Abstract
In this study, we describe an effective protocol for use in a multiplexed high-throughput antibody microarray with glycan binding protein detection that allows for the glycosylation profiling of specific proteins. Glycosylation of proteins is the most prevalent post-translational modification found on proteins, and leads diversified modifications of the physical, chemical, and biological properties of proteins. Because the glycosylation machinery is particularly susceptible to disease progression and malignant transformation, aberrant glycosylation has been recognized as early detection biomarkers for cancer and other diseases. However, current methods to study protein glycosylation typically are too complicated or expensive for use in most normal laboratory or clinical settings and a more practical method to study protein glycosylation is needed. The new protocol described in this study makes use of a chemically blocked antibody microarray with glycan-binding protein (GBP) detection and significantly reduces the time, cost, and lab equipment requirements needed to study protein glycosylation. In this method, multiple immobilized glycoprotein-specific antibodies are printed directly onto the microarray slides and the N-glycans on the antibodies are blocked. The blocked, immobilized glycoprotein-specific antibodies are able to capture and isolate glycoproteins from a complex sample that is applied directly onto the microarray slides. Glycan detection then can be performed by the application of biotinylated lectins and other GBPs to the microarray slide, while binding levels can be determined using Dylight 549-Streptavidin. Through the use of an antibody panel and probing with multiple biotinylated lectins, this method allows for an effective glycosylation profile of the different proteins found in a given human or animal sample to be developed.
Introduction
Glycosylation of protein, which is the most ubiquitous post-translational modification on proteins, modifies the physical, chemical, and biological properties of a protein, and plays a fundamental role in various biological processes1-6. Because the glycosylation machinery is particularly susceptible to disease progression and malignant transformation, aberrant glycosylation has been recognized as early detection biomarkers for cancer and other diseases 7-12. In fact, most current cancer biomarkers, such as the L3 fraction of α-1 fetoprotein (AFP) for hepatocellular carcinoma 13-15, and CA199 for pancreatic cancer 16, 17 are all aberrant glycan moieties on glycoproteins. However, methods to study protein glycosylation have been complicated, and not suitable for routine laboratory and clinical settings. Chen et al. has recently invented a chemically blocked antibody microarray with a glycan-binding protein (GBP) detection method for high-throughput and multiplexed profile glycosylation of native glycoproteins in a complex sample 18. In this affinity based microarray method, multiple immobilized glycoprotein-specific antibodies capture and isolate glycoproteins from the complex mixture directly on the microarray slide, and the glycans on each individual captured protein are measured by GBPs. Because all normal antibodies contain N-glycans which could be recognized by most GBPs, the critical step of this method is to chemically block the glycans on the antibodies from binding to GBP. In the procedure, the cis-diol groups of the glycans on the antibodies were first oxidized to aldehyde groups by using NaIO4 in sodium acetate buffer avoiding light. The aldehyde groups were then conjugated to the hydrazide group of a cross-linker, 4-(4-N-MaleimidoPhenyl)butyric acid Hydrazide HCl (MPBH), followed by the conjugation of a dipeptide, Cys-Gly, to the maleimide group of the MPBH. Thus, the cis-diol groups on glycans of antibodies were converted into bulky none hydroxyl groups, which hindered the lectins and other GBPs bindings to the capture antibodies. This blocking procedure makes the GBPs and lectins bind only to the glycans of captured proteins. After this chemically blocking, serum samples were incubated with the antibody microarray, followed by the glycans detection by using different biotinylated lectins and GBPs, and visualized with Cy3-streptavidin. The parallel use of an antibody panel and multiple lectin probing provides discrete glycosylation profiles of multiple proteins in a given sample 18-20. This method has been used successfully in multiple different labs 1, 7, 13, 19-31. However, stability of MPBH and Cys-Gly, complicated and extended procedure in this method affect the reproducibility, effectiveness and efficiency of the method. In this new protocol, we replaced both MPBH and Cys-Gly with one much more stable reagent glutamic acid hydrazide (Glu-hydrazide), which significantly improved the reproducibility of the method, simplified and shorten the whole procedure so that the it can be completed within one working day. In this new protocol, we describe the detailed procedure of the protocol which can be readily adopted by normal labs for routine protein glycosylation study and techniques which are necessary to obtain reproducible and repeatable results.
Keywords: Molecular Biology, Issue 63, Glycoproteins, glycan-binding protein, specific protein glycosylation, multiplexed high-throughput glycan blocked antibody microarray
Protocol
1. Print an Antibody Microarray for the Assay
Dilute all antibodies to 0.5 mg/ml in phosphate buffer saline, ph 7.2 (PBS).
