Abstract
Within the intestine reside unique populations of innate and adaptive immune cells that are involved in promoting tolerance towards commensal flora and food antigens while concomitantly remaining poised to mount inflammatory responses toward invasive pathogens1,2. Antigen presenting cells, particularly DCs and macrophages, play critical roles in maintaining intestinal immune homeostasis via their ability to sense and appropriately respond to the microbiota3-14. Efficient isolation of intestinal DCs and macrophages is a critical step in characterizing the phenotype and function of these cells. While many effective methods of isolating intestinal immune cells, including DCs and macrophages, have been described6,10,15-24, many rely upon long digestions times that may negatively influence cell surface antigen expression, cell viability, and/or cell yield. Here, we detail a methodology for the rapid isolation of large numbers of viable, intestinal DCs and macrophages. Phenotypic characterization of intestinal DCs and macrophages is carried out by directly staining isolated intestinal cells with specific fluorescence-labeled monoclonal antibodies for multi-color flow cytometric analysis. Furthermore, highly pure DC and macrophage populations are isolated for functional studies utilizing CD11c and CD11b magnetic-activated cell sorting beads followed by cell sorting.
Keywords: Immunology, Issue 63, intestine, immunology, APCs, dendritic cells, macrophages, cell culture
Protocol
1. Dissection and Dissociation of Intestinal Epithelial Cells
Preparation of reagents and equipment:
Warm Ca2+/Mg2+-free PBS (CMF PBS) to room temperature.
Warm Ca2+/Mg2+-free HBSS with 5% FBS (CMF HBSS/FBS) and 2mM EDTA to room temperature.
Warm Orbital shaker to 37 °C.
Note: Steps 1.1 to 1.7 must be performed as quickly as possible to minimize the extent of cell death and to achieve maximum cell yield.
Euthanize mice in a CO2 chamber and spray 70% ethanol onto the abdomen and thorax.
Make a small horizontal incision in the middle of the abdomen with a scissor and peel back the skin to expose the peritoneum.
Proceed to separate the stomach from the upper small intestine by cutting at the pyloric sphincter. Tease away the mesentery with forceps and cut again at the ileo-cecal valve to free the entire small intestine from the large intestine. Make a cut at the anal verge and again tease apart the mesentery until the large intestine is free.
Cut open the colon longitudinally using scissors and wash off fecal contents and mucus from the intestinal lumen in CMF PBS at room temperature.
Use scissors and forceps to macroscopically dissect out the Peyer's patches along the anti-mesenteric surface of the small intestine and cut open the small intestine longitudinally.
Wash the small intestine lumen of fecal contents and mucus in CMF PBS at room temperature.
Separately cut the small/large intestine into approximately 1.5 cm pieces and place into separate 50 ml conical tubes containing 30 ml of pre-warmed CMF HBSS/FBS and 2mM EDTA. Do not add more than 1 intestine per 50 ml conical tube.
Horizontally place each 50 ml conical tube into an orbital shaker and shake for at 250 rpm for 20 min at 37 °C.
Place a single mesh wire strainer over a waste bucket and pour the contents of each 50 ml conical tube through to recover the 1.5 cm pieces of intestine and place them in separate 50 ml conical tubes containing 30 ml of pre-warmed CHBSS/FBS with 2 mM EDTA.
Repeat 1.8.
2. Tissue Digestion and Isolation of Intestinal Cells
Preparations of reagents and equipment:
Collagenase solution:1.5 mg/ml Type VIII Collagenase dissolved in pre-warmed CMF HBSS/FBS with 40 μg/ml of DNase I.
Pre-warmed CMF HBSS/FBS and 2mM EDTA.
Pre-warmed orbital shaker at 37 °C.
Ice-cold CMF HBSS/FBS.
After the second round of shaking, pour the contents of each 50 ml conical tube through the strainer and transfer 1.5 cm pieces of intestine to a small plastic weigh boat after dabbing away excess media using a paper towel.
Rapidly mince the 1.5 cm pieces of intestine using scissors directly in the weight boat and add minced intestine to 20 ml of collagenase solution. Horizontally place each 50 ml conical tube into an orbital shaker and digest at 200 rpm for 10-20 min at 37 °C. Please see discussion below regarding optimization.
