Abstract
Msx1 is a key factor for the development of tooth and craniofacial skeleton and has been proposed to play a pivotal role in terminal cell differentiation. In this paper, we demonstrated the presence of an endogenous Msx1 antisense RNA (Msx1-AS RNA) in mice, rats, and humans. In situ analysis revealed that this RNA is expressed only in differentiated dental and bone cells with an inverse correlation with Msx1 protein. These in vivo data and overexpression of Msx1 sense and AS RNA in an odontoblastic cell line (MO6-G3) showed that the balance between the levels of the two Msx1 RNAs is related to the expression of Msx1 protein. To analyze the impact of this balance in the Msx-Dlx homeoprotein pathway, we analyzed the effect of Msx1, Msx2, and Dlx5 overexpression on proteins involved in skeletal differentiation. We showed that the Msx1-AS RNA is involved in crosstalk between the Msx-Dlx pathways because its expression was abolished by Dlx5. Msx1 was shown to down-regulate a master gene of skeletal cells differentiation, Cbfa1. All these data strongly suggest that the ratio between Msx1 sense and antisense RNAs is a very important factor in the control of skeletal terminal differentiation. Finally, the initiation site for Msx1-AS RNA transcription was located by primer extension in both mouse and human in an identical region, including a consensus TATA box, suggesting an evolutionary conservation of the AS RNA-mediated regulation of Msx1 gene expression.
Msx genes are homeobox genes related to the Drosophila msh (muscle segment homeobox)-like gene family. Msx homeogenes play an important role in inductive epithelio-mesenchymal interactions leading to vertebrate organogenesis (1). Among this family, Msx1 is a fundamental factor for craniofacial skeleton formation. In mouse, head Msx1 gene expression is located mainly in regions of cephalic neural crest cell migration and differentiation, as well as in the derived mesenchymal cells (2–4). Msx1 also is found in a variety of embryonic tissues requiring epithelio-mesenchymal interactions for their morphogenesis such as limb bud, embryonic tail, hair follicle, and tooth bud.
Msx1-deficient mice exhibit dental and craniofacial malformations, such as cleft palate, reduced mandible length, abnormalities of nasal, frontal, and parietal bones, as well as arrested tooth development, suggesting a role of Msx1 in outgrowth of these tissues (5, 6). In humans, mutations in Msx1 gene have been involved in tooth agenesis (7–9) and cleft palate (10), and the phenotype was proposed to be related to a dose effect of Msx1 protein (9). Interestingly, Msx1 down-regulation is associated with the terminal differentiation of several cell types such as cartilage (4, 11, 12) and muscle (13); indeed, in muscle cells, Msx1-forced expression results in a highly proliferative transformed phenotype and blocks myogenic terminal differentiation (14, 15) through the inhibition of a master gene expression, MyoD, by Msx1 (16). Thus, Msx1 is thought to prevent differentiation and enhance proliferation. Other factors are involved in the control of skeletal cell differentiation, such as Cbfa1 (core binding factor α1), a bone master gene (17). Cbfa1 and factors such as Msx2 or Dlx5, members of the homeodomain protein family, have been shown to control the expression of osteocalcin, an abundant bone matrix protein (17–20). The crosstalk between these transcription factors is merging during early patterning (21–23) but has not been investigated in vivo in later stages, because of early lethality of the corresponding null mutant mice. Furthermore, the mechanism of down-regulation of Msx1 gene expression associated with cell differentiation has not yet been established.
Involvement of endogenous antisense (AS) RNAs in the regulation of gene expression has been described for various genes in association with a down-regulation of their corresponding sense mRNA transcription and/or translation (24). AS RNAs also have been involved in parental imprinting (reviewed in ref. 25) and chromosome X inactivation (26). The mechanisms proposed for the regulation of gene expression by AS RNAs are numerous, and the discovery of novel sense/AS RNA interactions would be insightful in understanding this mechanism of gene expression down-regulation.
The aim of our study, based on the detection of abnormal high levels of Msx1 mRNA in Northern blots, was to explore such a mechanism of regulation for Msx1 protein expression by a finely tuned transcription of an Msx1-AS RNA. We demonstrate the existence of an endogenous Msx1-AS RNA in differentiated dental and craniofacial tissues of mice, rats, and humans. We also investigated the role of this AS RNA and show that it is able to block Msx1 protein expression and exhibits a reverse temporospatial distribution pattern with Msx1 protein both in vivo and in vitro. The impact of this regulation in the Msx-Dlx pathway was evaluated by analyzing the effect of Msx1, Msx2, and Dlx5 overexpression on proteins involved in skeletal differentiation. We show that Msx1-AS RNA is involved in crosstalk between the Msx and Dlx pathways because its expression was abolished by Dlx5. Finally, Msx1 was shown to down-regulate the master gene of osteoblastic determination, Cbfa1, strongly suggesting that the relative ratio between Msx1 sense and AS RNAs would be a key factor for cell differentiation and phenotypic expression in mineralized tissues.
Materials and Methods
Msx1-Deficient Mice.
Msx1-deficient mice were generated by insertion of the Lac-Z gene within exon 2 of the Msx1 gene (6). Heterozygous mice, phenotypically normal, were used for the detection of β-galactosidase activity, as described (27), at postnatal stages: after being embedded in paraffin, sagittal sections of the mandible were stained and observed with a light microscope (Leica, Deerfield, IL).
Msx1-AS RNA Isolation.