Aliquot 40 μl of each antibody into the 384-well source plate.
Load the 384-well source plate onto the Scienion sciFLEXARRAYER microarrayer.
Load 20 PATH microarray slides onto the microarrayer as target.
Set the microarrayer to print 48 identical subarrays, in which 27 antibodies and control proteins are spotted in triplicate in a 9x9 pattern (Figure 1E, 1F).
Start the microarrayer to print the antibody microarray slides.
Collect the antibody microarray slides, and store them in slides cassette with desiccant. Vacuum seal the cassette in a plastic bag by using vacuum sealer (Foodsaver).
Store the sealed microarray slides at 4 °C in refrigerator.
2. Chemically Block the Antibody Microarray to Prevent GBP Binding to the Capture Antibodies
The microarray assay starts once the microarray slides are chemically blocked and lasts for about 8 hours. Once started the microarray assay has to be completed (Steps 2 to 8).
Take the microarray slides out of the refrigerator, and equilibrate them to room temperature for 30 minutes.
Remove the slide from the storage box and briefly rinse them in phosphate buffer saline pH 7.2 with 0.1% Tween 20 (PBST0.1) once in a slide washing basin, and then in 15 mM sodium acetate buffer pH 5.0 with 0.1% Tween (CBT0.1) in a sequential fashion. Incubate the slides in CBT0.1 for 10 minutes in slide washing basin.
Prepare fresh 150 mM NaIO4 in 15 mM sodium acetate buffer pH 5.0 (CB), and keep it in into a slide washing basin in a refrigerator while avoiding light before use.
Remove the slide from the CB, and put it into the basin containing fresh NaIO4 with the antibody side facing up. Cover the basin with aluminum foil to avoid light, and incubate the slide basin for 2 hours with gentle shaking at 4 °C in a refrigerator.
Prepare 300 mL of 10 mM hydrazide glutamic acid (the blocker) in CB.
Remove the slide from the basin, and briefly rinse it in CB 3 times for 5 minutes each time in slide washing basin.
Incubate the slides in the blocker in a washing basin for 2 hours at room temperature with gentle shaking.
Remove the slides from the basin, and wash them with PBST0.1 for 3 minutes.
3. Block Non-specific Bindings to the Microarray with Bovine Serum Albumin (BSA)
Prepare 300 ml of 1% BSA in phosphate buffer saline pH 7.2 with 0.5% Tween (PBST0.5) in a slide washing basin, and incubate the microarray slide in the basin for 1 hour at room temperature with gently shaking.
Rinse the slides in PBST0.1 three times for 3 minutes each time.
Put the slide on a slide rack, and spin at 1,200 x g on a centrifugation for 2 minutes to dry the microarray slide.
4. Imprint Wax Grid onto the Microarray Slide to Separate each Subarray
Pre-heating the wax imprinter at 70 °C for 5 minutes.
Load the blocked microarray slide into the wax imprinter with antibody side facing to the wax. Gently pull the handle to imprint wax onto the slide evenly.
5. Apply Serum Samples onto the Microarray Slide
- During Step 2.4, prepare serum samples for either glyco profiling assay in one sample (5.1.1), or single glyco epiptope measurement among multiple samples (5.1.2).
- In an experiment for the glycan profilings of multiple serum glycoproteins in one serum sample by using multiple GBPs (see sample experiment 1), one serum samples will be applied onto all subarrays. In this case, 40 μl serum ample is diluted into 360 μl of PBS containing 0.1% Tween-20, 0.1% Brij 35, 100 μg/mL of mouse IgG, 100 μg/mL of rat IgG, 100 μg/mL of rabbit IgG, 100 μg/mL of goat IgG and 100 μg/ml of donkey IgG. This volume is sufficient for applying 6 μl of diluted serum solution onto each subarray.
- In an experiment for the one glycan measurement on multiple serum proteins among multiple serum samples by using one GBP detections (see sample experiment 2). In this case, 1 μl serum ample is diluted into 9 μl of PBS containing 0.1% Tween-20, 0.1% Brij 35, 100 μg/mL of mouse IgG, 100 μg/mL of rat IgG, 100 μg/mL of rabbit IgG, 100 μg/mL of goat IgG and 100 μg/mL of donkey IgG. This volume is sufficient for applying 6 μL of diluted serum solution onto each subarray.
After wax imprint in Step 4, carefully apply 6 μL of diluted sample or control samples (PBST0.1) to each subarray of the slide. Incubate the slide in a humidified cassette with wet paper towels at room temperature for 1 hour.
Rinse the slide with PBST0.1 three times for 3 minutes each time.