Briefly vortex to ensure thorough dissociation of any remaining intestinal tissue and filter through a 100 μm cell strainer directly into a 50 ml conical tube.
Top off each 50 ml conical tube with CMF HBSS/FBS and centrifuge at 1500 rpm for 5 min at 4 °C. If a solid pellet is not observed for colon samples after centrifugation, the samples should be centrifuged again for 3.5 min. Repeat this wash step once more.
Pour off the supernatant and resuspend the cell pellet in ice-cold CMF HBSS/FBS and place samples on ice.
Proceed to Section 3 for FACS acquisition/analysis or Section 4 for magnetic-bead enrichment for high-speed cell sorting.
3. Antibody Staining for Multi-Color Flow Cytometric Analysis of DCs and Macrophages
Preparations of reagents and equipment:
Ice-cold CMF PBS.
Ice-cold staining buffer (CMF PBS + 5% FBS).
Prepare dead cell stain in ice-cold CMF PBS at 1:1000 dilution using LIVE/DEAD Fixable Aqua Dead Cell Stain Kit.
Prepare the antibody staining cocktail by adding the following fluorescence-labeled monoclonal antibodies (mAbs) to the ice-cold staining buffer: CD45-PerCP, CD103-PE, CD11c-APC, MHC-II (I-Ab)-Alexa Fluor 700, CD11b-eFluor 450, F4/80-PE-Cy7.
Transfer cells into a 5 ml polystyrene round-bottom (FACS) tube.
Wash cells twice in ice-cold CMF PBS.
Incubate samples with dead cell stain for 15 min on ice in the dark.
Wash cells twice in ice-cold CMF PBS.
Block cells with 2.4G2 anti-FcγRIII/II in ice-cold staining buffer for 10 min on ice.
Wash cells in ice-cold staining buffer.
Incubate samples with antibody staining cocktail for 20 min on ice in the dark.
Wash cells with ice-cold staining buffer twice and resuspend samples in 400 μl of ice-cold staining buffer and pass through 40 μm filter cap on FACS tubes.
Acquire samples on LSR II cytometer (BD) as defined by gating strategy in Section 5 and Figure 1.
4. Enrichment of DCs and Macrophages from the Intestine
Preparations of reagents and equipment:
Ice-cold staining buffer (CMF PBS + 5% FBS).
Incubate single cell suspension obtained from step 2.5 with CD11b and CD11c MACS beads according to manufacturer's instructions.
Wash cells with ice-cold staining buffer followed by centrifugation.
Discard supernatant and resuspend the cell pellet in 1 mL ice-cold staining buffer and pass through a 100 μm cell strainer followed by a 40 μm cell strainer.
Enrich for magnetic bead-attached cells by positive selection using MACS LS magnetic column.
Repeat step 4.2 and discard supernatant.
Incubate cells with surface marker mAbs as described in step 3.7.
Wash magnetic bead-enriched cells twice with ice-cold staining buffer. Resuspend cell pellets in 500 μL ice-cold staining buffer without sodium azide, and pass through 40 μm cell strainer into a FACS tube.
Proceed to FACS-sorting on the BD ARIA II Cell Sorter to sort intestinal DC and/or macrophage subsets of interest.
5. Gating Strategy for LP APCs
Note: Please note that unstained intestinal cells may be utilized as a negative control to assist in the proper placement of the gates to separate positive and negative populations.
As shown in Figure 1, create a dot plot and exclude cells that are positive for the dead cell stain (Fig. 1A) followed by the exclusion of doublet events (Fig. 1B and C). Then, gate on the cells of interest according to forward and side scatter making sure to exclude debris (Fig. 1D).
Create another dot plot and further gate on CD45+ and I-Ab+ cells, which phenotypically characterizes APCs (Fig. 1E).
On a separate dot plot, analyze for CD11b and CD11c expression to distinguish specific DC and macrophage subsets (R1, R2, and R3; Fig. 1F). Cells of R1 are CD11c+ CD11bdull/- cells. The region, R2, is delineated such that these cells have similar levels of surface expression for CD11c as in cells of R1 but cells of R2 also express CD11b. Thereafter, the R3 region is designated for cells that are CD11b+ and CD11cdull/-.
