Abstract
Exit from mitosis in budding yeast requires inactivation of cyclin-dependent kinases through mechanisms triggered by the protein phosphatase Cdc14. Cdc14 activity, in turn, is regulated by a group of proteins, the mitotic exit network (MEN), which includes Lte1, Tem1, Cdc5, Cdc15, Dbf2/Dbf20, and Mob1. The direct biochemical interactions between the components of the MEN remain largely unresolved. Here, we investigate the mechanisms that underlie activation of the protein kinase Dbf2. Dbf2 kinase activity depended on Tem1, Cdc15, and Mob1 in vivo. In vitro, recombinant protein kinase Cdc15 activated recombinant Dbf2, but only when Dbf2 was bound to Mob1. Conserved phosphorylation sites Ser-374 and Thr-544 (present in the human, Caenorhabditis elegans, and Drosophila melanogaster relatives of Dbf2) were required for DBF2 function in vivo, and activation of Dbf2-Mob1 by Cdc15 in vitro. Although Cdc15 phosphorylated Dbf2, Dbf2–Mob1, and Dbf2(S374A/T544A)–Mob1, the pattern of phosphate incorporation into Dbf2 was substantially altered by either the S374A T544A mutations or omission of Mob1. Thus, Cdc15 promotes the exit from mitosis by directly switching on the kinase activity of Dbf2. We propose that Mob1 promotes this activation process by enabling Cdc15 to phosphorylate the critical Ser-374 and Thr-544 phosphoacceptor sites of Dbf2.
The cell cycle in Saccharomyces cerevisiae is controlled by a single cyclin-dependent kinase (Cdk), Cdc28. By associating with different cyclins, Cdc28 activity orchestrates cell cycle progression. After sister chromatid separation, mitotic Cdk activity is extinguished, and cells exit mitosis. Separation of sister chromatids and inactivation of mitotic Cdk are both regulated by proteolysis via the anaphase-promoting complex/cyclosome (APC/C) ubiquitin ligase. The APC/C in cooperation with the substrate specificity factor Cdc20 initiates anaphase by targeting the anaphase inhibitor Pds1 for degradation (1). Pds1 forms a complex with the separin Esp1, thereby preventing Esp1 from cleaving Scc1, a cohesin subunit that helps hold sister chromatids together (2, 3). Pds1 also impedes Cdk inactivation by preventing release of protein phosphatase Cdc14 from the nucleolus (4). Cdc14, which is tethered to the nucleolus in an inactive state by Net1/Cfi1 (5, 6), promotes cyclin degradation on its release from the nucleolus in anaphase by removing inhibitory phosphates from Cdh1/Hct1, which then targets Clb2 to the APC/C for ubiquitin-mediated proteolysis (7–9). Cdc14 also promotes the accumulation of the Cdk inhibitor Sic1 by dephosphorylating Sic1 and its transcription factor, Swi5, thereby stabilizing the former and allowing the latter to enter the nucleus (8).
Mobilization of Cdc14 activity during anaphase depends on a group of genes (LTE1, TEM1, CDC5, CDC15, DBF2/DBF20, and MOB1) that comprise the mitotic exit network (MEN). Loss of function of the MEN causes cells to arrest in late anaphase/telophase, similar to the arrest caused by overexpression of non-degradable B-type cyclin (10). Components of the MEN interact genetically with each other and act upstream of Cdc14 as suggested by epistasis studies (8, 11–17). Lte1, a putative guanine-nucleotide exchange factor (GEF), and Tem1, a GTP-binding protein, are likely to be at the top of this pathway (12, 15, 18). Lte1 is concentrated at the bud cortex, and Tem1 associates with the heterodimeric GTPase-activating protein (GAP) Bub2-Bfa1/Byr4 at the cytoplasmic face of the spindle pole body (SPB). Tem1 presumably becomes activated when the spindle pole is translocated into the bud, which brings it into proximity with Lte1 (19, 20).
The functional organization of the remaining components of the MEN is unclear. Protein kinase Cdc15 associates with Tem1 at the spindle pole in mitotic cells, and epistasis analyses suggest that it acts downstream of Lte1 and Tem1 (12, 13, 15, 19). Cdc5, Dbf2, and Dbf20 are also protein kinases (11, 21), but their functional relationship to other MEN components remains obscure. Cdc5 is implicated in a variety of processes in other organisms that do not appear to involve the MEN, including activation of both APC/CCdc20 and the protein phosphatase Cdc25 (22). Dbf2 and Dbf20 are functionally redundant homologues, but Dbf2, which is abruptly activated during anaphase, accounts for the majority of the Dbf2/Dbf20-associated kinase activity (21, 23). The biochemical function of Mob1 is unknown, but Mob1 binds Dbf2, and the lethality of a dbf2Δ/dbf20Δ is rescued by Mob1 overexpression (16, 17). To gain greater insight into the functional organization of the MEN, we examined the interaction between Dbf2, Mob1, and Cdc15. Here, we provide both genetic and biochemical evidence that Cdc15 directly phosphorylates and activates Dbf2. Surprisingly, this activation step depends on Mob1.
Materials and Methods
Yeast Methods.
The yeast strains used in this study are listed in Table 1. All strains are derivatives of W303 (can1-100, leu2-3, -112, and his3-11, -15, trp1-1, ura3-1, ade2-1). Chromosomal DBF2 in haploid strains was tagged with FlagHis6HA3 (FHH) by transformation with StuI-linearized p03-DBF2TS (RDB884), a pRS303 (24) plasmid with nucleotides −266 to +45 of DBF2, the PCR-generated FHH tag, followed by nucleotides +49 to +425 of DBF2 inserted into the PvuII site. Diploid strains were constructed by mating chromosomally tagged FHHDBF2 strains with untagged strains, with the exception of YAM2, in which the untagged strains were mated and the resulting diploid strain (YAM10) was transformed with p03-DBF2TS. The dbf2-2 strain, RJD1218, was transformed with pAM17, pAM18, pAM19, pAM20, pAM21, and pRS129, to obtain strains YAM4, YAM5, YAM6, YAM7, YAM8, and YAM9, respectively, and grown on minimal media (SD) supplemented with the appropriate nutrients, excluding tryptophan. All other strains were grown in 1% yeast extract, 2% peptone, 2% dextrose (YPD).
Table 1.
