Abstract
The embryonic spinal cord consists of cycling neural progenitor cells that give rise to a large percentage of the neuronal and glial cells of the central nervous system (CNS). Although much is known about the molecular mechanisms that pattern the spinal cord and elicit neuronal differentiation1, 2, we lack a deep understanding of these early events at the level of cell behavior. It is thus critical to study the behavior of neural progenitors in real time as they undergo neurogenesis.
In the past, real-time imaging of early embryonic tissue has been limited by cell/tissue viability in culture as well as the phototoxic effects of fluorescent imaging. Here we present a novel assay for imaging such tissue for long periods of time, utilizing a novel ex vivo slice culture protocol and wide-field fluorescence microscopy (Fig. 1). This approach achieves long-term time-lapse monitoring of chick embryonic spinal cord progenitor cells with high spatial and temporal resolution.
This assay may be modified to image a range of embryonic tissues3, 4 In addition to the observation of cellular and sub-cellular behaviors, the development of novel and highly sensitive reporters for gene activity (for example, Notch signaling5) makes this assay a powerful tool with which to understand how signaling regulates cell behavior during embryonic development.
Keywords: Neuroscience, Issue 62, Live imaging, chick, embryo, spinal cord, time-lapse, developing neuroepithelium
Protocol
1. Dish Preparation
Dishes used for slice culture are glass-bottomed (with a coverslip as the base) (WillCo dishes). These are placed on lens tissue in a 6 cm tissue culture dish to keep the glass bottom clean.
On the day before the experiment, add 2 ml 0.1% poly-L-lysine solution to the glass bottomed dish and incubate for 5min at room temperature to allow the poly-L-lysine to coat the glass bottom.
Remove poly-L-lysine solution and rinse three times in deionised water, and then once in 70% ethanol.
Leave to dry overnight. Dishes can also be dried by gently heating in a microwave at low power for 30-45 seconds.
2. Embryo Electroporation
Incubate eggs at 37 °C to Hamburger-Hamilton (HH) stage 10 (~36 hours) (or other desired stage).
Before beginning prepare glass needles (we use a Flaming/Brown model p87 microcapillary puller) and break off the tip of the needle using a fine forceps under a dissecting microscope. The end of the needle should be sharp enough to pierce the embryo while not being so narrow that it impedes injection of the DNA solution.
Window eggs and place electrodes (5mm apart) on either side of the embryo.
Inject DNA (~0.025 - 0.5 μg/μl in deionised water colored with a small amount of fast green) into the neural tube.
Apply current - 12-17 V three times, 50ms pulse length with 950 ms between pulses.
We use low concentrations of DNA and low electroporation voltages to achieve mosaic expression so that we can follow individual cells.
Cover window in the eggshell with cellotape and make sure it is sealed.
Allow embryos to recover for 3-4 hours or overnight at 37 °C.
3. Collagen and Slice Culture Medium Preparation
Prepare collagen mix and slice culture medium about an hour before slicing.
To 300 μl type 1 collagen add 100 μl 0.1% acetic acid solution and 100 μl 5 x L15 medium. Vortex thoroughly after each addition. The solution should turn yellow.
Now add 15-20 μl 7.5% sodium bicarbonate and thoroughly vortex. The solution should become slightly pink, and the volume of sodium bicarbonate required for this may differ. Keep on ice and make up fresh each time.
To 10 ml neurobasal medium add B-27 supplement and Glutamax, to a 1 x final concentration and 10 μl Gentamicin solution.
Place medium in a 37 °C incubator buffered with 5% CO2. Leave the top of the container loose to allow it to equilibrate with the CO2.
4. Slice Culture
Remove embryos from egg and wash in L15 medium.
Place in a tissue culture dish with a layer of sylgard at the bottom and pin them out through the surrounding extra-embryonic membranes so that they are stretched taut.
Using a microknife, slice embryo as straight as possible through region of interest. For spinal cord, slices should be between 1-2 somites thick. Leave slices attached to embryo while you slice other embryos so you don't lose them.