Analyze regions R1, R2, and R3 further for F4/80 and CD103 expression to differentiate macrophage and DC populations, respectively. The CD11c+ CD11bdull/- cells of R1 express high levels of the αE integrin, CD103, and low levels of F4/80 (Fig. 1G). CD11b+CD11c+ cells of R2 are composed of both DCs and macrophages based on their dichotomous expression of CD103 and F4/80 (Fig. 1H). Lastly, CD11b+CD11cdull/- cells of R3 constitute macrophages based on the phenotypic profile of F4/80+ and CD103- (Fig. 1I)16.
6. Representative Results
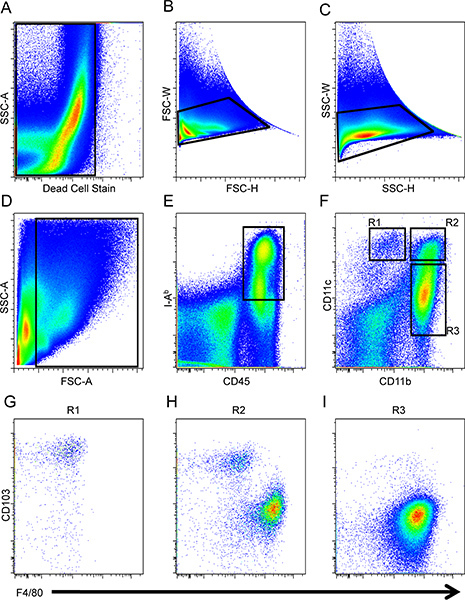 Figure 1. Gating strategy for intestinal DCs and macrophages. Dead cells (A) and doublets (B and C) were first excluded from the analysis and then small intestinal cells were gated accordingly to forward and side scatter (D), and APCs were defined as CD45+I-Ab+(E). Macrophages and DCs were identified by the expression of CD11b and CD11c (F). CD103 and F4/80 expression for cells pre-gated on R1 (G), R2 (H) and R3 (I) populations was analyzed.
Figure 1. Gating strategy for intestinal DCs and macrophages. Dead cells (A) and doublets (B and C) were first excluded from the analysis and then small intestinal cells were gated accordingly to forward and side scatter (D), and APCs were defined as CD45+I-Ab+(E). Macrophages and DCs were identified by the expression of CD11b and CD11c (F). CD103 and F4/80 expression for cells pre-gated on R1 (G), R2 (H) and R3 (I) populations was analyzed.
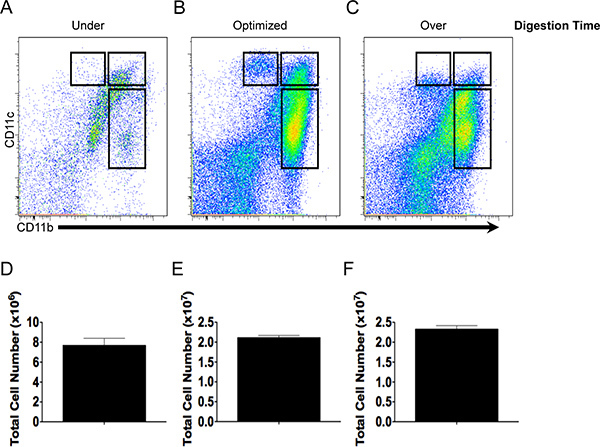 Figure 2. Cell yield and antibody staining quality depends on digestion time. CD11b and CD11c staining pattern and total cell yield of under- (A, D), optimally- (B, E) or over-digested (C, F) intestinal tissue.
Figure 2. Cell yield and antibody staining quality depends on digestion time. CD11b and CD11c staining pattern and total cell yield of under- (A, D), optimally- (B, E) or over-digested (C, F) intestinal tissue.