Yeast strains used in this study
| Strain* | Relevant genotype |
|---|---|
| YAM1 | MATa/MATα DBF2∷FHHDBF2 (HIS3)/+ |
| YAM2 | MATa/MATα mob1-77/mob1-77 DBF2∷FHHDBF2 (HIS3)/+ |
| YAM3 | MATa/MATα cdc15-2/cdc15-2 pep4∷TRP1/pep4∷TRP1 bar1∷LEU2/+ DBF2∷FHHDBF2 (HIS3)/+ |
| YAM4 | MATα dbf2-2 [pGAL1-FHHDBF2/TRP1] |
| YAM5 | MATα dbf2-2 [pGAL1-FHHDBF2(N305A)/TRP1] |
| YAM6 | MATα dbf2-2 [pGAL1-FHHDBF2(S374A)/TRP1] |
| YAM7 | MATα dbf2-2 [pGAL1-FHHDBF2(T544A)/TRP1] |
| YAM8 | MATα dbf2-2 [pGAL1-FHHDBF2(S374A/T544A)/TRP1] |
| YAM9 | MATα dbf2-2 [pRS129] |
| RJD361 | MATα |
| RJD381 | MATa/MATα |
| RJD1218 | MATα dbf2-2 |
| RJD1224 | MATa DBF2∷FHHDBF2 (HIS3) |
| RJD1225 | MATα cdc5-1 DBF2∷FHHDBF2 (HIS3) |
| RJD1228 | MATα cdc14-1 DBF2∷FHHDBF2 (HIS3) |
| RJD1264 | MATa tem1-3 bar1∷hisG DBF2∷FHHDBF2 (HIS3) |
| RJD1265 | MATa cdc15-2 bar1∷LEU2 pep4∷TRP1 DBF2∷FHHDBF2 (HIS3) |
All strains are in the W303 background (ura3, leu2, trp1, his3, ade2).
Plasmid and Baculovirus Constructions.
The plasmids constructed for this study are listed in Table 2. Sequences of oligonucleotides and details of constructs are available on request. All PCR-amplified coding sequences were verified by DNA sequencing.
Table 2.
Plasmids constructed for this study
| Plasmid | Construct* | Alias |
|---|---|---|
| RDB884 | pRS303-FHHDBF2(nt+49→425) | |
| RDB1267 | pFastBac1-FHHDBF2 | AM1 |
| RDB1331 | pFastBacHTa(-XhoI)-H6Mob1 | AM4 |
| RDB1335 | pFastBacHTa(-XhoI)-H6Mob1TM9 | AM5 |
| RDB1337 | pFastBac1-FHHDBF2(N305A) | AM7 |
| RDB1351 | pFastBac1-FHHDBF2(S374A) | AM8 |
| RDB1352 | pFastBac1-FHHDBF2(T544A) | AM9 |
| RDB1354 | pFastBac1-FHHDBF2(S374A/T544A) | AM10 |
| RDB1414 | pRS129-FHHDBF2 | AM17 |
| RDB1415 | pRS129-FHHDBF2(N305A) | AM18 |
| RDB1416 | pRS129-FHHDBF2(S374A) | AM19 |
| RDB1417 | pRS129-FHHDBF2(T544A) | AM20 |
| RDB1418 | pRS129-FHHDBF2(S374A/T544A) | AM21 |
Details of constructs are available upon request.
FHH and the N-terminal portion of DBF2 (nucleotides +49 to +437) were amplified by PCR from p03DBF2TS with primers that generated RsrII and EcoRI sites at the 5′ and 3′ ends, respectively. The remainder of DBF2 was obtained as an EcoRI fragment from pRS304-DBF2-myc (a gift from C. L. Denis, University of New Hampshire). The two fragments were ligated into the RsrII/EcoRI sites of pFastBac1 (GIBCO/BRL) to yield pAM1. DBF2 was subjected to site-directed mutagenesis using pAM1 as a template, to create a kinase-inactive mutant (pAM7) and the phosphorylation site mutants (pAM8, pAM9, and pAM10).
MOB1 was cloned into pFastBacHTa(-XhoI), which contains a His6 (H6) tag and lacks the XhoI site, removed by XhoI cleavage, followed by Klenow fill-in before religation. MOB1 was amplified from pRSETA-MOB1(1-314) (a gift from F. Luca, University of Pennsylvania) by PCR, creating a BamHI site at the 5′ end and XhoI and EcoRI sites at the 3′ end. MOB1 was ligated into the BamHI/EcoRI sites of pFastBacHTa(-XhoI) to yield pAM4. An XhoI-EcoRI fragment that contained the TEV2Myc9 (TM9) tagging cassette (two tobacco etch virus protease sites followed by nine copies of the Myc epitope and a stop codon) was ligated into the XhoI/EcoRI sites of pAM4 to yield pAM5. The Bac-to-Bac baculovirus system (GIBCO/BRL) was subsequently used to generate recombinant baculoviruses.
DBF2 was subcloned into pRS129, a pRS314-based (CEN ARS) plasmid with a GAL1 promoter, for RJD1218 transformations. pAM1 was cleaved with RsrII, the ends blunted, and then digested with SpeI. FHHDBF2 was ligated into the SmaI/SpeI sites of pRS129, yielding pAM17. The same procedure was used for obtaining pAM18, pAM19, pAM20, and pAM21 (Table 2).
Yeast Extract Immunoprecipitation and Kinase Assay.
Yeast strains were grown in YPD at 25°C to an OD600 of 0.5, and half of each culture was shifted to 37°C for ≈3 h, until greater than 90% of the cells at 37°C had arrested. Approximately twenty-five OD600 units of cells were harvested and washed with Solution A (150 mM NaCl/50 mM Tris, pH 7.4) and resuspended in 200 μl Lysis Buffer (150 mM NaCl/50 mM Tris, pH 7.4/2 mM EDTA, pH 8.0/1% Triton X-100/10% glycerol/2 mM DTT/25 μg/ml aprotinin/10 μg/ml pepstatin/10 μg/ml chymostatin/10 μg/ml leupeptin/1 mM benzamidine/1 mM PMSF/10 mM NaF, 60 mM β-glycerophosphate/10 mM sodium pyrophosphate/2 mM sodium vanadate). An equal volume of glass beads was added, and the cells were lysed by eight cycles of vortexing and icing, at 30-s intervals. The lysates were clarified by centrifugation and supplemented with 1 μl 12CA5 (anti-HA monoclonal antibody), incubated on ice for 1 h, then added to 10 μl of Protein A beads (Sigma). The samples were rotated for 1 h at 4°C, and the beads were washed three times with Lysis Buffer, twice with solution B (150 mM NaCl/50 mM Tris, pH 7.4/2 mM EDTA, pH 8.0/1% Triton X-100/10% glycerol/2 mM DTT), and once with solution A, then washed with Dbf2 kinase buffer (DKB: 50 mM Tris, pH 7.4/60 mM potassium acetate/10 mM magnesium chloride/1 mM DTT/10 μM ATP) before incubation at room temperature for 30 min in 10 μl of DKB with 2 μCi of [γ-32P]ATP and 5 μg of calf thymus H1 histone (Sigma). Reactions were stopped with 2× SDS/PAGE sample buffer and analyzed by SDS/PAGE. FHHDbf2 levels were detected by Western blotting using the primary antibody 12CA5, followed by goat anti-mouse horse radish peroxidase (HRP)-conjugate (Bio-Rad), and ECL+ (Amersham Pharmacia). The H1 kinase assay was evaluated by PhosphorImager (Molecular Dynamics).