Prepare a 200 μl micropipette tip by cutting off ~1 mm of the tip and attaching this to a p2 or p10. This tip will be used to transfer the spinal cord slices to the glass bottom dish. A 10 μl tip is not broad enough to pick up slices.
Coat the inside of the tip with collagen by pipetting 1 μl collagen mix prepared earlier. Leave for 1-2 mins and then rinse with L15 medium. This will prevent the tissue from sticking to the inside of the tip.
Detach slice from embryo using a microknife and remove from the dish using a p2 or p10 fitted with a 200 μl tip and set to 1 μl. Try to take up as little medium as possible.
Vortex the collagen mix and drop 5-8 μl of this on to the poly-l-lysine coated dish.
Immediately put embryo slice into collagen and position into place with a pair of fine forceps. Slices should be positioned so that the side to be imaged is flush with the coverslip. The tissue should adhere to the poly-l-lysine coating on the coverslip.
Repeat this until you have several slices on the coverslip. We can usually place 6-9 slices on one dish.
Once all the slices are in place, cover the dish and allow the collagen to set for 20 minutes. Some of the earliest placed collagen may already have started to dry out. To prevent this add a small amount of L15 to these before covering.
Once set, carefully add 2ml slice culture medium that has been equilibrated in 5% CO2 at 37 °C for at least an hour. Care must be taken to not dislodge the collagen from the coverslip.
Place in a 37 °/ 5% CO2 incubator and allow slices to recover for at least 3 hours before imaging. If there is no humidity, place dish in a perspex box with a bit of damp tissue paper in one corner.
5. Imaging Embryo Slices
We use a DeltaVision Core wide-field microscope fitted with a WeatherStation environmental chamber to image slices. The chamber is constantly maintained at 37 °C, with a CO2 perfusion apparatus to maintain the microscope stage at 5% CO2 / 95% air.
Imaging is normally carried out using a 40 x / 1.30 NA oil immersion lens and images are captured with a CoolSnap HQ2 cooled CCD camera.
Z-sections are captured every 1.5 μm through 45 μm tissue. Exposure time should be kept as low as possible. We normally expose for 5-50 ms for each Z-section. Images are captured every 7 minutes and up to 9 slices can be visited using the DeltaVision system's precise point visiting function.
6. Representative Results
An example time-lapse sequence of a spinal cord progenitor cell is shown in Fig. 2a and corresponding Movie 1. Imaging was started on a spinal cord slice from a two-day-old (HH stage 12) embryo. This cell was transfected with a construct expressing GFP-αTubulin. During this early stage, neural progenitor cells undergo predominantly progenitor-progenitor mode divisions during which the cells divide to generate two further cycling progenitor cells. Fig. 2b and corresponding Movie 2 shows a cell transfected with GFP-GPI (GPI anchored GFP), marking the cell membrane. This cell undergoes a division during which the basal process splits into two and is equally inherited by the daughter cells.
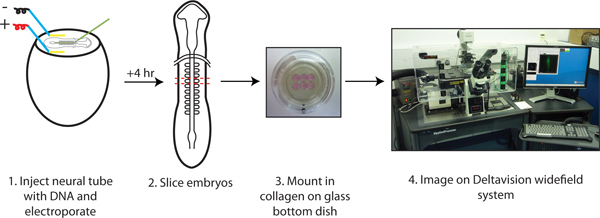 Figure 1. Slice culture protocol for time-lapse imaging.
Figure 1. Slice culture protocol for time-lapse imaging.