Intestinal cells were isolated from a C57BL/6 mouse small intestine and DCs and macrophages were analyzed by FACS on the BD LSR II. Voltage and compensation were set using unstained and single fluorochrome-stained splenocytes. Dead cells (Fig. 1A) and doublets (Fig. 1B and C) were first excluded from the analysis. Cells of interest were then analyzed according to forward and side scatter (Fig. 1D) followed by gating on CD45+ and I-Ab+ cells (Fig. 1E). Thereafter, CD11b and CD11c expression was assessed among the CD45+I-Ab+ cells to delineate three regions (R1, R2, and R3; Fig. 1F). CD103 and F4/80 expression in the three regions was evaluated to distinguish between DCs and macrophages, respectively. The CD11c+ CD11bdull/- cells of R1 expressed high levels of the αE integrin, CD103, and low levels of F4/80 (Fig. 1G). CD11b+CD11c+ cells of R2 were composed of both DCs and macrophages based on their dichotomous expression of CD103 and F4/80 (Fig. 1H) while CD11b+CD11cdull/- cells of R3 constitute macrophages based on the phenotypic profile of F4/80+ and CD103- (Fig. 1I)16. Macrophages within the R2 gate and macrophages in the R3 gate have similar forward and side scatter properties and are distinguishable by CD11c expression. The functional dichotomy of these subsets remains incompletely understood.
The relationship between the duration of tissue digestion on total cell yield and the expression of CD11b and CD11c is illustrated in Figure 2. Intestinal tissue that was digested for 3 min (under-digestion) yielded low total cell number (Fig. 2D) and thus few DCs and macrophages available for characterization (Fig. 2A). Tissue digestion for 11 min produced a robust yield of live cells (Fig. 2E) with populations of DCs and macrophages that expressed high levels of CD11b and CD11c and were phenotypically distinct (Fig. 2C). In contrast, digestion for 50 min (over-digestion) resulted in a similar cell yield when compared to optimized digestion (Fig. 2E and F), however, delineation of different cell populations using CD11b and CD11c became more obscure as the expression of CD11c diminished (Fig. 2C) and the number of dead cells increased (data not shown).
Discussion
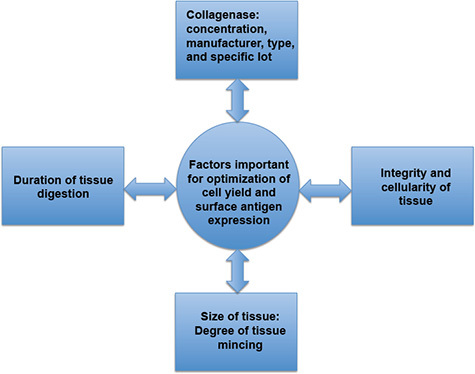 Figure 3. Factors important for the optimization of cell yield and surface antigen expression. Cell yield and surface antigen expression are directly affected by the duration of tissue digestion, the specific characteristics of the collagenase, the degree of tissue mincing, and the presence or absence of inflammation, which may affect tissue integrity and cellularity. Prolonged tissue digestion may result in decreased cell viability and surface antigen expression while inadequate tissue digestion may result in a paucity of cells for analysis.
Figure 3. Factors important for the optimization of cell yield and surface antigen expression. Cell yield and surface antigen expression are directly affected by the duration of tissue digestion, the specific characteristics of the collagenase, the degree of tissue mincing, and the presence or absence of inflammation, which may affect tissue integrity and cellularity. Prolonged tissue digestion may result in decreased cell viability and surface antigen expression while inadequate tissue digestion may result in a paucity of cells for analysis.
Here, we detailed a methodology for the rapid isolation of mouse intestinal DCs and macrophages for phenotypic characterization using multi-color flow cytometry and for enrichment using MACS beads and cell sorting to conduct functional studies on purified cells. Optimizing the concentration of collagenase and the duration of tissue digestion is necessary to produce a robust cell yield without compromising viability and surface antigen expression. Under-digestion yields a low total cell number and a paucity of DCs and macrophages for characterization (Fig. 2A and D). On the other hand, over-digestion yields a total cell number similar to optimized conditions (Fig. 2F) but the number of dead cells are appreciably increased (data not shown) and the quality of the staining is compromised. Similar to under-digestion, the over-digestion of tissue would complicate phenotypic characterization and purification.