Baculovirus Protein Expression and Purification.
Hi5 insect cells were infected with the appropriate baculovirus at a multiplicity of infection (moi) of 10. Cells infected with the recombinant Cdc15H6 or Cdc15(K54L)H6 baculoviruses (gifts from D. O. Morgan, University of California, San Francisco) were collected after 72 h. FHHDbf2 baculovirus-infected cells were harvested 48 h postinfection, as were cells coinfected (at a 1:1 ratio) with H6Mob1TM9 baculovirus. Insect cells treated with okadaic acid were incubated for 3 h in 0.1 μM okadaic acid (GIBCO/BRL) before harvesting. Harvested cells were washed once in 1× PBS and resuspended in 500 μl of cold buffer A (10 mM Hepes-KOH, pH 7.5/150 mM NaCl/20 mM β-glycerophosphate/5 mM EGTA/5 mM β-mercaptoethanol/1% 3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonate (CHAPS)/1 mM PMSF/10 μg/ml pepstatin/10 μg/ml leupeptin/10 μg/ml chymostatin; 0.1 mM sodium orthovanadate and 3 μM microcystin were supplemented in buffer A for okadaic acid-treated cells) per 107 cells, then incubated on ice for 15 min.
Cdc15H6 was purified by adding the clarified lysate to 100 μl Ni-NTA beads (Qiagen, Chatsworth, CA) and incubated at 4°C for 1 h on a rotator. The beads were washed six times with buffer B (10 mM Hepes-KOH, pH 7.5/500 mM NaCl/20 mM β-glycerophosphate/5 mM EGTA/5 mM β-mercaptoethanol/0.1% CHAPS; 0.1 mM sodium orthovanadate was supplemented in buffer B for okadaic acid-treated cells), then eluted with 100 μl of buffer C (10 mM Hepes-KOH, pH 7.5/25 mM NaCl/20 mM β-glycerophosphate/5 mM EGTA/5 mM β-mercaptoethanol/200 mM imidazole, pH 7.5) for 10 min at 4°C. The eluted protein was dialyzed twice for 2 h in 500 ml HBS (10 mM Hepes-KOH, pH 7.5/150 mM NaCl) supplemented with 1 mM DTT and 1 mM EDTA. The same procedure was used in the purification of Cdc15(K54L)H6. Purification of FHHDbf2 and FHHDbf2-H6Mob1TM9 was as for Cdc15H6, but Anti-FLAG M2 beads (Sigma) were used instead and not eluted.
Protein Kinase Assays with Recombinant Proteins.
Bead-bound FHHDbf2 and FHHDbf2-H6Mob1TM9 were freeze-thawed once and washed three times with TBST (50 mM Tris, pH 7.6/150 mM NaCl/0.2% Triton X-100) before being used in kinase assays. H1 kinase assays with baculovirus-expressed FHHDbf2 were performed as described in Yeast Extract Immunoprecipitation and Kinase Assay. Phosphorylation of various FHHDbf2-based substrates by Cdc15H6 was performed by washing beads with Cdc15 kinase buffer (CKB: 50 mM Hepes-KOH, pH 7.5/5 mM MgCl2/2.5 mM MnCl2/5 mM β-glycerophosphate/1 mM DTT/20 μM ATP) and incubating in CKB and 2 μCi of [γ-32P]ATP for 30 min at room temperature. To monitor activation of FHHDbf2 substrates by Cdc15H6, immobilized FHHDbf2 substrates were washed with CKB and then incubated with CKB (with 1 mM ATP) and Cdc15H6 for 30 min at room temperature. The beads were washed three times with TBST to remove cold ATP and Cdc15H6, washed once with DKB, then incubated with DKB, 2 μCi of [γ-32P]ATP, and 5 μg histone H1 for 30 min at room temperature. HBS was substituted for Cdc15H6 in controls. Kinase reactions were stopped with 2× SDS/PAGE sample buffer and analyzed by SDS/PAGE. Protein levels were detected by Western blotting with anti-His6 (Santa Cruz Biotechnology) followed by goat anti-rabbit HRP-conjugate (Bio-Rad) and ECL+ (Amersham Pharmacia). PhoshorImager analysis was used to evaluate kinase assays.
Phosphopeptide Mapping.
FHHDbf2-H6Mob1TM9 and the FHHDbf2 mutants were retrieved from insect cells on anti-Flag beads and were incubated with Cdc15H6 in CKB containing 1 μM of ATP and 2 μCi of [γ-32P]ATP for 30 min at room temperature. The samples were fractionated by SDS/PAGE and transferred to nitrocellulose. γ-32P-labeled FHHDbf2 was detected by autoradiography and excised. The samples were then digested with trypsin and prepared for phosphopeptide mapping as described (25). The digested peptides were resuspended in 2.2% formic acid, 7.8% glacial acetic acid (pH 1.9 buffer) and resolved on thin-layer cellulose plates (EM Science) by electrophoresis in the first dimension (pH 1.9 buffer) and chromatography (37.5% 1-butanol, 25% pyridine, 7.5% glacial acetic acid) in the second dimension. Peptides were visualized by autoradiography.
Results
Dbf2 Kinase Activity Is Dependent on Tem1, Cdc15, and Mob1.
The proteins Tem1, Cdc15, Cdc5, Dbf2, and Mob1 form a poorly understood regulatory network that controls exit from mitosis (5, 6, 8, 11–17). To gain insight into the regulation of exit from mitosis, we sought to position Dbf2 within this network by measuring Dbf2 kinase activity in various mutant strains.