Total RNA from the murine odontoblastic MO6-G3 cell line immortalized with the simian virus 40 T antigen (28) was extracted according to Chomczinski and Sacchi (29). Reverse transcription was performed by following the manufacturer's protocol (Invitrogen) with either an oligo(dT) primer or the P1f primer, allowing specific synthesis of Msx1-AS cDNA (Fig. 1). Overlapping Msx1-AS cDNA fragments then were amplified by PCR: the PCR was performed with either primers AS4 and P1r or AS7 and P1r for cDNA obtained with the oligo(dT) primer. For cDNA obtained with the P1f primer, the PCR was performed with either primers P1f and P2r or primers P2f and AS10 (Fig. 1). PCR on RNA served as control for genomic DNA contamination. These overlapping PCR fragments were sequenced according to the Sanger procedure (Sequenase; Amersham Pharmacia). The sequences were compared with sequence databanks with the blast algorithm (30) and aligned with the CLUSTAL-W algorithm (31).
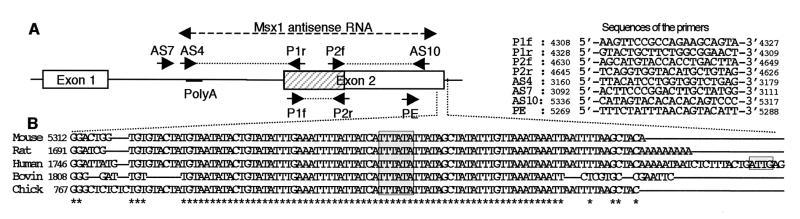
Figure 1.
Structure of the Msx1-AS RNA. (A) The sequence of Msx1-AS cDNA was determined by sequencing overlapping PCR fragments obtained with the different sets of primer indicated on the figure, which represents the murine Msx1 genomic DNA sequence. The homeobox located in exon 2 is hatched. A putative polyadenylation signal is indicated. (B) The 3′ ends of exon 2 of Msx1 cDNA sequences from mouse, rat, human, bovine, and chicken (accession nos. S73812, D83036, M76732, D30750, and X65791, respectively) were aligned with the clustal-w algorithm. Stars indicate conserved nucleotides. The putative TATA and CAAT boxes are underlined.
In Situ Hybridization.
Distribution of Msx1 RNAs during tooth and bone formation was analyzed at Theiler stage embryonic day 14.5 (E14.5) and E16.5 and 1 day after birth, as described (32) in C57BL6 mice (Charles River Breeding Laboratories). Msx1 sense and AS RNA digoxigenin-labeled probes were synthesized from a Bluescript-SK(+) plasmid containing 350 bp of exon 2 of the mouse Msx1 gene (6) after linearization with BamHI or HindIII endonucleases by using T7 and T3 RNA polymerases, respectively (Roche Diagnostics). In situ hybridization was performed as described (32) with minor modifications: cryostat sections were hybridized with 30 μl of digoxigenin-labeled probes diluted 1:200, and the reaction was revealed by an antidigoxigenin Fab alkaline phosphatase conjugate (Roche Diagnostics). The color-development reactions were performed for 2–18 h depending on the tissues and the stage of development. The sections were dehydrated, mounted under a coverslip, and photographed with a Leica photomicroscope.
Determination of the Msx1-AS RNA Transcription Starting Point.
The PE primer (5′-TTTCTATTTAACAGTACATT-3′, Fig. 1) was designed after sequence alignment of the region located downstream of the Msx1-AS TATA box (clustal-w algorithm) in a region conserved between mouse and human. Ten picomoles of this primer was 5′-labeled with [γ-32P]dATP by using T4-polynucleotide kinase (33). Two micrograms of total RNA from the MO6-G3 cells or from 9-week-old human embryonic whole orofacial tissues, excluding brain, was reverse-transcribed with this primer. Product of the reaction was analyzed by electrophoresis on denaturing 8% acrylamide gel and autoradiography. Sequencing products served as size markers. The human sample was recruited from R. Debré's Hospital, Paris (P. Blot, Obstetrical Service) under the French ethical rules and the approval of the National Consulting Ethics Committee.
Construction of an Msx1-AS cDNA Expression Vector.
Mouse Msx1-AS cDNA was obtained from MO6-G3 cells RNA by reverse transcription (RT)–PCR amplification with primers AS4 and AS10. This fragment was inserted in the pDEST 12.2 plasmid under the control of the cytomegalovirus promoter according to the manufacturer's protocol (Gateway cloning system; Life Technologies, Gaithersburg, MD).
Cell Cultures and Transfection Assays.
MO6-G3 cells were plated out in MEM supplemented with 15% FCS at 8 × 104 cells/cm2. After 72 h, cells were transfected with 3 μg of the Msx1, Msx2, or Dlx5 expression vectors or with the Msx1-AS cDNA expression vector at the indicated concentrations and 12.5 μl of lipofectamine per dish during 5 h in 1 ml of serum-free medium, according to the manufacturer's procedure (Life Technologies). The reaction was stopped by 1 ml of medium containing 30% FCS. After 72 h, the medium was removed, and the cells were rinsed with PBS and used for RNA and protein analysis. Control cells were transfected with a plasmid containing no insert. Each experiment was made in triplicate.
RNA Analysis.
