here we also show an example of the negative consequence of nicking the cerebellum. The resulting artifactual staining is indicated with red asterisks. Scale bar = 1 mm (LS = lobulus simplex; PML = paramedian lobule; COP = copula pyramidis).
Figure 2. A wholemount cerebellum stained for ZebrinII/aldolaseC showing Purkinje cell stripes in images of the anterior (AZ), central (CZ), and posterior (PZ) zones of the cerebellum. Here we also show an example of the negative consequence of nicking the cerebellum. The resulting artifactual staining is indicated with red asterisks. Scale bar = 1 mm (LS = lobulus simplex; PML = paramedian lobule; COP = copula pyramidis).
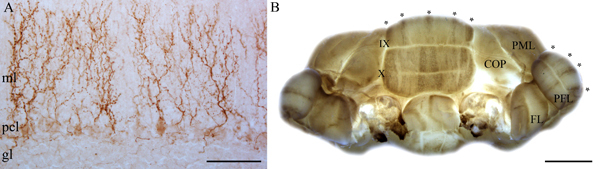
Figure 3. A. CART is expressed in climbing fiber terminals in the molecular layer. Scale bar = 100 μm (ml = molecular layer; pcl = Purkinje cell layer; gl = granular layer). B. Wholemount immunohistochemistry of CART expression in the NZ. Scale bar = 2 mm (COP = copula pyramidis; PML = paramedian lobule; PFL = paraflocculus; FL = flocculus).
Discussion
We have described the technical details required for successful wholemount staining using a versatile immunohistochemical approach for revealing protein expression in the developing and adult brain. By using this approach, complex molecular expression patterns can be analyzed and brain topography appreciated without the need for laborious and time consuming tissue sectioning procedures.
This protocol has been used to reveal the patterned expression of several Purkinje cell proteins in both the adult1,2,8,9 and early postnatal mouse cerebellum10,11,12. Our original studies also demonstrated the utility of the wholemount approach for examining granule cell antigen expression1 and anterogradely traced mossy fibers2, which reside ~250 μm below the pial surface in granular layer. In addition, several studies have made modifications to the original protocol for successful staining of proteins in Purkinje cells1,13 (antigen retrieval) and afferent fibers7,14 (signal amplification; Fig. 2). Moreover, wholemount immunohistochemical staining has also been applied to the cerebella of multiple mammalian4,15 and avian16,17 species, used to map patterned Purkinje cell loss in mouse models of neurodegenerative diseases18,19, and modified to stain heart20, lung21, cranial nerves22 and corneal nerves23. The major departure of the current protocol from previous descriptions of the wholemount method1,2 is that here we provide not only updated details for better success when using the procedure, but also a complete guide for fixing the tissue, dissecting the tissue and an in depth video illustration of the technique.
There are several limitations of wholemount staining. First, only surface patterns are typically visualized without further dissection and staining or without increasing the length of time that the tissue is incubated in antibodies. Further, staining will not reach deep within the cortex if it is hidden due to foliation1,2. However, wholemount stained tissue can be sectioned after processing and restained, with the same antibody or another, to reveal cellular expression profiles deeper within the tissue1,2. Second, the basic wholemount staining protocol is long. We suggest that each experimenter empirically determine the staining efficiency of their antibodies and shorten the post-fixation time, shorten the number and length of washes, and/or shorten the length of incubation in the primary antibodies. With these adjustments the protocol may be completed in a considerably shorter time. Third, for large cerebella the wholemount staining protocol requires large volumes of the antibody solutions, which can be expensive and/or only available in limited quantities. However, it is possible to save the primary antibodies after each run by freezing the solution at -20°C. Each experimenter will have to determine whether their antibody can withstand freezing before it is re-used. We have successfully stained tissue using previously frozen aliquots of zebrinII. Fourth, not all antibodies are compatible with the basic wholemount approach. For example, the antibody commonly used to detect the striped expression of the small heat shock protein, HSP25, requires that the tissue be first processed for antigen retrieval13. Regardless, as for staining tissue sections, traditional methods of troubleshooting antibody-antigen binding have also proven useful for optimizing wholemount staining.