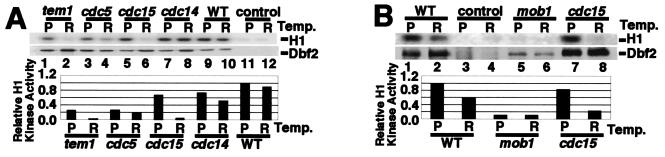
The DBF2 locus in the temperature sensitive mutants tem1-3, cdc5-1, cdc15-2, and cdc14-1 was modified to encode a protein tagged at its amino terminus with the tripartite Flag-His6-HA3 epitope. The FHHDBF2 mob1-77 combination was synthetic lethal, and therefore a mob1-77/mob1-77 FHHDBF2/+ diploid strain was generated. FHHDbf2 immunoprecipitated from mutants grown at the permissive and nonpermissive temperatures was assayed for kinase activity by using histone H1 as an artificial substrate. In the tem1-3 and cdc15-2 mutants, FHHDbf2 activity was sharply decreased at the restrictive temperature although, at the permissive temperature of tem1-3, kinase activity was also reduced in comparison to wild-type (Fig. 1A). FHHDbf2 kinase activity was also significantly diminished in both the cdc5-1 and mob1-77/mob1-77 mutants, but the reduction was observed at both permissive and restrictive temperatures (Fig. 1). The latter result was unexpected, because Mob1 has been proposed to function downstream of Dbf2 (16). It was also noted in repeated experiments that the amount of FHHDbf2 immunoprecipitated from the mob1-77 diploid strain was reduced in comparison to other strains. In contrast to the above mutants, FHHDbf2 activity remained unchanged in cdc14-1 and wild-type cells. Taken together, these observations suggested that Dbf2 required signaling by MEN proteins to become activated, and that Dbf2 acted at or near the terminus of the MEN, before the mobilization of Cdc14.
Figure 1.
Dbf2 kinase activity in MEN mutants. (A) Dbf2 kinase activity is decreased in the tem1-3 and cdc5-1 strains at the permissive temperature, but severely reduced in the tem1-3 and cdc15-2 strains at the restrictive temperature. The temperature sensitive strains tem1-3, cdc5-1, cdc15-2, and cdc14-1 with chromosomally tagged DBF2 (FHHDBF2) were grown at the permissive temperature of 25°C, and then half of each culture was shifted to 37°C, the restrictive temperature, for ≈3 h. An untagged strain was used as a negative control. Cells were lysed with glass beads, FHHDbf2 was immunoprecipitated with anti-HA monoclonal antibody 12CA5 bound to protein A beads, and bead-bound histone H1 kinase activity was evaluated (Upper) and quantitated by PhosphorImager. PhosphorImager analysis of an anti-HA immunoblot (Lower) was used to estimate FHHDbf2 levels. The ratios of histone H1 kinase activity/FHHDbf2 antigen level are plotted in the graph. (B) In the mob1-77/mob1-77 mutant, Dbf2 kinase activity is greatly diminished. Immunoprecipitations, H1 kinase assays, and quantitations were performed as in A, but with diploid strains.
Mob1 Is Required for Obtaining Active Dbf2 from Okadaic Acid-Treated Insect Cells.
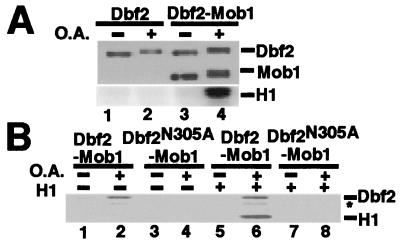
Tem1 is thought to act near the top of the MEN, and a combination of genetic and biochemical evidence suggests strongly that Cdc15 acts directly downstream of Tem1 (12, 13, 15, 19). Thus, we reasoned that Cdc15, Cdc5, and/or Mob1 might directly activate Dbf2. To distinguish between these possibilities, we sought to reconstitute Dbf2 activation in vitro with recombinant proteins. Because Cdc15 and Cdc5 are protein kinases, we first tested the possibility that Dbf2 is activated by phosphorylation. FHHDbf2 was expressed in Hi5 insect cells by infection with a recombinant baculovirus, and cells were incubated either with or without the phosphatase inhibitor okadaic acid before harvesting. FHHDbf2 recovered on anti-Flag resin from okadaic acid-treated cells migrated more slowly on SDS/PAGE, suggesting that okadaic acid promoted accumulation of phosphate on FHHDbf2 (Fig. 2A, Upper, lanes 1 and 2). Nevertheless, this material did not exhibit kinase activity (Lower).
Figure 2.
Active Dbf2 is obtained from okadaic acid-treated insect cells when coexpressed with Mob1. (A) Okadaic acid- and Mob1-dependent activation of Dbf2 expressed in insect cells. Hi5 insect cells were infected with FHHDbf2 or coinfected with FHHDbf2 and H6Mob1TM9 baculoviruses. Okadaic acid (O.A.) treatment (lanes 2 and 4) was for 3 h before harvesting cells. Anti-FLAG beads were used for immunoprecipitation, and bead-bound FHHDbf2 and FHHDbf2-H6Mob1TM9 were subjected to an H1 kinase assay with [γ-32P]ATP, which was evaluated by phosphorimaging (Lower). FHHDbf2 and H6Mob1TM9 proteins were evaluated by immunoblotting with anti-His6 (Upper). The samples shown in lanes 1 and 2 (Upper) were overloaded 6-fold to compensate for the reduced accumulation of Dbf2 in insect cells. (B) H1 phosphorylation is attributed to Dbf2. The indicated FHHDbf2 and H6Mob1TM9 complexes were isolated from insect cells treated with (even-numbered lanes) or without (odd-numbered lanes) okadaic acid, and evaluated for autophosphorylation (lanes 1–4) or histone H1 kinase activity (lanes 5–8) as described in A. The identity of the band marked with the asterisk is not known.