RNA from the transfected cells or from dental tissues was extracted according to Chomczinski and Sacchi (29). It was analyzed either by Northern blotting or RT-PCR. For Northern blot analysis, 10 μg of total RNA was electrophoresed on 1% agarose gel and transferred onto a nylon membrane (Hybond N+; Amersham Pharmacia). The membrane then was hybridized with [α-32P]UTP-labeled Msx1 sense or AS iboprobe, prepared as described in the in situ hybridization section, and autoradiographed. For RT-PCR analysis, 2 μg of total RNA was reverse-transcribed with an oligo(dT) primer according to the manufacturer's protocol (Invitrogen). The PCR was performed in 50 μl with 1 μl of the RT reaction and 10 pmol of the following primers for 30 cycles, except GAPDH and osteocalcin (25 cycles): Msx2, 5′-CCTCGGTCAAGTCGGAAAATTC-3′ and 5′-CGTATATGGATGCTGCTTGCAG-3′; Dlx5, 5′-TGGCAAACCAAAGAAAGTTC-3′ and 5′-AATAGAGTGTCCCGGAGG-3′; osteocalcin, 5′-CTCACTCTGCTGGCCCTG-3′ and 5′-CCGTAGATGCGTTTGTAGGC-3′; type-I collagen α1 chain, 5′-AAGATGTGCCACTCTGACTG-3′ and 5′-ATAGGTGATGTTCTGGGAGG-3′; Cbfa1, 5′-GGACGAGGCAAGAGTTTCAC-3′ and 5′-TGCCTGCCTGGGATCTGTAA-3′; GAPDH, 5′-TTCCAGTATGATTCCACTCA-3′ and 5′-CTGTAGCCATATTCATTGTC-3′; Msx1 sense, 5′-CTCATGGCCGATCACAGGAA-3′ (specific of the sense transcript, as it is located in exon 1 to which AS RNA does not extend) and P2r; and Msx1-AS, 5′-CTCTCTTTAACTCCTTGCTT-3′ (specific of the AS transcript, as it is located in intron 1) and P2r. Under these conditions, preliminary experiments showed that all reactions remained within the exponential phase (data not shown). In all analysis, RNA without prior RT served as a control for genomic DNA contamination.
Western Blot Analysis.
Proteins from the transfected cells were subjected to one-dimensional SDS/PAGE according to Laemmli (34) on 10% polyacrylamide gel under reducing conditions, in a minislab gel system (MiniProtean II Electrophoresis cell; Bio-Rad). Proteins were transferred onto nitrocellulose membranes (Hybond ECL; Amersham Pharmacia) by using a semidry transblot apparatus (Bio Trans Midi, Ann Arbor, MI). Ponceau red staining of the membranes allowed verification that the same protein amount was deposited in the different lanes. The membranes were incubated with polyclonal rabbit antibodies directed against mouse Msx1 protein (Babco, Richmond, CA) diluted at 1:1,000 and then with horseradish peroxidase-F(ab′)2 goat anti-mouse antibodies diluted at 1:20,000. After washing, the membranes were incubated for 1 min with the Super Signal Substrate (WB; Pierce) and chemiluminescence was detected by exposure of the membranes to a Hyperfilm ECL (Amersham Pharmacia) for 1 min.
Immunocytochemical Analysis.
Msx1 protein expression was analyzed in the transfected cells. Immunofluorescent labeling was performed by using the same antibodies as in the Western blot analysis and fluorescein-labeled secondary antibodies (Amersham Pharmacia). In the control, the primary antibodies were replaced by nonrelevant rabbit antibodies.
Results
Identification of an Endogenous Msx1-AS RNA.
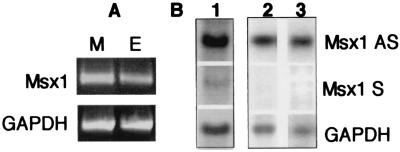
The presence of an Msx1-AS RNA first was suspected while studying Msx1 distribution in murine mesenchymal and epithelial dental tissues in 56-day-old mice. In heterozygous Msx1 (+/−) transgenic mice bearing an inserted Lac-Z gene within exon 2 of the Msx1 gene (6), no β-galactosidase expression was detected after birth in dental tissues (27). Surprisingly, an Msx1 transcript was detected by oligo(dT)-primed RT-PCR with primers P1f and P2r in both tissues (Fig. 2A), suggesting the existence of a translational inhibition of Msx1 and/or the presence of an Msx1-AS RNA.
Figure 2.
Identification of an Msx1-AS RNA. (A) RNA from a 56-day-old mouse dental mesenchyme (M) and epithelium (E) was analyzed by oligo(dT)-primed RT-PCR by using the P1f-P2r set of primers. GAPDH PCR amplification served as internal control. (B) RNA from the murine MO6-G3 cell line (lane 1) or from dental epithelium (lane 2) and mesenchyme (lane 3) of a 56-day-old rat was analyzed by Northern blot and hybridized with riboprobes corresponding to both strands of the homeobox of Msx1 gene. GAPDH mRNA hybridization served as internal control.
To test the existence of such an RNA, we used a murine odontoblastic cell line, MO6-G3. This cell line expresses tooth-specific markers such as dentin sialophosphoprotein, as well as dentin and bone markers such as osteocalcin (28). RNA from these cells was analyzed by Northern blot by using riboprobes corresponding to both strands of exon 2 of Msx1 cDNA. Fig. 2B shows the hybridization signal with both probes, confirming the existence of an Msx1-AS RNA in these cells.
An Msx1-AS transcript also was detected by Northern blot analysis of RNA from dental epithelium and mesenchyme from a 56-day-old rat incisor (Fig. 2B). The sense RNA was not detectable at this stage. These data suggest that the expression of an Msx1-AS RNA is not restricted to mice.
In Vivo Expression of Msx1-AS RNA in Mice.
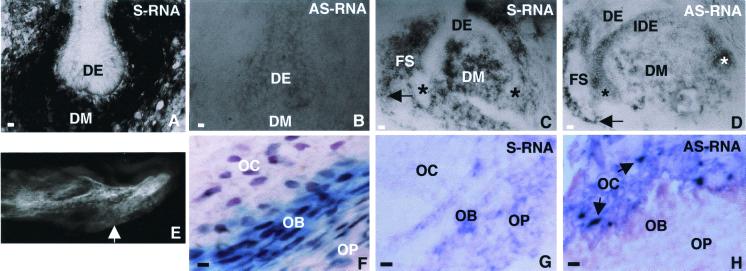
In vivo expression of Msx1 was investigated by in situ hybridization during tooth morphogenesis and bone craniofacial formation. As reported previously (4–6), Msx1 sense transcripts were detected early during tooth development (Fig. 3A), whereas Msx1-AS RNA was not evidenced (Fig. 3B). In contrast, at stage E16.5, both transcripts were present but with distinct distribution: sense mRNA was restricted to the dental mesenchyme and follicular sac (Fig. 3C) whereas AS RNA was present in the dental mesenchyme, the mesenchyme surrounding the follicular sac, and the epithelium (Fig. 3D).