Several additional parameters for consideration using this protocol are the manufacturer, type and specific lot of collagenase, the integrity and cellularity of the intestine, and degree of tissue mincing. As variability in collagenase activity may exist between different manufacturers, types of collagenase, and production lots, the potency of digestion may vary greatly and requires optimization. Hence, selection of the most appropriate type of collagenase is critical when designing an experiment as the quality and reproducibility of the data may be affected. In our experience, collagenase type VIII obtained from Sigma-Aldrich has provided the best results, however, we have also had success using collagenase type IV from Sigma-Aldrich.
While the above detailed protocol has been optimized for use on healthy small and large intestines, it can be successfully used for the isolation of DCs and macrophages from inflamed tissue exhibiting increased cellularity and architectural distortion associated with inflammation. The presence of intestinal inflammation may dramatically affect the rate of digestion—often increasing the sensitivity of the intestinal tissue to the action of proteolytic enzymes based on our experience. Therefore, the duration of digestion or the concentration of collagenase must be adapted accordingly to changes in tissue integrity associated with inflammation.
Furthermore, the degree of mincing the tissue will affect the duration of tissue digestion. Mincing the tissue into smaller pieces increases the surface area for digestion and yields more cells but precautions should be taken, as the tissue may be more susceptible to over-digestion. Consequently, the duration of digestion may need to be decreased. In contrast, poor mincing will result in larger pieces of tissue that will digest poorly resulting in a low total cell yield. Increasing the duration of digestion may compensate for poor mincing to a certain extent.
Upon optimizing the parameters for intestinal tissue digestion with collagenase, a robust cell yield can be rapidly achieved. As a result, intestinal DC and macrophage populations can be more accurately characterized and purified for functional studies to assess cytokine production, antigen presentation, and the regulation of immune cells.
Disclosures
No conflicts of interest declared.
Acknowledgments
We thank Aaron Rae (Emory University Department of Pediatrics and Children's Healthcare of Atlanta Flow Core) for cell sorting. This work was supported by NIH grant AA01787001, a Career Development Award from the Crohn's and Colitis Foundation of America, and an Emory-Egleston Children's Research Center seed grant to T.L.D.
References
- Maloy KJ, Powrie F. Intestinal homeostasis and its breakdown in inflammatory bowel disease. Nature. 2011;474:298–306. doi: 10.1038/nature10208. [DOI] [PubMed] [Google Scholar]
- Nagler-Anderson C, Terhoust C, Bhan AK, Podolsky DK. Mucosal antigen presentation and the control of tolerance and immunity. Trends Immunol. 2001;22:120–122. doi: 10.1016/s1471-4906(00)01830-5. [DOI] [PubMed] [Google Scholar]
- Abraham C, Medzhitov R. Interactions between the host innate immune system and microbes in inflammatory bowel disease. Gastroenterology. 2011;140:1729–1737. doi: 10.1053/j.gastro.2011.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macdonald TT, Monteleone I, Fantini MC, Monteleone G. Regula tine. Gastroenterology. 2011;140:1768–1775. doi: 10.1053/j.gastro.2011.02.047. [DOI] [PubMed] [Google Scholar]
- Rescigno M. Intestinal dendritic cells. Adv. Immunol. 2010;107:109–138. doi: 10.1016/B978-0-12-381300-8.00004-6. [DOI] [PubMed] [Google Scholar]
- Platt AM, Bain CC, Bordon Y, Sester DP, Mowat AM. An independent subset of TLR expressing CCR2-dependent macrophages promotes colonic inflammation. J. Immunol. 2010;184:6843–6854. doi: 10.4049/jimmunol.0903987. [DOI] [PubMed] [Google Scholar]