Because Dbf2 kinase activity depended on Mob1 in vivo (Fig. 1B), we tested whether Mob1 influenced the recovery of Dbf2 kinase activity from insect cells. Hi5 cells were coinfected with baculoviruses that expressed FHHDbf2 and H6Mob1TM9, and FHHDbf2 was retrieved on anti-Flag resin. H6Mob1TM9 was recovered in association with FHHDbf2, indicating that the recombinant proteins form a complex (Fig. 2A, Upper, lane 3). H6Mob1TM9 association with FHHDbf2 was specific as the Flag resin did not bind H6Mob1TM9 alone (data not shown). Importantly, the FHHDbf2–H6Mob1TM9 complex retrieved from okadaic acid-treated Hi5 cells had histone H1 kinase activity, whereas the protein complex from untreated cells was inactive (Fig. 2A, Lower, lanes 3 and 4). To determine whether the kinase activity was due to FHHDbf2, a mutant Dbf2 lacking an asparagine residue that is conserved in protein kinase active sites (26), FHHDbf2(N305A), was coexpressed with H6Mob1TM9. Whereas Dbf2 has both autophosphorylation (16) and histone H1 kinase activities, FHHDbf2(N305A)-H6Mob1TM9 was inactive in both assays (Fig. 2B). These results suggest that FHHDbf2 was activated only when bound to H6Mob1TM9, and that activation of the FHHDbf2–H6Mob1TM9 complex was promoted by phosphorylation.
Baculovirus-Expressed Cdc15 Activates Dbf2 in an ATP- and Mob1-Dependent Manner.
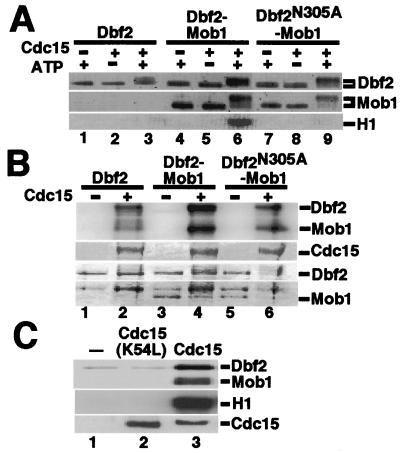
To determine the identity of the putative Dbf2–Mob1-activating kinase, H6HA3Cdc5 and Cdc15H6 were expressed in insect cells from recombinant baculoviruses, purified, and incubated in the presence of ATP with various FHHDbf2 substrates (FHHDbf2, FHHDbf2-H6Mob1TM9, and FHHDbf2(N305A)-H6Mob1TM9) retrieved from untreated insect cells on anti-Flag resin. After incubation, soluble H6HA3Cdc5 and Cdc15H6 were washed away, and the kinase activity of immobilized FHHDbf2 was measured. Recombinant H6HA3Cdc5 had no effect on the activity of recombinant FHHDbf2 substrates (data not shown). In contrast, both FHHDbf2 and H6Mob1TM9 were modified in an ATP-dependent manner on incubation with Cdc15H6 (Fig. 3A, lanes 3, 6, and 9). Moreover, Cdc15H6 activated FHHDbf2 in an ATP-dependent manner, but only when FHHDbf2 was bound to H6Mob1TM9 (lane 6).
Figure 3.
Dbf2 kinase activity depends on Mob1 and is activated by Cdc15 protein kinase. (A) Cdc15 phosphorylates Dbf2, activating its Mob1-dependent kinase activity in the presence of ATP. Baculovirus-expressed FHHDbf2, FHHDbf2-H6Mob1TM9, and FHHDbf2(N305A)-H6Mob1TM9 attached to beads were incubated with (+) or without (−) Cdc15H6 purified from insect cells, in CKB containing no ATP (−) or 1 mM ATP (+). The beads were then washed, and half were subjected to immunoblotting with anti-His6 (Top and Middle). The other half was incubated with DKB containing histone H1 and [γ-32P]ATP. Reaction products were detected by PhosphorImager (Bottom). (B) Dbf2 and Mob1 are phosphorylated by Cdc15. Same as A, except that FHHDbf2 substrates were incubated with Cdc15H6 in the presence of 20 μM ATP plus [γ-32P]ATP. (Top) PhosphorImager analysis of the kinase reaction. The Bottom three panels are the protein levels as detected by staining with Coomassie Blue. (C) Kinase-inactive Cdc15 neither phosphorylates nor activates Dbf2. FHHDbf2-H6Mob1TM9 was incubated with buffer (lane 1), Cdc15(K54L)H6, or Cdc15H6 as in B (Top), or subjected to an H1 kinase assay as in A (Middle). Reaction products were visualized by phosphorimaging. (Bottom) Cdc15H6 levels as detected by immunoblotting with anti-His6.
To address whether Cdc15H6 phosphorylated FHHDbf2 and H6Mob1TM9, we incubated Cdc15H6 with FHHDbf2, FHHDbf2-H6Mob1TM9, and FHHDbf2(N305A)-H6Mob1TM9 in the presence of [γ-32P]ATP. Cdc15H6 incorporated label into all three substrates (Fig. 3B). The most intense incorporation was obtained with FHHDbf2-H6Mob1TM9, suggesting that, on its activation by Cdc15, FHHDbf2 may phosphorylate itself and its Mob1 partner. To confirm that Cdc15H6 activated FHHDbf2-H6Mob1TM9 by phosphorylation, we obtained from D. Morgan (University of California, San Francisco) a recombinant baculovirus that expressed a Cdc15H6 mutant previously shown to lack kinase activity, in which the conserved lysine was mutated to leucine (15). As shown in Fig. 3C, Cdc15(K54L)H6 neither phosphorylated nor activated FHHDbf2-H6Mob1TM9. We conclude that Cdc15 phosphorylated both Dbf2 and Mob1, and that phosphorylation of one or both of these proteins switched on the kinase activity of Dbf2.
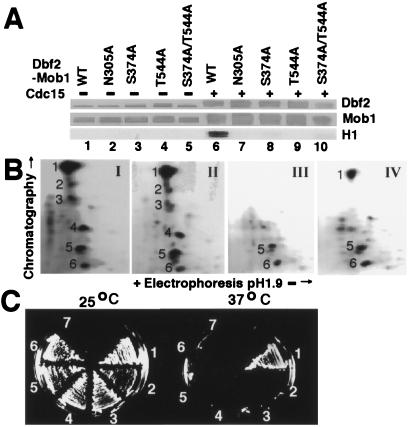
Cdc15 Phosphorylates Dbf2 on Ser-374 and Thr-544.