Figure 3.
In situ hybridization analysis of Msx1 sense and AS RNAs during mouse craniofacial development. Expression of the two Msx1 mRNAs was analyzed in mouse frontal incisor serial sections. At bud stage (E14.5), sense mRNA (S-RNA) was restricted (A) to the dental mesenchyme (DM). Msx1-AS RNA (AS-RNA) was not detected (B) in the mesenchyme (DM) or in the epithelium (DE). At late cap stage (E16.5), AS RNA (D) was present in the cervical loops (*) and the inner dental epithelium (IDE). In the mesenchyme (DM) both RNAs were present, and in the follicular sac (FS), the AS RNA-positive cells (D, arrow) surrounded the sense RNA-positive cells (C, arrow). Periosteum from half-mandibles indicated in the microradiograph (E, arrow) from newborn heterozygous Msx1-deficient mice were analyzed by β-galactosidase histoenzymology (F), and in situ hybridization for Msx1 sense (G) and AS (H) RNAs. β-Galactosidase activity was restricted to osteoprogenitor cells (OP) and osteoblasts (OB). Sense RNA (G) was detected in the osteoprogenitor cells. In osteocytes (OC), only AS RNA was detected (H) but not sense RNA (G) or Msx1/β-galactosidase protein (F). [Bars = 1.7 μm (A, B, F–H) and 3 μm (C and D).]
In Msx1 (+/−) heterozygous mice, the expression of β-galactosidase was driven by the Msx1 promoter. The merging basal convexity of the mandible (Fig. 3E) is associated with an aboral-oral gradient of Msx1-driven β-galactosidase (Fig. 3F). This gradient is stable from E15.5 until 3 months (data not shown). The distribution of Msx1 RNAs also was studied during these stages of bone formation in the half-mandible (Fig. 3 G and H). Progenitor bone cells highly expressed Msx1 sense transcript (Fig. 3G) and Msx1-driven β-galactosidase (Fig. 3F), whereas differentiated and entrapped osteocytes expressed Msx1-AS transcript but not the protein (Fig. 3 F and H). A transient coexpression of Msx1 sense mRNA and β-galactosidase also was observed in the epithelium of hair follicles, and their nondetection was coincident with the presence of Msx1-AS RNA (data not shown). Thus, Msx1-AS RNA is found in differentiated cells and is not restricted to dental tissue but also is expressed in the entire orofacial complex.
Msx1-AS cDNA Structure.
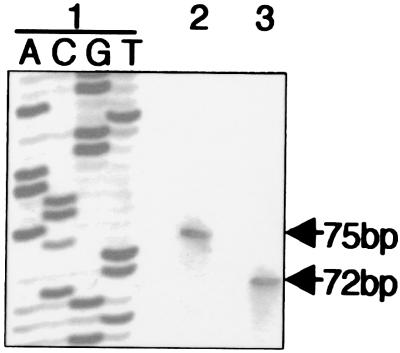
The murine Msx1-AS cDNA sequence was determined through overlapping PCR fragments, based on a previously published sequence (accession no. S73812) (Fig. 1A). This Msx1-AS cDNA is about 2.2 kb in length and fully complementary to the region extending from of the 3′ end of exon 2 to the middle of intron 1 of the genomic Msx1 DNA sequence. No significant ORFs were found in the Msx1-AS cDNA sequence (National Center for Biotechnology Information ORF Finder tool), indicating that it is most likely not translated. A consensus polyadenylation signal (AATAAA) was detected at position 2141–2146 of the Msx1-AS cDNA (accession no. AF267728). Furthermore, RT-PCR analysis of the 3′ extremity of the Msx1-AS RNA showed an amplified fragment with primers AS4 and P1r but not AS7 and P1r, suggesting the location of the RNA 3′ end to be between primers AS4 and AS7 (data not shown). Comparison of the Msx1 sense cDNA sequences from five species (Fig. 1B) identified a strictly conserved, 66-bp region at the 3′ noncoding end that contains the predicted AS initiation site. This conserved sequence was characterized by the presence of a consensus TATA box (TATAAA) located on the AS strand and surrounded by less-conserved regions. Furthermore, the only available 3′ genomic sequence of Msx1 (human, accession no. M76732) revealed a consensus CAAT box on the AS strand 62 bp upstream of the identified consensus TATA box sequence. To validate the function of this TATA box, a primer-extension analysis was performed on RNA isolated from the murine MO6-G3 cells or from orofacial tissues from a 9-week-old human with the PE primer (Fig. 4). The results show an extended fragment of 75 bp in the murine sample, indicating that the AS RNA transcription starts 32 bp downstream of the putative TATA box sequence. In humans, an extended fragment of 72 bp was detected, revealing the presence of an Msx1-AS RNA. These data strongly suggest that the consensus sequence probably corresponds to a functional AS TATA box. They also confirm that an Msx1-AS RNA is expressed in a number of species, suggesting an evolutionary conservation.
Figure 4.
Determination of the Msx1-AS RNA starting point. Primer extension analysis of RNA isolated from the murine MO6-G3 cells (lane 2) and from orofacial tissues of a 9-week-old human embryo (lane 3) was performed by using the [γ-32P]dATP-labeled PE primer. Sanger sequencing product served as the size marker (lane 1). Extended fragments of 75 bp in mouse and 72 bp in human were obtained.