Dry the slide by spinning it at 1200 x g for 2 minute.
6. Apply Biotinylated GBP (Lectin or Anti-glycan Antibody) onto the Slide
- During Step 2.4, prepare 10μg/ml of biotinylated lectins/GBPs in PBST0.1.
- In glycan profiling experiment that probe one sample with multiple lectins (sample experiment 1), prepare 350 μL of biotinylated lectin that is sufficient for all subarrays.
- In single glycan epitope/biomarker screening in multiple samples by using multiple lectins, prepare 10 μl of each biotinylated lectin that is sufficient for one subarray.
Apply 6 μL of the diluted biotinylated lectin(s) to each subarray of the slide, and incubate in the humidified slide box with wet paper towels at room temperature for 1 hour.
Rinse the slides with PBST0.1 three times for 3 minutes each time.
Dry the slide by spinning it at 1200 x g in centrifuge for 2 minute.
7. Apply Dye Labeled NeutrAvidin for Fluorescence Detection
Prepare 350 μL of Dylight 549 labeled NeutrAvidin that is sufficient for all subarrays.
Apply 6 μL of Dylight 549 labeled NeutrAvidin onto each subarray, and incubate the slide in the humidified slide cassette at room temperature for 1 hour.
Rinse the slide with PBST0.1 three times for 3 minutes each time.
Dry the slide by spinning the it at 1200 x g in centrifuge for 2 minute.
8. Obtain Microarray Slide Image by Scanning the Slide
Scan the slide by using a fluorescence microarray scanner at 10 μm resolution. The laser and PMT settings should be as strong as possible, but no saturation spot is observed.
9. Data Extraction and Analysis
Open the image in ArrayPro 3.2.
Set up the array template according to the array map that shows the antibody spots locations. Carefully align each template circle onto the corresponding spot in the image.
Extract the intensity of each spot into an Excel file for further analysis.
10. Representative Results
Sample Experiment 1
Glycosylation profiling of multiple serum glycoproteins in hepatocellular carcinoma patient serum sample by using chemically blocked antibody microarray with multiple lectins detection.
The goal of this experiment is to explore the individual glycosylation profile of 20 glycoproteins in hepatocellular carcinoma (HCC) patient serum sample by using chemically blocked antibody microarray with lectin detection. An antibody microarray, that contains 48 identical subarrays which include 26 antibodies and biotin-BSA, was designed and manufactured as described in Step 1. These 26 antibodies were against 20 serum glycoproteins that identified as promising early diagnosis value for HCC patients by using lectin based immunoprecipitation combined with mass spectrometric protein identification12, 32 as shown in Table 1. The pattern and arrangement of the antibody spots printed in triplicate in one representative subarray are shown in Figure 1E and 1F, respectively. Two identical microarray slides, one was not chemically blocked (Figure 1A), while the other one was (Figure 1B), were used to carry out the same glycosylation profiling experiment in order to demonstrate the importance of the chemically blocking procedure to the analysis. For the chemically blocked slide (Figure 1B), the experiment started at the Step 2; for the none chemically blocked slide (Figure 1A), the experiment started from Step 3. The experiment was carried by following all the steps described in the protocol except for Step 5.1.2 and 6.1.2. In the Step 5.2, a PBST0.1 control sample was applied onto subarrays in column 1 and 3, and a pooled HCC serum sample was applied onto subarrays in column 2 and 4, respectively (as shown in Figure 1G). This comparison is to show the effectiveness, efficiency of the procedure, as well as the antigen binding affinity of the antibodies after chemically blocking. 22 biotinylated lectins (as shown in Table 1) that specific to different glycans18, 20 were applied onto each subarray as shown in Figure 1G for glycosylation profiling. Images of the chemically blocked (Figure 1B) and non-chemically blocked (Figure 1A) microarrays after the glycosylation profiling assay by following the protocol. As shown in the subarrays in column 1 and 3 in non-chemically blocked microarrays (Figure 1A and Figure 1C), on which only PBST0.1 was applied, most lectins bound to capture antibodies, and showed very high background that comparable to those subarrays in column 2 and 4, on which the serum sample was applied. It is impossible to obtain glycan profile information from this microarray slide. On the contrary, when the same experiment was done on a chemically blocked antibody microarray slide, the subarrays in column 1 and 3, on which only PBST0.1 was applied, most lectins showed no or very low bindings to capture antibodies; while high antigen bindings were still observed in subarrays in column 2 and 4, on which serum sample was applied (Figure 1B and 1D). These result showed the chemically blocking procedure was a critical step fore the measurement of glycans on antibody-captured glycoproteins. By following the protocol, glycosylation profiles of 22 glycoproteins in HCC serum can be obtained.