Activation of the human serine/threonine protein kinase Ndr (nuclear, Dbf2-related) requires its phosphorylation on Ser-281 and Thr-444 (27). These residues are conserved in the relatives of Ndr, including Dbf2 (28). To test whether these residues might correspond to sites of phosphorylation by Cdc15, we mutated the corresponding residues in FHHDbf2 (Ser-374 and Thr-544) to alanine to create single and double phosphorylation site mutants that were coexpressed with H6Mob1TM9 in insect cells. All three mutants were incapable of being activated by Cdc15H6 (Fig. 4A, Bottom, lanes 8, 9, and 10). The double mutant, FHHDbf2(S374A/T544A), was still upshifted in molecular weight (lane 10, Top) and incorporated [γ-32P]ATP (data not shown; see Fig. 4B) when treated with Cdc15H6, suggesting that Cdc15H6 phosphorylated FHHDbf2 on multiple sites. To address whether S374 and T544 were phosphorylated by Cdc15H6, FHHDbf2–H6Mob1TM9 complexes were treated with Cdc15H6 plus [γ-32P]ATP and fractionated by SDS/PAGE. Radiolabeled FHHDbf2 was excised from the gel, digested with trypsin, and resolved into phosphopeptides by TLC (Fig. 4B). Six major phosphopeptides were recovered from wild-type FHHDbf2, all of which were also recovered from the kinase mutant, FHHDbf2(N305A) (panels I and II). By contrast, the phosphopeptide map of FHHDbf2(S374A/T544A) (panel III) lacked four of the major phosphopeptides (peptides 1, 2, 3, and 4). To test whether Mob1 promotes activation of Dbf2 by enabling phosphorylation of the critical Ser-374 and Thr-544 residues, we prepared in parallel a phosphopeptide map for FHHDbf2 treated with Cdc15H6 in the absence of H6Mob1TM9 (panel IV). Interestingly, the resulting pattern was reminiscent of that obtained for the FHHDbf2(S374A/T544A)–H6Mob1TM9 complex, in that peptides 2, 3, and 4 were absent, whereas peptide 1 was reduced in amount. Taken together, these data suggest that binding of Mob1 to Dbf2 enables Cdc15 to phosphorylate Dbf2 on Ser-374 and Thr-544, and that phosphorylation of these residues underlies the mobilization of Dbf2 kinase activity.
Figure 4.
Direct phosphorylation of Dbf2 by Cdc15 is required for Dbf2 kinase activity and DBF2 function. (A) S374A and T544A mutations prevent Dbf2 activation by Cdc15. Baculovirus-expressed wild-type FHHDbf2, kinase-inactive FHHDbf2(N305A), and the phosphorylation site mutants FHHDbf2(S374A), FHHDbf2(T544A), and FHHDbf2(S374A/T544A) were coexpressed with H6Mob1TM9, purified on Flag resin, treated with Cdc15H6, and assayed for H1 kinase activity as in Fig. 3A. The Top two panels are anti-His6 Western blots and the Bottom panel displays the kinase activity, detected by autoradiography. (B) Tryptic map of Dbf2 phosphorylation sites. Bead-bound (I) FHHDbf2-H6Mob1TM9, (II) FHHDbf2(N305A)-H6Mob1TM9, (III) FHHDbf2(S374A/T544A)-H6Mob1TM9, and (IV) FHHDbf2 from insect cells were phosphorylated by Cdc15H6 as in Fig. 3B with [γ-32P]ATP. The proteins were transferred onto nitrocellulose, and FHHDbf2 was excised and digested into peptides, which were separated in two dimensions and detected by autoradiography. I and II were exposed for 1 day, III and IV for 5 days, although the intensity was normalized with photoshop using peptide 6. (C) Dbf2 phosphorylation sites S374 and T544 are required in vivo. Plasmids containing (1) FHHDBF2, (2) FHHDBF2(N305A), (3) FHHDBF2(S374A), (4) FHHDBF2(T544A), (5) FHHDBF2(S374A/T544A), or (6) vector alone were introduced into a dbf2-2 strain, and transformants were grown on SD-TRP plates at 25°C and 37°C. Sector 7 was streaked with the untransformed strain.
Dbf2 Phosphorylation Sites Ser-374 and Thr-544 Are Required in Vivo.
To determine whether Ser-374 and Thr-544 were important for Dbf2 function in vivo, we examined whether DBF2 phosphorylation site mutants, on expression from the GAL1 promoter, were able to rescue the conditional lethal dbf2-2 strain. Wild type FHHDBF2, but not the single or double phosphorylation site mutants, nor the kinase-dead FHHDBF2(N305A) mutant, restored growth of dbf2-2 on both glucose- (Fig. 4C) and galactose- (not shown) based media at the nonpermissive temperature. Thus, Ser-374 and Thr-544 were essential for Dbf2 function in vivo, presumably because Cdc15 must phosphorylate these sites to activate Dbf2 kinase activity.
Discussion
Organizing the MEN.
Spindle pole body-associated Tem1 is presumably activated when, on spindle elongation, it contacts guanine nucleotide exchange factor Lte1 in the cortex of the bud (19, 20). Activated Tem1 may enable recruitment of Cdc15 to the SPB, possibly through other proteins such as Nud1 (29). Mob1 would also need to be recruited to the SPB, if as in Schizosaccharomyces pombe, Mob1 localizes to the SPB only during mitosis (30, 31). Cdc15 would then be able to activate Dbf2, which appears to be present at the SPB throughout most of the cell cycle (32). Activated Dbf2 presumably phosphorylates one or more substrates, ultimately leading to release of Cdc14 from the nucleolus and exit from mitosis.
It remains completely unclear how Cdc5 fits into the mitotic exit network. Dbf2 kinase activity was reduced in cdc5-1 cells grown at the permissive temperature but, in contrast to tem1-3 and cdc15-1 mutants, was not significantly reduced on shift to the nonpermissive temperature. Whereas our observations are consistent with several scenarios, analysis of Cdc5 homologues that regulate cytokinesis place the polo-like kinase upstream in the pathway (22). Further genetic, biochemical, and cytological studies will be required to order Cdc5 function within the MEN.
What Does Mob1 Do?
Our data point to Mob1 as playing a key role in the activation of the Dbf2. Dbf2 kinase activity was dramatically lowered at both the permissive and restrictive temperatures in the mob1-77/mob1-77 strain, and Mob1 was absolutely required for the Cdc15-dependent activation of Dbf2 in a reconstituted system that used purified recombinant proteins. How does Mob1 promote Dbf2 activation? Although Cdc15 phosphorylated Dbf2 in the absence of Mob1, phosphorylation of the functionally critical Ser-374 or Thr-544 (or both) residues of Dbf2 was strongly stimulated by Mob1. In addition, Mob1 appears to stabilize Dbf2, in that Dbf2 levels were significantly reduced in mob1 mutant cells, and accumulation of Dbf2 in insect cells was significantly improved on coexpression with Mob1 (data not shown). On binding to Dbf2, Mob1 may promote a change in conformation that both stabilizes Dbf2 and facilitates its phosphorylation on Ser-374 and Thr-544 by Cdc15. By analogy, binding of cyclin A causes major conformational changes in Cdk2 (33) that are thought to enable phosphorylation of the critical T160 residue of Cdk2 by the Cdk-activating kinase p40(MO15) (34).