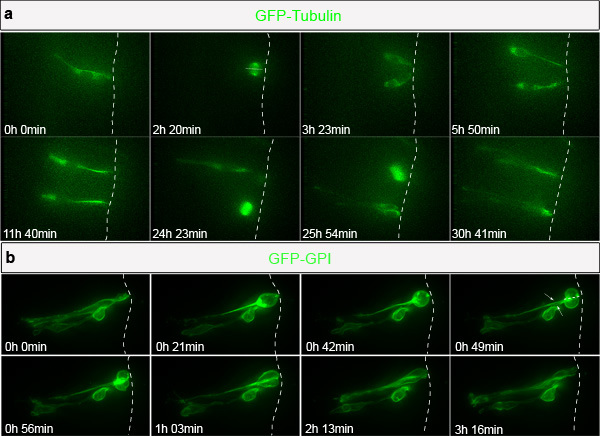 Figure 2. Cell behavior during normal spinal cord development. (a) Selected frames from Movie 1. Cell expressing GFP-tubulin divides with a cleavage plane that is perpendicular to the apical surface (2 h 20 min), generating two daughter cells that divide once again (24 h 23 min and 25 h 54 min). (b) Selected frames from Movie 2. Cell transfected with GFP-GPI undergoes cell division during which the basal process is split (white arrows) and equally inherited by the daughter cells.
Figure 2. Cell behavior during normal spinal cord development. (a) Selected frames from Movie 1. Cell expressing GFP-tubulin divides with a cleavage plane that is perpendicular to the apical surface (2 h 20 min), generating two daughter cells that divide once again (24 h 23 min and 25 h 54 min). (b) Selected frames from Movie 2. Cell transfected with GFP-GPI undergoes cell division during which the basal process is split (white arrows) and equally inherited by the daughter cells.
Movie 1. Click here to view movie.
Movie 2. Click here to view movie.
Discussion
We present here a novel time-lapse imaging assay to monitor cell behavior in chicken embryonic slice culture. This assay enables high resolution imaging of living tissue for up to 70 hours, although time frames between 24-48 hours are easier to capture. The use of a high NA oil objective and relatively short intervals between time-points enables image acquisition at high resolution, as well as allowing us to monitor cell behavior that often occurs rapidly, and can be easily missed during conventional time-lapse imaging using confocal microscopy.
The major advantage of this assay is the ability to image cells over long periods of time. The use of wide-field6 instead of confocal laser scanning7 microscopy is critical for this. Confocal microscopy has traditionally offered several advantages over wide-field microscopy, including the ability to take optical sections and eliminate out of focus information directly7, but the use of a pinhole leads to a loss of light to the detector8, excluding a significant amount of information, and necessitating longer exposure times. Wide-field microscopy, on the other hand, uses full field illumination and all the light passing through the objective is sent to the detector. This coupled with a high quantum efficiency cooled coupled device (CCD) camera ensures imaging with a high signal to noise ratio compared to confocal microscopy9, with very fast exposure times. In our application, we must image for long periods, record 3D stacks and ultimately resolve small—near-diffraction limited structures. Although confocal microscopy prevents out-of-focus light from ever reaching the detector and thus significantly reduces background in thick, densely-labeled samples7, 8, it only achieves a usable signal-to-noise ratio with much larger light input than wide-field microscopy10. For very light-sensitive sparsely-labeled samples like our electroporated spinal cord slices, therefore, wide-field microscopy performs better than confocal microscopy. When combined with image restoration by deconvolution, which removes out of focus information and improves contrast, wide-field is then particularly good for detection of small, dim objects11. While not appropriate for every tissue imaging experiment, this approach has been very effective for our live cell imaging application.
The choice of fluorescent protein is an important factor that can influence cell survival. We find that the best results are obtained from constructs that use green fluorescent protein (GFP)12 as a marker. We are currently assessing a range of red fluorescent proteins that can be effectively used in combination with GFP for dual channel time-lapses. There are a variety of such proteins available13. Although many proteins are stable as fusions with a fluorescent protein, some proteins may be rendered unstable by fusion, resulting in reduced cell viability. In such cases, the use of constructs containing an internal ribosome entry site (IRES) is useful to separate the protein of interest from the fluorescent protein.