Experiment 2
Screen for altered fucosylation on specific serum glycoproteins as biomarkers to discriminated liver cirrhosis and hepatocellular carcinoma patients.
The goal of this experiment is to screen for altered fucosylation on specific serum glycoproteins as biomarkers that discriminate liver cirrhosis and hepatocellular carcinoma (HCC) patients. Different from the Experiment 1, in which only one serum sample was applied onto every subarrays and probed with various lectins, in this assay, total 40 different serum samples from HCC and cirrhosis patients were applied onto each subarray, and probed with one lectin (AAL). Statistical analysis, such as T test, receiver-operating characteristic (ROC) curve, was done to evaluate the distributions or diagnostic performance of the glycan epiptope/biomarker on each individual protein in all serum samples. We used the same antibody microarray manufactured in Experiment 1 except for anti-CA19-9 and anti-Lewis X antibodies in this study. The experiment was carried out from Sept 2 to Step 9 except for the Step 5.1.1 and 6.1.1. Total 40 serum samples from 20 cirrhosis and 20 HCC patients were applied at random subarray of the 48 subarrays along with the control PBS samples as negative control. Fucosylation of each the captured proteins was then detected by using biotinylated Fucose-specific lectin. The microarray image shown in Figure 1 demonstrated the lectin AAL only bound to serum proteins captured on the microarray (Figure 2D) instead of captured antibodies (Figure 2E). The AAL binding intensities of all the spots were then extracted, and analyzed by using T test and ROC curves to evaluate the performance of the fucosylation (AAL binding intensity) of each serum protein on the discrimination between the HCC and cirrhosis groups. The results showed that the fucosylation of GP73 protein gave the best discrimination between the two groups with a p = 0.03 and area-under-curve of ROC curve equals to 0.72. This experiment demonstrated this procedure is a rapid, efficient method for the glycan epitope/biomarker screening on multiple samples within multiple proteins.
| ID | Name of the reagent | Abbreviation | Company | Catalog # |
| L1 | Biotinylated Concanavalin A | ConA | Vector Laboratories | BK-1000 |
| L2 | Biotinylated Sambucus Nigra Lectin | SNA | Vector Laboratories | B-1305 |
| L3 | Biotinylated Lens Culinaris Agglutinin | LCA | Vector Laboratories | BK-2000 |
| L4 | Biotinylated Ricinus Communis Agglutinin I | RCA | Vector Laboratories | BK-1000 |
| L5 | Biotinylated Aleuria Aurantia Lectin | AAL | Vector Laboratories | B-1395 |
| L6 | Biotinylated Erythrina Cristagalli Lectin | ECL | Vector Laboratories | BK-3000 |
| L7 | Biotinylated Griffonia (Bandeiraea) Simplicifolia Lectin II | GSL II | Vector Laboratories | BK-3000 |
| L8 | Biotinylated Wheat Germ Agglutinin | WGA | Vector Laboratories | BK-1000 |
| L9 | Biotinylated Phaseolus vulgaris Erythroagglutinin | PHA-E | Vector Laboratories | BK-2000 |
| L10 | Biotinylated Phaseolus vulgaris Leucoagglutinin | PHA-L | Vector Laboratories | BK-2000 |
| L11 | Biotinylated Peanut Agglutinin | PNA | Vector Laboratories | BK-1000 |
| L12 | Biotinylated Pisum Sativum Agglutinin | PSA | Vector Laboratories | BK-2000 |
| L13 | Biotinylated Dolichos Biflorus Agglutinin | DBA | Vector Laboratories | BK-1000 |
| L14 | Biotinylated Datura Stramonium Lectin | DSL | Vector Laboratories | BK-3000 |
| L15 | Biotinylated Sophora Japonica Agglutinin | SJA | Vector Laboratories | BK-2000 |
| L16 | Biotinylated Soybean Agglutinin | SBA | Vector Laboratories | BK-1000 |
| L17 | Biotinylated Solanum Tuberosum (Potato) Lectin | STL | Vector Laboratories | BK-3000 |