Contrary to our results, previous studies have shown that overexpression of Mob1 rescues the lethality of dbf2Δ/dbf20Δ (16), which would place Mob1 downstream of Dbf2. Mob1 may be a downstream substrate for Dbf2, because phosphorylation of Mob1 was reduced in the kinase-dead Dbf2(N305A) complex (Fig. 3B). Phosphorylation of Mob1 by Dbf2 may enhance, but may not be essential for, a downstream effector function of Mob1, such that overexpression of Mob1 can overcome the normally essential requirement for Dbf2/Dbf20. Studies in S. pombe, however, have shown that mob1 mutants are not rescued by overexpression of Sid2 (the S. pombe ortholog of Dbf2), and vice versa (30, 31). Our data, while arguing that Mob1 functions upstream of Dbf2, do not exclude a downstream function. A final resolution of this issue will require the identification of direct physiological targets of Dbf2-Mob1, which to date have remained elusive.
MEN and SIN.
The septation initiation network (SIN) in S. pombe is an analogous pathway to the MEN. The SIN includes Plo1, Spg1, Cdc7, Sid2, and Mob1, which correspond to the S. cerevisiae homologues Cdc5, Tem1, Cdc15, Dbf2, and Mob1 (30). As in budding yeast, the fission yeast Spg1 is regulated by a two-component GTPase-activating protein consisting of Byr4-Cdc16, which holds Spg1 in an inactive GDP-state at the SPB (35).
The SIN contains several components with no known counterparts in the MEN, including Sid1, Cdc14, Sid4, and Cdc11 (30). Sid1, associated with Cdc14 (no relation to S. cerevisiae Cdc14), localizes to the SPB with Cdc7-Spg1 during late anaphase, and epistasis analysis places it upstream of Sid2 (36). Taken together, these observations suggest that unidentified Sid1 and Cdc14 homologues in S. cerevisiae act between Cdc15 and Dbf2 (22). It remains possible, albeit unlikely, that an insect cell protein kinase binds tightly to either Cdc15 or Dbf2-Mob1, and is directly responsible for the activation of Dbf2. Such a hypothetical kinase would need to be activated in a Cdc15-dependent manner, because kinase-dead Cdc15 was unable to sustain activation of Dbf2-Mob1 (Fig. 3C). Another possibility is that the in vitro activation of Dbf2-Mob1 by Cdc15 is incomplete, and that Sid1/Cdc14 homologues or other factors (e.g., Sid4, Cdc11) may be required for optimal activation. This possibility is supported by quantitative considerations: the maximal kcat we observed for active Dbf2-Mob1 produced in our reconstituted system was 0.15 min−1 (R. Azzam, unpublished data), although this observation was based on histone H1 and not a physiological substrate, which remain unknown.
We wish to stress that the placement of Sid1 upstream of Sid2 was based on evaluating the localization of Sid2 in sid1 mutant strains, and not on direct biochemical analysis (36). In budding yeast, similar logic would place Cdc14 upstream of Dbf2 (32), even though it is well-accepted that Dbf2 is upstream of Cdc14 in the sense that it is required for the release of active Cdc14 from the nucleolus during anaphase. Interpreting the results of such experiments may be confounded by the operation of positive feedback loops within the MEN (37), so perhaps localization studies alone do not warrant placement of Sid1-Cdc14 upstream of Sid2.
At first glance the MEN and SIN pathways regulate different processes: mitotic exit in budding yeast, and cytokinesis in fission yeast. However, various lines of evidence point to a role for the MEN in cytokinesis, including the prominent cell separation defect of net1-1 tem1Δ and net1-1 cdc15Δ strains (5, 38), and the re-localization of MEN components from the SPB to the bud neck during late anaphase (32, 39, 40). An intriguing hypothesis is that the mobilization of Cdc14 activity by the MEN helps ensure the sequential progression of mitotic exit and cytokinesis. Specifically, the liberated Cdc14 dephosphorylates and hyperactivates Cdc15 (37), which may serve to re-direct the activity of Cdc15/Mob1/Dbf2 from the spindle pole body to the cleavage furrow.
Acknowledgments
We thank C. L. Denis and F. Luca for plasmids. We are particularly indebted to S. L. Jaspersen and D. O. Morgan for unpublished Cdc15 and Cdc15(K54L) baculoviruses. A.S.M. thanks W. Shou for her insight and encouragement, R. Azzam for helpful discussions, and R. Verma for help with peptide mapping. This work was supported by a grant from the National Institutes of Health (GM59940) and a Beckman Young Investigator Award (to R.J.D.). A.S.M. and R.J.D. are supported by the Howard Hughes Medical Institute.
