Abstract
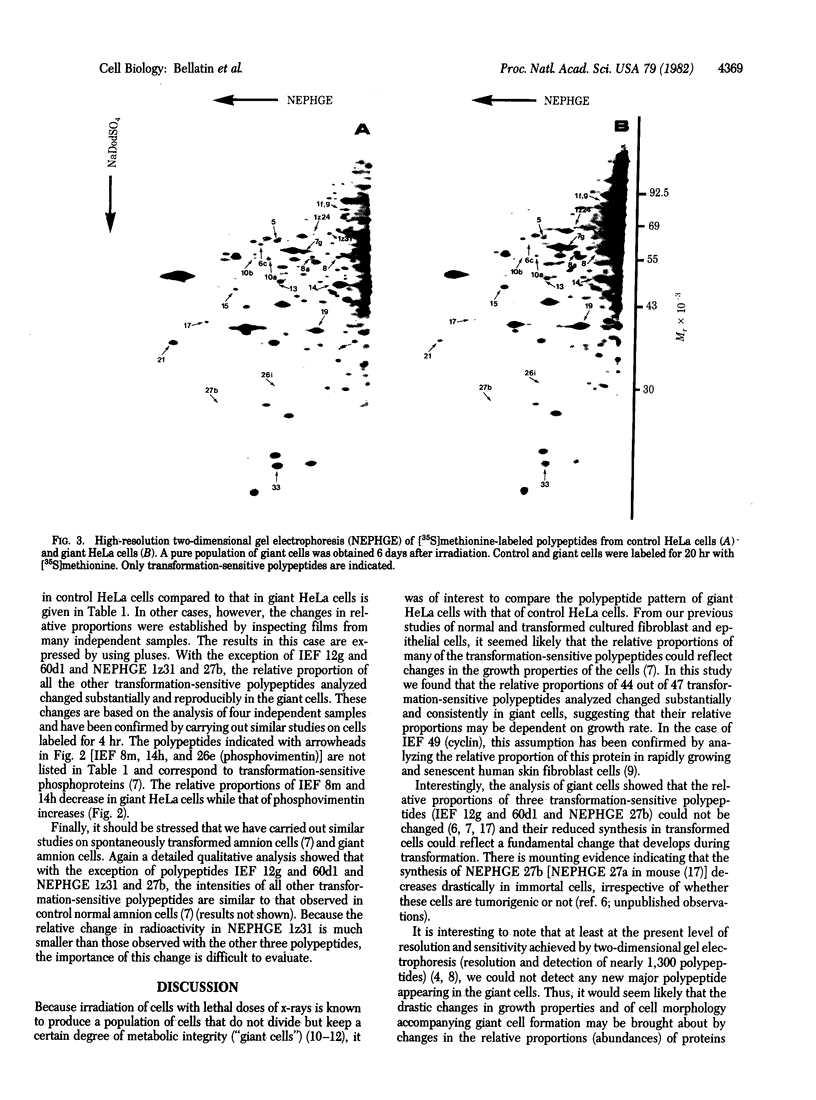
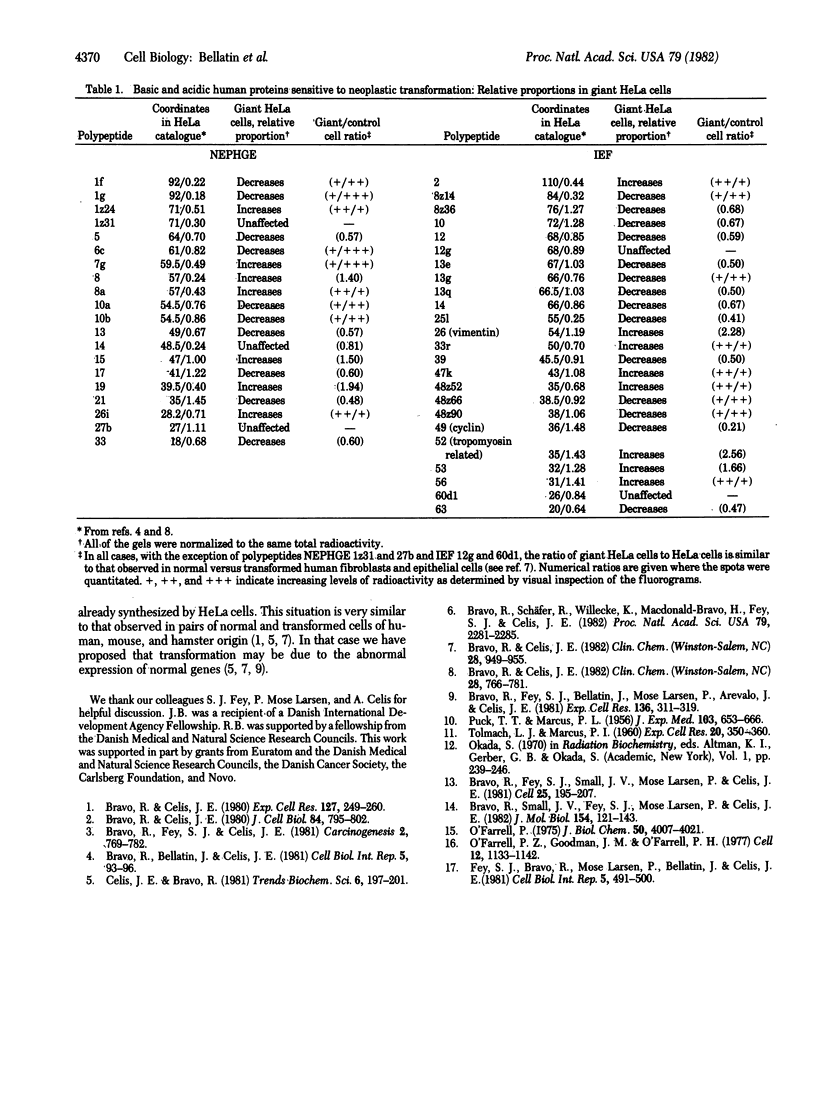
Irradiation of HeLa cells with 1,100 rads (1 rad = 0.01 J/kg = 0.01 Gy) of x-rays yielded a pure population of giant cells 5-7 days after irradiation. These cells do not divide but go through an intermittent DNA synthetic phase. The population of giant cells in S phase (8%) is considerably lower than that of control asynchronous HeLa cells (30%), but 80% of the giant cells go through S phase as determined by 48-hr labeling with [3H]thymidine. Previous studies with high-resolution two-dimensional gel electrophoresis identified 58 [35S]methionine-labeled polypeptides common to human epithelial amnion cells and lung fibroblasts, whose rate of synthesis is sensitive to neoplastic transformation [Bravo, R. & Celis, J. (1982) Clin. Chem. (Winston-Salem, NC) 28, 949-955]. These polypeptides also have been identified in HeLa cells and other transformed human cells such as Detroit 98, Chang liver, Fl-amnion, and WISH-amnion [Bravo, R. & Celis, J. (1982) Clin. Chem. (Winston-Salem, NC) 28, 949-955]. After irradiation of HeLa cells and giant cell formation, the relative proportions of most of the transformation-sensitive polypeptides (43 of 47) reverted to levels similar to those observed in non-tumorigenic cells. This suggests that their relative proportions are dependent on the growth properties of the cells. In particular, the relative proportions of three polypeptides (designated 12g and 60d1 in isoelectric focusing and 27b in nonequilibrium pH gradient electrophoresis) were not affected, indicating that their reduced amounts in transformed cells could reflect a fundamental change that develops during transformation.