- Coombes JL, Powrie F. Dendritic cells in intestinal immune regulation. Nat. Rev. Immunol. 2008;8:435–446. doi: 10.1038/nri2335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelsall B. Recent progress in understanding the phenotype and function of intestinal dendritic cells and macrophages. Mucosal Immunol. 2008;1:460–469. doi: 10.1038/mi.2008.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pulendran B, Tang H, Denning TL. Division of labor, plasticity, and crosstalk between dendritic cell subsets. Curr. Opin. Immunol. 2008;20:61–67. doi: 10.1016/j.coi.2007.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denning TL, Wang YC, Patel SR, Williams IR, Pulendran B. Lamina propria macrophages and dendritic cells differentially induce regulatory and interleukin 17-producing T cell responses. Nat. Immunol. 2007;8:1086–1094. doi: 10.1038/ni1511. [DOI] [PubMed] [Google Scholar]
- Niess JH. CX3CR1-mediated dendritic cell access to the intestinal lumen and bacterial clearance. Science. 2005;307:254–258. doi: 10.1126/science.1102901. [DOI] [PubMed] [Google Scholar]
- Milling SW, Cousins L, MacPherson GG. How do DCs interact with intestinal antigens. Trends Immunol. 2005;26:349–352. doi: 10.1016/j.it.2005.04.010. [DOI] [PubMed] [Google Scholar]
- Bilsborough J, Viney JL. Gastrointestinal dendritic cells play a role in immunity, tolerance, and disease. Gastroenterology. 2004;127:300–309. doi: 10.1053/j.gastro.2004.01.028. [DOI] [PubMed] [Google Scholar]
- Stagg AJ, Hart AL, Knight SC, Kamm MA. The dendritic cell: its role in intestinal inflammation and relationship with gut bacteria. Gut. 2003;52:1522–1529. doi: 10.1136/gut.52.10.1522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medina-Contreras O. CX3CR1 regulates intestinal macrophage homeostasis, bacterial translocation, and colitogenic Th17 responses in mice. J. Clin. Invest. 2011;121:4787–4795. doi: 10.1172/JCI59150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denning TL. Functional Specializations of Intestinal Dendritic Cell and Macrophage Subsets That Control Th17 and Regulatory T Cell Responses Are Dependent on the T Cell/APC Ratio, Source of Mouse Strain, and Regional Localization. J. Immunol. 2011. pp. 187–733. [DOI] [PMC free article] [PubMed]
- Kim YG. The Nod2 sensor promotes intestinal pathogen eradication via the chemokine CCL2-dependent recruitment of inflammatory monocytes. Immunity. 2011;34:769–780. doi: 10.1016/j.immuni.2011.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulz O. Intestinal CD103+, but not CX3CR1+, antigen sampling cells migrate in lymph and serve classical dendritic cell functions. J. Exp. Med. 2009;206:3101–3114. doi: 10.1084/jem.20091925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaensson E. Small intestinal CD103+ dendritic cells display unique functional properties that are conserved between mice and humans. J. Exp. Med. 2008;205:2139–2149. doi: 10.1084/jem.20080414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uematsu S. Regulation of humoral and cellular gut immunity by lamina propria dendritic cells expressing Toll-like receptor 5. Nat. Immunol. 2008;9:769–776. doi: 10.1038/ni.1622. [DOI] [PubMed] [Google Scholar]
- Schenk M, Bouchon A, Seibold F, Mueller C. TREM-1--expressing intestinal macrophages crucially amplify chronic inflammation in experimental colitis and inflammatory bowel diseases. J. Clin. Invest. 2007;117:3097–3106. doi: 10.1172/JCI30602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun CM. Small intestine lamina propria dendritic cells promote de novo generation of Foxp3 T reg cells via retinoic acid. J. Exp. Med. 2007;204:1775–1785. doi: 10.1084/jem.20070602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamada N. Abnormally differentiated subsets of intestinal macrophage play a key role in Th1-dominant chronic colitis through excess production of IL-12 and IL-23 in response to bacteria. J. Immunol. 2005;175:6900–6908. doi: 10.4049/jimmunol.175.10.6900. [DOI] [PubMed] [Google Scholar]
- Denning TL. CD4+ Th cells resembling regulatory T cells that inhibit chronic colitis differentiate in the absence of interactions between CD4 and class II MHC. J. Immunol. 2003;171:2279–2286. doi: 10.4049/jimmunol.171.5.2279. [DOI] [PubMed] [Google Scholar]