Ratio of Msx1 AS and Sense RNAs and Msx1 Protein Expression.
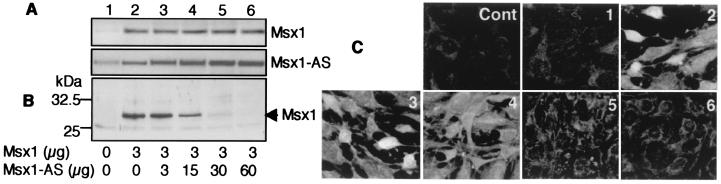
Expression of Msx1 sense and AS RNAs was analyzed by Northern blot (Fig. 2B) and PCR amplification of specifically reverse-transcribed sense or AS RNAs from the MO6-G3 cells (Fig. 5A). Both RNAs were detected, but the intensity of the hybridization signals or PCR products suggests that the AS RNA is more abundant than the sense RNA. Under these conditions, Msx1 protein is not detectable by Western blot or immunocytochemical analysis (Fig. 5 B and C, lane 1). However, after transfection of an Msx1 expression vector, the ratio between the two RNAs appeared to change, the sense RNA becoming more abundant than the AS RNA (Fig. 5A), resulting in the detection of Msx1 protein (Fig. 5 B and C, lane 2). This effect was reversed by coexpression of increasing concentrations of Msx1 AS RNA, in a dose-dependent manner, leading to the nondetection of Msx1 protein (Fig. 5 A–C, lanes 3–6). These results strongly support a negative regulatory effect of the Msx1-AS RNA on the expression of Msx1 protein dependent on the relative ratios of sense and AS transcripts.
Figure 5.
Msx1 expression in the MO6-G3 cells. MO6-G3 cells either were not transfected (lane 1) or were transfected with 3 μg of control (not shown as giving results identical to those of nontransfected cells) or 3 μg of Msx1 (lane 2) expression vector or with both Msx1 (3 μg) and Msx1-AS (3–60 μg) expression vectors (lanes 3–6). (A) RNA from these cells was reverse-transcribed with either primer P2r or P1f, allowing specific synthesis of the sense (Msx1 S) or AS (Msx1 AS) cDNA, respectively. The cDNA was analyzed by RT-PCR with primers P1f–P2r. PCR on RNA served as a control for the absence of genomic contamination (not shown). (B) Proteins from transfected cells were analyzed by Western blot with polyclonal rabbit antibodies directed against murine Msx1. (C) Transfected cells were analyzed by immunocytochemistry with polyclonal rabbit antibodies directed against mouse Msx1. Cells incubated with nonrelevant rabbit antibodies as primary antibodies served as control (Cont).
Msx1 Protein Expression Related to Cbfa1 and Osteocalcin Expression.
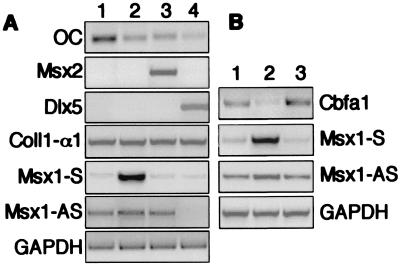
Osteocalcin is a major, noncollagenous protein common to bone and dentin. Analysis of its expression in bone cells has defined two main regulatory regions in its promoter. The first corresponds to a binding region for Cbfa1, a potent activator of osteocalcin expression (17–20, 35). The second is a binding domain for inhibitory factors related to Msx1, such as Msx2 and Dlx5, a member of the distal, less-related gene family (36–38). To analyze whether the balance of the two Msx1 RNAs and, consequently, Msx1 protein expression level could contribute to the control of cell activity, the expression of both Cbfa1 and its major target—osteocalcin—was analyzed in the MO6-G3 cell line. Both were expressed (Fig. 6, lanes 1), whereas Msx2 or Dlx5 mRNA was not detectable. Overexpression of Msx1, Msx2, or Dlx5 resulted in the decrease in the level of osteocalcin RNA, normalized to GAPDH (Fig. 6A). This reduction is specific to osteocalcin because it is not observed for the type I collagen α1-chain mRNA, another extracellular protein of the mineralized tissues. Interestingly, Dlx5 overexpression also resulted in undetectable levels of Msx1-AS RNA (Fig. 6A, lane 4), suggesting that Dlx5 negatively regulates Msx1-AS RNA transcription.
Figure 6.
Msx1 expression and genes involved in mineralization. The MO6-G3 cells were transfected with a vector containing no insert, as control (lane 1), or with expression vectors corresponding to Msx1 (lane 2), Msx2 (lane 3), or Dlx5 (lane 4). (A) RNA isolated from the transfected cells were analyzed by RT-PCR for osteocalcin (OC), α1 chain of type I collagen (Coll1-α1), Msx2, Dlx5, GAPDH, Msx1 sense (Msx1-S), and AS (Msx1-AS) RNA expression. (B) RNA from the transfected cells also was analyzed by RT-PCR for Cbfa1 expression. RNA served as control for the absence of genomic contamination (not shown).
Overexpression of Msx1 also induced a decrease of Cbfa1 transcripts (Fig. 6B). This decrease was not observed after Msx2-forced expression. Thus, these data suggested that Cbfa1 expression is inhibited by Msx1.
Discussion
Msx1 Sense–AS RNA Interactions.
Although gene expression regulation is classically explained through mechanisms involving the binding of transcription factors to cis-regulatory elements, other mechanisms such as interactions of endogenous AS RNAs with the corresponding sense transcripts are emerging (for review, see ref. 24). The present study demonstrates the existence of an endogenous Msx1-AS RNA in mice, rats, and humans. The extensive overlapping complementary region between the sense and AS RNAs (more than 1,000 bp) highly suggests a physical interaction between them. In addition, a consensus polyadenylation signal is present at the 3′ end of the Msx1-AS RNA, and this RNA is detectable by RT-PCR analysis by using an oligo(dT) primer, indicating that it is polyadenylated.