Full text
PDF

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bravo R., Bellatin J., Celis J. E. [35S]-methionine labelled polypeptides from HELA cells. Coordinates and percentage of some major polypeptides. Cell Biol Int Rep. 1981 Jan;5(1):93–96. doi: 10.1016/0309-1651(81)90162-4. [DOI] [PubMed] [Google Scholar]
- Bravo R., Celis J. E. A search for differential polypeptide synthesis throughout the cell cycle of HeLa cells. J Cell Biol. 1980 Mar;84(3):795–802. doi: 10.1083/jcb.84.3.795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bravo R., Celis J. E. Gene expression in normal and virally transformed mouse 3T3b and hamster BHK21 cells. Exp Cell Res. 1980 Jun;127(2):249–260. doi: 10.1016/0014-4827(80)90430-9. [DOI] [PubMed] [Google Scholar]
- Bravo R., Celis J. E. Human proteins sensitive to neoplastic transformation in cultured epithelial and fibroblast cells. Clin Chem. 1982 Apr;28(4 Pt 2):949–954. [PubMed] [Google Scholar]
- Bravo R., Celis J. E. Up-dated catalogue of HeLa cell proteins: percentages and characteristics of the major cell polypeptides labeled with a mixture of 16 14C-labeled amino acids. Clin Chem. 1982 Apr;28(4 Pt 2):766–781. [PubMed] [Google Scholar]
- Bravo R., Fey S. J., Bellatin J., Larsen P. M., Arevalo J., Celis J. E. Identification of a nuclear and of a cytoplasmic polypeptide whose relative proportions are sensitive to changes in the rate of cell proliferation. Exp Cell Res. 1981 Dec;136(2):311–319. doi: 10.1016/0014-4827(81)90009-4. [DOI] [PubMed] [Google Scholar]
- Bravo R., Fey S. J., Celis J. E. Gene expression in murine hybrids exhibiting different morphologies and tumorigenic properties. Carcinogenesis. 1981;2(8):769–782. doi: 10.1093/carcin/2.8.769. [DOI] [PubMed] [Google Scholar]
- Bravo R., Fey S. J., Small J. V., Larsen P. M., Celis J. E. Coexistence of three major isoactins in a single sarcoma 180 cell. Cell. 1981 Jul;25(1):195–202. doi: 10.1016/0092-8674(81)90244-0. [DOI] [PubMed] [Google Scholar]
- Bravo R., Schafer R., Willecke K., MacDonald-Bravo H., Fey S. J., Celis J. E. More than one-third of the discernible mouse polypeptides are not expressed in a Chinese hamster-mouse embryo fibroblast hybrid that retains all mouse chromosomes. Proc Natl Acad Sci U S A. 1982 Apr;79(7):2281–2285. doi: 10.1073/pnas.79.7.2281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bravo R., Small J. V., Fey S. J., Larsen P. M., Celis J. E. Architecture and polypeptide composition of HeLa cytoskeletons. Modification of cytoarchitectural polypeptides during mitosis. J Mol Biol. 1982 Jan 5;154(1):121–143. doi: 10.1016/0022-2836(82)90421-1. [DOI] [PubMed] [Google Scholar]
- Fey S. J., Bravo R., Larsen P. M., Bellatin J., Celis J. E. [35S]-methionine labelled polypeptides from secondary mouse kidney fibroblasts: coordinates and one dimensional peptide maps of some major polypeptides. Cell Biol Int Rep. 1981 May;5(5):491–500. doi: 10.1016/0309-1651(81)90176-4. [DOI] [PubMed] [Google Scholar]
- O'Farrell P. H. High resolution two-dimensional electrophoresis of proteins. J Biol Chem. 1975 May 25;250(10):4007–4021. [PMC free article] [PubMed] [Google Scholar]
- O'Farrell P. Z., Goodman H. M., O'Farrell P. H. High resolution two-dimensional electrophoresis of basic as well as acidic proteins. Cell. 1977 Dec;12(4):1133–1141. doi: 10.1016/0092-8674(77)90176-3. [DOI] [PubMed] [Google Scholar]
- PUCK T. T., MARCUS P. I. Action of x-rays on mammalian cells. J Exp Med. 1956 May 1;103(5):653–666. doi: 10.1084/jem.103.5.653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- TOLMACH L. J., MARCUS P. I. Development of x-ray induced giant HeLa cells. Exp Cell Res. 1960 Aug;20:350–360. doi: 10.1016/0014-4827(60)90163-4. [DOI] [PubMed] [Google Scholar]