When imaging spinal cord slices, care must be taken to minimize exposure to fluorescent light. The microscope eyepieces must be used only with transmitted light (brightfield) to locate and position slices. Fluorescent images should always be acquired using the microscope software using minimal exposure times. These should be kept in the range of 5-50 ms and the use of neutral density filters should be experimented with. Although longer exposure times may initially produce clearer images, the phototoxic effects of fluorescent light exposure may lead to cell death. The first 5-10 μm of tissue that is closest to the coverslip should not be imaged as this region contains tissue that may have been damaged during slicing.
Although the examples presented here are of embryos electroporated at HH stage 10 and imaged 6-7 hours later, this method can be used for embryos up to HH stage 18. At later stages, the spinal cord tissue is much larger and so is difficult to slice by hand. In these cases, embedding the embryos in low melting point agarose and sectioning with a vibratome may yield better results. Even though we present this assay as a method for imaging cells in the developing spinal cord, this approach has also been modified to image other embryonic tissues, including the sensory placodes3.
Disclosures
No conflicts of interest declared.
References
- Ulloa F, Briscoe J. Morphogens and the Control of Cell Proliferation and Patterning in the Spinal Cord. Cell Cycle. 2007;6:2640–2649. doi: 10.4161/cc.6.21.4822. [DOI] [PubMed] [Google Scholar]
- Briscoe J, Novitch BG. Regulatory pathways linking progenitor patterning, cell fates and neurogenesis in the ventral neural tube. Philosophical Transactions of the Royal Society B: Biological Sciences. 2008;363:57–70. doi: 10.1098/rstb.2006.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiau C, Das R, Storey K. An effective assay for high cellular resolution time-lapse imaging of sensory placode formation and morphogenesis. BMC Neuroscience. 2011;12:37. doi: 10.1186/1471-2202-12-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilcock AC, Swedlow JR, Storey KG. Mitotic spindle orientation distinguishes stem cell and terminal modes of neuron production in the early spinal cord. Development. 2007;134:1943–1954. doi: 10.1242/dev.002519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vilas-Boas F, Fior R, Swedlow JR, Storey KG, Henrique D. A novel reporter of notch signalling indicates regulated and random notch activation during Vertebrate neurogenesis. BMC Biology. 2011;9:58. doi: 10.1186/1741-7007-9-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herman B. Fluorescence microscopy. Bios Scientific Publishers. 1998.
- Conchello J-A, Lichtman JW. Optical sectioning microscopy. Nat. Meth. 2005;2:920–931. doi: 10.1038/nmeth815. [DOI] [PubMed] [Google Scholar]
- Pawley JB. In: Fundamental Limits in Confocal Microscopy Handbook Of Biological Confocal. Pawley JB, editor. Springer US; 2006. [Google Scholar]
- Swedlow JR, Hu K, Andrews PD, Roos DS, Murray JM. Measuring tubulin content in Toxoplasma gondii: A comparison of laser-scanning confocal and wide-field fluorescence microscopy. Proceedings of the National Academy of Sciences. 2002;99:2014–2019. doi: 10.1073/pnas.022554999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray JM, Appleton PL, Swedlow JR, Waters JC. Evaluating performance in three-dimensional fluorescence microscopy. Journal of Microscopy. 2007;228:390–405. doi: 10.1111/j.1365-2818.2007.01861.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biggs DSC. Current Protocols in Cytometry. John Wiley & Sons, Inc; 2001. 3D Deconvolution Microscopy. [DOI] [PubMed] [Google Scholar]
- Chalfie M, Tu Y, Euskirchen G, Ward W, Prasher D. Green fluorescent protein as a marker for gene expression. Science. 1994;263:802–805. doi: 10.1126/science.8303295. [DOI] [PubMed] [Google Scholar]
- Wiedenmann J, Oswald F, Nienhaus GU. Fluorescent proteins for live cell imaging: Opportunities, limitations, and challenges. IUBMB Life. 2009;61:1029–1042. doi: 10.1002/iub.256. [DOI] [PubMed] [Google Scholar]