Short endogenous AS RNAs generally are produced from a locus different from that of the sense RNA, whereas the transcription of long AS RNA occurs from the same locus as the sense RNA (24). The Msx1-AS RNA is more than 2 kb in length, fully complementary to part of the Msx1 genomic DNA sequence, and, probably, a functional TATA box was located upstream from the Msx1-AS cDNA sequence, strongly suggesting that both Msx1 RNAs are produced from the same locus, on opposite strands.
Numerous sense–AS RNA interactions have been reported that affect the transcription of sense RNA (24). Volk et al. (39) have demonstrated the presence of bFGF AS RNA involved in the regulation of the bFGF mRNA expression and turnover, probably through the regulation of mRNA polyadenylation. Down-regulation of p53 has been associated with nuclear accumulation of an AS RNA complementary to intron 1 of the p53 gene likely inhibiting the transport of the sense RNA to the cytoplasm (40). N-myc AS RNAs have been shown to form duplexes with the sense transcript in a region including the exon 1 donor splice site and have been implicated in the modulation of N-myc mRNA level, through splicing inhibition of the first intron (41). In the same manner, the complementary region between the two Msx1 RNAs extends across intron 1 and exon 2, including the splice site. Therefore, the interaction between the two Msx1 RNAs could protect Msx1 sense RNA from splicing, controlling the level of Msx1 sense RNA.
AS RNAs also have been involved in parental imprinting (reviewed in ref. 25). In all cases, the expression of the AS RNA from the repressed allele seems exclusive with accumulation of the sense transcript. In a similar manner, an AS to Xist, a transcript associated with the inactive X chromosome, is expressed from both X chromosomes before the onset of inactivation, and its expression persists on the future, active X until Xist is turned off (26), suggesting that the AS RNA is involved in the repression of the RNA produced from the complementary strand. Msx1 is not functionally imprinted, as exemplified by the mutant allele we have generated: no mutant phenotype is observed in heterozygotes whether the mutant allele is transmitted by the father or by the mother (D. Houzelstein, P. Pommier, and B.R., unpublished data). This is in good agreement with the late and spatially restricted expression observed for Msx1-AS RNA: association with imprinting would lead to an early and ubiquitous expression. However, the same mechanisms that regulate imprinted genes via the production of AS RNAs might work for the Msx1 gene.
Although our results do not allow definition of the precise mechanism for Msx1 sense–AS RNA interactions, they suggest a negative regulation of Msx1 expression by Msx1-AS RNA. During mouse development, Msx1 is expressed strongly in the proliferative progenitor cells of dental mesenchyme and bone and down-regulated in the terminal differentiated tissues (2, 4, 6). By contrast, our results showed an inverse distribution of the Msx1-AS RNA (Fig. 3). As shown in the MO6-G3 cells, when the AS transcript is more abundant, Msx1 protein is undetectable, and, conversely, overexpression of the sense RNA results in production of the Msx1 protein (Fig. 5). This suggests that the outcome of Msx1 sense and AS interactions probably depends on their relative ratio. Thus, our results strongly suggest a down-regulation of Msx1 by the Msx1-AS RNA both in vivo and in vitro.
Msx1 and Mineralized Tissue Formation.
Studies concerning the regulation of mineralized tissue formation have focused on the regulation of matrix protein expression such as osteocalcin. Osteocalcin is expressed specifically in mature matrix forming cells such as osteoblasts and odontoblasts. Cbfa1, involved in osteocalcin gene regulation, has been shown to be essential for the osteoblast differentiation (17). Mice lacking this factor exhibit an absence of bone formation (35), and mutations of Cbfa1 in humans are responsible for cleidocranial dysplasia (42). The MO6-G3 odontoblastic cells reproduce the regulation previously established for osteocalcin gene expression by Msx1, Msx2, and Dlx5 in osteoblasts (36–38), suggesting common mechanisms of regulation between these two mineralized, matrix-forming cells and, thus, provide a convenient model for studying regulation of genes involved in mineralized tissue formation. In this model, Msx1 was able to decrease Cbfa1 RNA levels, which, therefore, represents a second master gene down-regulated by Msx1. Indeed, Msx1 has been shown to inhibit MyoD gene expression in differentiating muscle cells in vitro (15, 16) and in vivo (43). This ability of Msx1 to down-regulate expression of genes essential for differentiation provides insights on its negative effect on cell differentiation.
The crosstalk between Dlx and Msx signalization pathways has been reported at early stages of tooth development by analysis of null mutants in mice (21, 22). We have chosen an alternative strategy by using an immortalized cell line that allows one to study these interactions at later stages, i.e., in differentiated cells. In vivo, Dlx5 and Msx1 proteins are present at the initial stages of tooth and bone differentiation but are not detected in more mature differentiated cells (27, 44, 45), whereas Cbfa1 and Msx1-AS RNA display an inverse distribution. In the MO6-G3 cells, on one hand, Dlx5 dramatically blocked Msx1-AS RNA expression, and, on the other hand, Cbfa1, the main activator of osteocalcin gene expression, was negatively regulated by Msx1 (Fig. 6). Thus, according to the stage of differentiation and the factors present in the cells, osteocalcin gene expression could be submitted to (i) a direct regulation at the promoter level, activated by Cbfa1 or inhibited by the Msx/Dlx factors as described previously (17–20, 36–38) and (ii) an indirect mechanism involving interregulations between these regulatory factors, i.e., a negative control of Msx1-AS RNA by Dlx5 and a negative control of Cbfa1 expression by Msx1 or a decrease of Msx1 protein controlled by the balance between the two Msx1 RNAs. Thus, these effects on osteocalcin gene, considered as a marker for mineralized tissue differentiation, suggest an important role for the Msx1 sense–AS RNA levels in controlling the transition from proliferation toward terminal differentiation. The persistence of Msx1-AS RNA in stages in which the sense RNA has disappeared could reflect a locking mechanism of Msx1 expression by overexpression of Msx1-AS RNA. Furthermore, Msx1-AS was found in several species (human, mouse, and rat), indicating an evolutionary conservation, as well as in a variety of craniofacial mineralizing structures. Interestingly, the Msx1-AS putative TATA box is conserved in Msx2 cDNA sequences (data not shown), suggesting a more general regulation pathway of the Msx gene family.