We are currently exploring the possibility of adapting our wholemount protocol for use with fluorescent-tagged secondary antibodies. With the appropriate microscope equipped with the ability to image different colored fluorophores under low-power, one would be able to not only analyze the organization of multiple Purkinje cell maps in the same animal, but also examine the relationship between Purkinje cell stripe patterns and the topography of WGA-Alexa labeled afferent fibers in three-dimensions2,4.7,24. Furthermore, the recent availability of large scale gene expression databases (Allen Brain Atlas, Genepaint, Brain Gene Expression Map) have opened new avenues for merging our high throughput wholemount approach with whole genome analyses for examining the molecular topography of the entire brain.
Disclosures
We have nothing to disclose.
Acknowledgments
RVS is supported by new investigator start-up funds from Albert Einstein College of Medicine of Yeshiva University.
References
- Sillitoe RV, Hawkes R. Whole-mount Immunohistochemistry: A high-throughput screen for patterning defects in the mouse cerebellum. J. Histochem. Cytochem. 2002;50:235–244. doi: 10.1177/002215540205000211. [DOI] [PubMed] [Google Scholar]
- Kim S-H, Che P, Chung S-H, Doorn D, Hoy M, Larouche M, Marzban H, Sarna J, Zahedi S, Hawkes R. Whole-Mount Immunohistochemistry of the Brain. Current Protocols in Neuroscience. 2006. [DOI] [PubMed]
- Dent JA, Polson AG, Klymkowsky MW. A whole-mount immunocytochemical analysis of the expression of the intermediate filament protein vimentin in Xenopus. Development. 1989;105:61–74. doi: 10.1242/dev.105.1.61. [DOI] [PubMed] [Google Scholar]
- Sillitoe RV, Malz CR, Rockland K, Hawkes R. Antigenic compartmentation of the primate and tree shrew cerebellum: a common topography of zebrin II in Macaca mulatta and Tupaia belangeri. J. Anat. 2004;204:257–269. doi: 10.1111/j.0021-8782.2004.00282.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozol K, Hayden JM, Oberdick J, Hawkes R. Transverse zones in the vermis of the mouse cerebellum. J. Comp. Neurol. 1999;412:95–111. [PubMed] [Google Scholar]
- Apps R, Hawkes R. Cerebellar cortical organization: a one-map hypothesis. Nat. Rev. Neurosci. 2009;10:670–681. doi: 10.1038/nrn2698. [DOI] [PubMed] [Google Scholar]
- Reeber SL, Sillitoe RV. Patterned expression of a cocaine- and amphetamine-regulated transcript peptide reveals complex circuit topography in the rodent cerebellar cortex. J. Comp. Neurol. 2011;519:1781–1796. doi: 10.1002/cne.22601. [DOI] [PubMed] [Google Scholar]
- Sarna JR, Marzban H, Watanabe M, Hawkes R. Complementary stripes of phospholipase Cbeta3 and Cbeta4 expression by Purkinje cell subsets in the mouse cerebellum. J. Comp. Neurol. 2006;496:303–313. doi: 10.1002/cne.20912. [DOI] [PubMed] [Google Scholar]
- Demilly A, Reeber SL, Gebre SA, Sillitoe RV. Neurofilament heavy chain expression reveals a unique parasagittal stripe topography in the mouse cerebellum. Cerebellum. 2011;10:409–421. doi: 10.1007/s12311-010-0156-y. [DOI] [PubMed] [Google Scholar]
- Larouche M, Hawkes R. From clusters to stripes: the developmental origins of adult cerebellar compartmentation. Cerebellum. 2006;5:77–88. doi: 10.1080/14734220600804668. [DOI] [PubMed] [Google Scholar]
- Marzban H, Chung S, Watanabe M, Hawkes R. Phospholipase Cbeta4 expression reveals the continuity of cerebellar topography through development. J. Comp. Neurol. 2007;502:857–871. doi: 10.1002/cne.21352. [DOI] [PubMed] [Google Scholar]