Abbreviations
- MEN
mitotic exit network
- SIN
septation initiation network
- SPB
spindle pole body
- Cdk
cyclin-dependent kinase
- APC/C
anaphase-promoting complex/cyclosome
- FHH
FlagHis6HA3
- CKB
Cdc15 kinase buffer
- DKB
Dbf2 kinase buffer
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Morgan D O. Nat Cell Biol. 1999;1:E47–E53. doi: 10.1038/10039. [DOI] [PubMed] [Google Scholar]
- 2.Uhlmann F, Lottspeich F, Nasmyth K. Nature (London) 1999;400:37–42. doi: 10.1038/21831. [DOI] [PubMed] [Google Scholar]
- 3.Uhlmann F, Wernic D, Poupart M A, Koonin E V, Nasmyth K. Cell. 2000;103:375–386. doi: 10.1016/s0092-8674(00)00130-6. [DOI] [PubMed] [Google Scholar]
- 4.Shirayama M, Toth A, Galova M, Nasmyth K. Nature (London) 1999;402:203–207. doi: 10.1038/46080. [DOI] [PubMed] [Google Scholar]
- 5.Shou W, Seol J-H, Shevchenko A, Baskerville C, Moazed D, Shevchenko A, Charbonneau H, Deshaies R. Cell. 1999;97:233–244. doi: 10.1016/s0092-8674(00)80733-3. [DOI] [PubMed] [Google Scholar]
- 6.Visintin R, Hwang E, Amon A. Nature (London) 1999;398:818–823. doi: 10.1038/19775. [DOI] [PubMed] [Google Scholar]
- 7.Zachariae W, Schwab M, Nasmyth K, Seufert W. Science. 1998;282:1721–1724. doi: 10.1126/science.282.5394.1721. [DOI] [PubMed] [Google Scholar]
- 8.Visintin R, Craig K, Hwang E S, Prinz S, Tyers M, Amon A. Mol Cell. 1998;2:709–718. doi: 10.1016/s1097-2765(00)80286-5. [DOI] [PubMed] [Google Scholar]
- 9.Jaspersen S L, Charles J F, Morgan D O. Curr Biol. 1999;9:227–236. doi: 10.1016/s0960-9822(99)80111-0. [DOI] [PubMed] [Google Scholar]
- 10.Hoyt M A. Cell. 2000;102:267–270. doi: 10.1016/s0092-8674(00)00031-3. [DOI] [PubMed] [Google Scholar]
- 11.Kitada K, Johnson A L, Johnston L H, Sugino A. Mol Cell Biol. 1993;13:4445–4457. doi: 10.1128/mcb.13.7.4445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shirayama M, Matsui Y, Toh-e A. Mol Cell Biol. 1994;14:7476–7482. doi: 10.1128/mcb.14.11.7476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shirayama M, Matsui Y, Toh-e A. Mol Gen Genet. 1996;251:176–185. doi: 10.1007/BF02172916. [DOI] [PubMed] [Google Scholar]
- 14.Grandin N, de Almeida A, Charbonneau M. Mol Gen Genet. 1998;258:104–116. doi: 10.1007/s004380050712. [DOI] [PubMed] [Google Scholar]
- 15.Jaspersen S L, Charles J F, Tinker-Kulberg R L, Morgan D O. Mol Biol Cell. 1998;9:2803–2817. doi: 10.1091/mbc.9.10.2803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Komarnitsky S I, Chiang Y C, Luca F C, Chen J, Toyn J H, Winey M, Johnston L H, Denis C L. Mol Cell Biol. 1998;18:2100–2107. doi: 10.1128/mcb.18.4.2100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Luca F C, Winey M. Mol Biol Cell. 1998;9:29–46. doi: 10.1091/mbc.9.1.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shirayama M, Matsui Y, Tanaka K, Toh-e A. Yeast. 1994;10:451–461. doi: 10.1002/yea.320100404. [DOI] [PubMed] [Google Scholar]
- 19.Bardin A J, Visintin R, Amon A. Cell. 2000;102:21–31. doi: 10.1016/s0092-8674(00)00007-6. [DOI] [PubMed] [Google Scholar]
- 20.Pereira G, Hofken T, Grindlay J, Manson C, Schiebel E. Mol Cell. 2000;6:1–10. [PubMed] [Google Scholar]
- 21.Toyn J H, Johnston L H. EMBO J. 1994;13:1103–1113. doi: 10.1002/j.1460-2075.1994.tb06359.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nigg E A. Nat Rev Mol Cell Biol. 2001;2:21–32. doi: 10.1038/35048096. [DOI] [PubMed] [Google Scholar]
- 23.Toyn J H, Araki H, Sugino A, Johnston L H. Gene. 1991;104:63–70. doi: 10.1016/0378-1119(91)90465-n. [DOI] [PubMed] [Google Scholar]
- 24.Sikorski R S, Hieter P. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hunter T, Sefton B M. In: Methods in Enzymology. Abelson J N, Simon M I, editors. Vol. 201. San Diego: Academic; 1991. p. 547. [Google Scholar]
- 26.Hanks S K, Quinn A M, Hunter T. Science. 1988;241:42–52. doi: 10.1126/science.3291115. [DOI] [PubMed] [Google Scholar]
- 27.Millward T, Cron P, Hemmings B A. Proc Natl Acad Sci USA. 1995;92:5022–5026. doi: 10.1073/pnas.92.11.5022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Millward T A, Hess D, Hemmings B A. J Biol Chem. 1999;274:33847–33850. doi: 10.1074/jbc.274.48.33847. [DOI] [PubMed] [Google Scholar]
- 29.Gruneberg U, Campbell K, Simpson C, Grindlay J, Schiebel E. EMBO J. 2000;19:6475–6488. doi: 10.1093/emboj/19.23.6475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hou M C, Salek J, McCollum D. Curr Biol. 2000;10:619–622. doi: 10.1016/s0960-9822(00)00492-9. [DOI] [PubMed] [Google Scholar]
- 31.Salimova E, Sohrmann M, Fournier N, Simanis V. J Cell Sci. 2000;113:1695–1704. doi: 10.1242/jcs.113.10.1695. [DOI] [PubMed] [Google Scholar]
- 32.Frenz L M, Lee S E, Fesquet D, Johnston L H. J Cell Sci. 2000;113:3399–3408. doi: 10.1242/jcs.113.19.3399. [DOI] [PubMed] [Google Scholar]
- 33.Jeffrey P D, Russo A A, Polyak K, Gibbs E, Hurwitz J, Massague J, Pavletich N P. Nature (London) 1995;376:313–320. doi: 10.1038/376313a0. [DOI] [PubMed] [Google Scholar]
- 34.Kaldis P, Russo A A, Chou H S, Pavletich N P, Solomon M J. Mol Biol Cell. 1998;9:2545–2560. doi: 10.1091/mbc.9.9.2545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Balasubramanian M K, McCollum D, Surana U. J Cell Sci. 2000;113:1503–1513. doi: 10.1242/jcs.113.9.1503. [DOI] [PubMed] [Google Scholar]
- 36.Guertin D A, Chang L, Irshad F, Gould K L, McCollum D. EMBO J. 2000;19:1803–1815. doi: 10.1093/emboj/19.8.1803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jaspersen S L, Morgan D O. Curr Biol. 2000;10:615–618. doi: 10.1016/s0960-9822(00)00491-7. [DOI] [PubMed] [Google Scholar]
- 38.Lippincott J, Shannon K B, Shou W, Deshaies R J, Li R. J Cell Sci. 2001;114:1379–1386. doi: 10.1242/jcs.114.7.1379. [DOI] [PubMed] [Google Scholar]
- 39.Xu S, Huang H K, Kaiser P, Latterich M, Hunter T. Curr Biol. 2000;10:329–332. doi: 10.1016/s0960-9822(00)00382-1. [DOI] [PubMed] [Google Scholar]
- 40.Song S, Grenfell T Z, Garfield S, Erikson R L, Lee K S. Mol Cell Biol. 2000;20:286–298. doi: 10.1128/mcb.20.1.286-298.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]