In conclusion, the structural and functional data presented in this study demonstrate that the production of an Msx1-AS RNA is a fundamental and conserved mechanism that regulates expression of Msx1, a key transcription factor. This mechanism may be essential for the differentiation of craniofacial structures, especially those associated with mineralized matrices.
Acknowledgments
We thank Prof. P. Blot (Paris) for the human samples. We are grateful to N. Mauro for her technical help in cell culture. We thank Dr. J. C. Scimeca (Nice, France) for his help in Western blot analysis. Dlx5 expression vector is a gift from Dr. S. Harris (San Antonio, TX) and Dr. J. Rubeinstein (San Francisco). Msx1 and Msx2 expression vectors are a gift from Dr. C. Abate-Shen (Piscataway, NJ). S.O.C. was supported by Coordenadoria de Aperfeiçoamento de Pessoal de Nível Superior (Brazil). This work was supported by grants from the Program Hospitalier de Recherche Clinique (AOM96067).
Abbreviations
- AS
antisense
- E
embryonic day
- RT
reverse transcription
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
Data deposition: The sequence reported in this paper has been deposited in the GenBank database [accession no. AF267728 (Msx1-AS cDNA)].
References
- 1.Thesleff I, Vaahtojkari A, Partanem A. Int J Dev Biol. 1995;39:35–50. [PubMed] [Google Scholar]
- 2.Robert B, Sassoon D, Jacq B, Gehring W, Buckingham M. EMBO J. 1989;8:91–100. doi: 10.1002/j.1460-2075.1989.tb03352.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hill R, Jones P, Ress A, Sime C, Justice M, Copeland N, Jenkins N, Graham E, Davidson D. Gene Dev. 1989;3:26–37. doi: 10.1101/gad.3.1.26. [DOI] [PubMed] [Google Scholar]
- 4.Mackenzie A, Leeming G, Jowett A, Ferguson W, Sharpe P. Development. 1991;111:269–285. doi: 10.1242/dev.111.2.269. [DOI] [PubMed] [Google Scholar]
- 5.Satokata I, Maas R. Nat Genet. 1994;6:348–356. doi: 10.1038/ng0494-348. [DOI] [PubMed] [Google Scholar]
- 6.Houzelstein D, Cohen A, Buckingham M, Robert B. Mech Dev. 1997;65:123–133. doi: 10.1016/s0925-4773(97)00065-8. [DOI] [PubMed] [Google Scholar]
- 7.Padanilam B, Stadler H, Mills K, McLeod L, Solursh M, Lee B, Ramirez F, Buetow K, Murray J. Hum Mol Genet. 1992;1:407–410. doi: 10.1093/hmg/1.6.407. [DOI] [PubMed] [Google Scholar]
- 8.Vastardis H, Karimbux N, Guthua S, Seidman J, Seidman C. Nat Genet. 1996;13:417–421. doi: 10.1038/ng0896-417. [DOI] [PubMed] [Google Scholar]
- 9.Hu G, Vastardis H, Bendall A, Wang Z, Logan M, Zhang H, Nelson C, Stein S, Greenfield N, Seidman C, et al. Mol Cell Biol. 1998;18:6044–6051. doi: 10.1128/mcb.18.10.6044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Van Den Boogaard M, Dorland M, Beemer F, van Amstel H. Nat Genet. 2000;24:342–343. doi: 10.1038/74155. [DOI] [PubMed] [Google Scholar]
- 11.Coelho C, Upholt W, Kosher R. Differentiation. 1993;52:129–137. doi: 10.1111/j.1432-0436.1993.tb00623.x. [DOI] [PubMed] [Google Scholar]
- 12.Mina M, Gluhak J, Upholt W, Kollar E, Rogers B. Dev Dyn. 1995;202:195–214. doi: 10.1002/aja.1002020211. [DOI] [PubMed] [Google Scholar]
- 13.Houzelstein D, Auda-Boucher G, Cheraud Y, Rouaud T, Blanc I, Tajbakhsh S, Buckingham M, Fontaine-Perus J, Robert B. Development. 1999;126:2689–2701. doi: 10.1242/dev.126.12.2689. [DOI] [PubMed] [Google Scholar]
- 14.Song K, Wang Y, Sasson D. Nature (London) 1992;360:477–481. doi: 10.1038/360477a0. [DOI] [PubMed] [Google Scholar]
- 15.Odelberg S J, Kollhoff A, Keating M T. Cell. 2000;103:1099–1109. doi: 10.1016/s0092-8674(00)00212-9. [DOI] [PubMed] [Google Scholar]
- 16.Woloshin P, Song K, Degnin C, Killary A, Goldhamer D, Sassoon D, Thayer M. Cell. 1995;82:611–620. doi: 10.1016/0092-8674(95)90033-0. [DOI] [PubMed] [Google Scholar]
- 17.Ducy P, Zhang R, Geoffroy V, Ridali A, Karsenty G. Cell. 1997;89:747–754. doi: 10.1016/s0092-8674(00)80257-3. [DOI] [PubMed] [Google Scholar]
- 18.Geoffroy V, Ducy P, Karsenty G. J Biol Chem. 1995;270:30973–30979. doi: 10.1074/jbc.270.52.30973. [DOI] [PubMed] [Google Scholar]