- Blank MC, Grinberg I, Aryee E, Laliberte C, Chizhikov VV, Henkelman RM, Millen KJ. Multiple developmental programs are altered by loss of Zic1 and Zic4 to cause Dandy-Walker malformation cerebellar pathogenesis. Development. 2011;138:1207–1216. doi: 10.1242/dev.054114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawada K, Sakata-Haga H, Fukui Y. Alternating array of tyrosine hydroxylase and heat shock protein 25 immunopositive Purkinje cell stripes in zebrin II-defined transverse zone of the cerebellum of rolling mouse. Nagoya. Brain Res. 2010;1343:46–53. doi: 10.1016/j.brainres.2010.04.062. [DOI] [PubMed] [Google Scholar]
- Sawada K, Fukui Y, Hawkes R. Spatial distribution of corticotropin-releasing factor immunopositive climbing fibers in the mouse cerebellum: Analysis by whole mount immunohistochemistry. Brain Res. 2008;1222:106–117. doi: 10.1016/j.brainres.2008.05.029. [DOI] [PubMed] [Google Scholar]
- Marzban H, Hawkes R. On the architecture of the posterior zone of the cerebellum. Cerebellum. 2011;10:422–434. doi: 10.1007/s12311-010-0208-3. [DOI] [PubMed] [Google Scholar]
- Pakan JM, Graham DJ, Wylie DR. Organization of visual mossy fiber projections and zebrin expression in the pigeon vestibulocerebellum. J. Comp. Neurol. 2010;518:175–198. doi: 10.1002/cne.22192. [DOI] [PubMed] [Google Scholar]
- Iwaniuk AN, Marzban H, Pakan JM, Watanabe M, Hawkes R, Wylie DR. Compartmentation of the cerebellar cortex of hummingbirds (Aves: Trochilidae) revealed by the expression of zebrin II and phospholipase C beta 4. J. Chem. Neuroanat. 2009;37:55–63. doi: 10.1016/j.jchemneu.2008.10.001. [DOI] [PubMed] [Google Scholar]
- Sarna JR, Larouche M, Marzban H, Sillitoe RV, Rancourt DE, Hawkes R. Patterned Purkinje cell degeneration in mouse models of Niemann-Pick type C disease. J. Comp. Neurol. 2003;456:279–291. doi: 10.1002/cne.10522. [DOI] [PubMed] [Google Scholar]
- Sarna JR, Hawkes R. Patterned Purkinje cell loss in the ataxic sticky mouse. Eur. J. Neurosci. 2011;34:79–86. doi: 10.1111/j.1460-9568.2011.07725.x. [DOI] [PubMed] [Google Scholar]
- El-Bizri N, Guignabert C, Wang L, Cheng A, Stankunas K, Chang CP, Mishina Y, Rabinovitch M. SM22alpha-targeted deletion of bone morphogenetic protein receptor 1A in mice impairs cardiac and vascular development, and influences organogenesis. Development. 2008;135:2981–2991. doi: 10.1242/dev.017863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mondrinos MJ, Koutzaki S, Lelkes PI, Finck CM. A tissue-engineered model of fetal distal lung tissue. Am. J. Physiol. Lung Cell Mol. Physiol. 2007;293:639–650. doi: 10.1152/ajplung.00403.2006. [DOI] [PubMed] [Google Scholar]
- Coppola E, Rallu M, Richard J, Dufour S, Riethmacher D, Guillemot F, Goridis C, Brunet JF. Epibranchial ganglia orchestrate the development of the cranial neurogenic crest. Proc. Nat. Acad. Sci. 2010;107:2066–2071. doi: 10.1073/pnas.0910213107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubilus JK, Linsenmayer TF. Developmental guidance of embryonic corneal innervation: roles of Semaphorin3A and Slit2. Dev. Biol. 2010;344:172–184. doi: 10.1016/j.ydbio.2010.04.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reeber SL, Gebre SA, Sillitoe RV. Fluorescence mapping of afferent topography in three dimensions. Brain Struct. Funct. 2011;216:159–169. doi: 10.1007/s00429-011-0304-2. [DOI] [PubMed] [Google Scholar]
- Davis CA. Whole-mount immunohistochemistry. Methods Enzymol. 1993;225:502–516. doi: 10.1016/0076-6879(93)25034-y. [DOI] [PubMed] [Google Scholar]