- 19.Ducy P, Geoffroy V, Karsenty G. Connect Tissue Res. 1996;35:7–14. doi: 10.3109/03008209609029169. [DOI] [PubMed] [Google Scholar]
- 20.Banerjee C, Hiebert S, Stein J, Lian J, Stein G. Proc Natl Acad Sci USA. 1996;93:4968–4973. doi: 10.1073/pnas.93.10.4968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bei M, Maas R. Development. 1998;125:4325–4333. doi: 10.1242/dev.125.21.4325. [DOI] [PubMed] [Google Scholar]
- 22.Chen Y, Bei M, Woo I, Satokata I, Maas R. Development. 1996;122:3035–3044. doi: 10.1242/dev.122.10.3035. [DOI] [PubMed] [Google Scholar]
- 23.Vainio S, Karavanova I, Jowett A, Thesleff I. Cell. 1993;75:45–58. [PubMed] [Google Scholar]
- 24.Kumar M, Carmichael G. Microbiol Mol Biol Rev. 1998;62:1415–1434. doi: 10.1128/mmbr.62.4.1415-1434.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sleutels F, Barlow D P, Lyle R. Curr Opin Genet Dev. 2000;10:229–233. doi: 10.1016/s0959-437x(00)00062-9. [DOI] [PubMed] [Google Scholar]
- 26.Lee J T, Davidow L S, Warshawsky D. Nat Genet. 1999;21:400–404. doi: 10.1038/7734. [DOI] [PubMed] [Google Scholar]
- 27.Lezot F, Thomas B, Hotton D, Forest N, Orestes-Cardoso S, Robert B, Sharpe P, Berdal A. J Bone Miner Res. 2000;15:430–441. doi: 10.1359/jbmr.2000.15.3.430. [DOI] [PubMed] [Google Scholar]
- 28.MacDougall M, Thiemann F, Ta H, Hsu P, Chen L, Snead M. Connect Tissue Res. 1995;33:67–103. doi: 10.3109/03008209509016988. [DOI] [PubMed] [Google Scholar]
- 29.Chomczinski P, Sacchi M. Anal Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- 30.Atlschul S, Gish W, Miller W, Myers E, Lipman D. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 31.Higgins D, Sharp P. CABIOS. 1989;5:151–153. doi: 10.1093/bioinformatics/5.2.151. [DOI] [PubMed] [Google Scholar]
- 32.Hotton D, Davideau J, Bernaudin J, Berdal A. Connect Tissue Res. 1995;32:137–143. doi: 10.3109/03008209509013716. [DOI] [PubMed] [Google Scholar]
- 33.Sambrook J, Fritsch E, Maniatis T. Molecular Cloning: A Laboratory Manual. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 34.Laemli U. Nature (London) 1970;15:680. [Google Scholar]
- 35.Komori T, Yagu H, Nomura S, Yamaguchi A, Sasaki K, Deguchi K, Shimizu Y, Bronson R, Gao Y, Inada M, et al. Cell. 1997;89:755–764. doi: 10.1016/s0092-8674(00)80258-5. [DOI] [PubMed] [Google Scholar]
- 36.Ryoo H, Hoffmann H, Beumer T, Frenkel B, Towler D, Stein G, Stein J, vanVijnen A, Lian J. Mol Endocrinol. 1997;11:1681–1692. doi: 10.1210/mend.11.11.0011. [DOI] [PubMed] [Google Scholar]
- 37.Newberry E, Latifi T, Towler D. Biochemistry. 1998;37:16361–16368. doi: 10.1021/bi981878u. [DOI] [PubMed] [Google Scholar]
- 38.Towler O, Rutledge S, Rodan G. Mol Endocrinol. 1994;8:1484–1493. doi: 10.1210/mend.8.11.7877617. [DOI] [PubMed] [Google Scholar]
- 39.Volk R, Koster M, Poting A, Hartmann L, Knochel W. EMBO J. 1989;8:2983–2988. doi: 10.1002/j.1460-2075.1989.tb08448.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Khochbin S, Brocard P, Grumwald D, Lawrence J. Ann NY Acad Sci. 1992;660:77–87. doi: 10.1111/j.1749-6632.1992.tb21060.x. [DOI] [PubMed] [Google Scholar]
- 41.Krystal G, Armstrong B, Battey J. Mol Cell Biol. 1990;10:4180–4191. doi: 10.1128/mcb.10.8.4180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mundlos S, Otto F, Mundlos C, Muliken J, Aylsworth A, Albright S, Lindhout D, Cole W, Henn W, Knoll J, et al. Cell. 1997;89:773–779. doi: 10.1016/s0092-8674(00)80260-3. [DOI] [PubMed] [Google Scholar]
- 43.Bendall A, Ding J, Hu G, Shen M, Abate-Shen C. Development. 1999;126:4965–4976. doi: 10.1242/dev.126.22.4965. [DOI] [PubMed] [Google Scholar]
- 44.Acampora D, Merlo G, Paleari L, Zerega B, Postiglione M, Mantero S, Bober E, Barbieri O, Simeone A, Levi G. Development. 1999;126:3795–3809. doi: 10.1242/dev.126.17.3795. [DOI] [PubMed] [Google Scholar]
- 45.Davideau J, Demri P, Gu T, Simmons D, Nessman C, Forest N, MacDougall M, Berdal A. Mech Dev. 1999;81:183–186. doi: 10.1016/s0925-4773(98)00227-5. [DOI] [PubMed] [Google Scholar]