| L18 | Biotinylated Griffonia (Bandeiraea) Simplicifolia Lectin I | GSL I | Vector Laboratories | BK-2000 |
| L19 | Biotinylated Vicia Villosa Lectin | VVL | Vector Laboratories | BK-2000 |
| L20 | Biotinylated Lycopersicon Esculentum (Tomato) Lectin | LEL | Vector Laboratories | BK-3000 |
| L21 | Biotinylated Ulex Europaeus Agglutinin I | UEA I | Vector Laboratories | BK-1000 |
| L22 | Biotinylated Jacalin | JACALIN | Vector Laboratories | BK-3000 |
| A1 | Goat F(ab')2 Fragment anti-human IgM, Fc5μ antibody | IgM | Jackson Immuno Research | 109-006-129 |
| A2 | Donkey F(ab')2 Frag anti-human IgG (H+L) antibody | AB1 | Jackson Immuno Research | 709-006-149 |
| A3 | Mouse anti-human IgG F(ab')2 monoclonal antibody | AB3 | Jackson Immuno Research | 209-005-097 |
| A4 | Goat anti-human alpha 2 macroglobulin polyclonal antibody | A2M | GeneTex | GTX62924 |
| A5 | Rabbit anti-human alpha-1-antitrypsin polyclonal antibody | A1AT | Lee Biosiences | CA1T-80A |
| A6 | Mouse anti-human alpha-1-antitrypsin monoclonal antibody | A1AT | Sigma Aldrich | SAB4200198 |
| A7 | Rabbit anti-human alpha-1-antitrypsin polyclonal antibody | ACT | NeoMarkers | RB-367-A1 |
| A8 | Rabbit anti-human alpha-1-antichymotrypsin polyclonal antibody | ACT | Fisher Scientific | RB9213R7 |
| A9 | Mouse anti-human transferrin monoclonal antibody | Transferrin | GeneTex | GTX101035 |
| A10 | Rabbit anti-human transferrin polyclonal antibody | Transferrin | GeneTex | GTX77130 |
| A11 | Goat anti-human apolipoprotein J polyclonal antibody | ApoJ | Abcam | ab7610 |
| A12 | Mouse anti-human GP73 monoclonal antibody | GP73 | Abbott | 14H4-23 |
| A13 | Mouse anti-human GP73 monoclonal antibody | GP73 | SANTA CRUZ BIOTECHNOLOGY INC | sc-101275 |
| A14 | Rabbit anti-human alpha-1 fetoprotein polyclonal antibody | AFP | GenWay | GWB-41C966 |
| A15 | Mouse anti-human alpha-1 fetoprotein monoclonal antibody | AFP | Fitzgerald | 10-A05A |
| A16 | Mouse anti-human hemopexin monoclonal antibody | Hemopexin | Assaypro | 60190-05011 |
| A17 | Mouse anti-human glypican-3(1G12) monoclonal antibody | GPL3 | Santa Cruz Bio | sc-65443 |
| A18 | Mouse anti-human Kininogen (LMW) monoclonal antibody | Kininogen | Assaypro | 20333-05011 |
| A19 | Rabbit anti-human MMP-21 monoclonal antibody | MMP21 | Epitomic | 1955-1 |
| A20 | Mouse anti-human CEACAM-1 monoclonal antibody | CEACAM | R&D Systems | MAB1180 |
| A21 | Rat anti-human DPPIV/CD26 monoclonal antibody | DPPIV | R&D Systems | MAB22441 |
| A22 | Mouse anti-human PIVKA II monoclonal antibody | PIVICA | Crystal chem | 8040 |
| A23 | Mouse anti-carcinoembryonic antigen | CEA | US biological | C1300 |
| A24 | Mouse anti-CA125 Cancer Antigen | CA125 | US biological | C0050-01D |
| A25 | Mouse anti -CA19-9 Cancer antigen | CA19-9 | US biological | C0075-18 |
| A26 | Mouse anti-Lewis x monoclonal antibody | Lewis X | Calbiochem | 434631 |
| bio | Biotinylated BSA (positive control) | Bio | Home-made | N/A |
Table 1. List of lectins and antibodies used in this protocol.
| Name of the reagent s/equipments | Company | Catalogue number |
| Non contact microarrayer | BioDot Inc | sciFLEXARRAYER |
| 384 microplate | Fisher | 14-230-243 |
| FoodSaver | FoodSaver | V3835 |
| Ultrathin nitrocellulose coate microarray slides | Gentel | PATH |
| Slide Imprinter (optional) | The Gel Company | WSP60-1 |
| Shaker | Fisher | 15-453-211 |
| Centrifuge | Eppendorf | 5804 000.013 |
| Slide washing basin/Slide Staining Dish with Removable Rack | Fisher | 08-812 |
| Slide incubation chamber/microscope slide box | Fisher | 03-448-5 |
| Brij 35, 30 w/v% solution in water | Acros Organics | AC32958-0025 |
| Tween-20 | Fisher | P337-100 |
| Sodium Periodate (NaIO4) | Sigma | 311448 |
| L-Glutamic acid γ-hydrazide | Sigma | G-7257 |
| Sodium Acetate Anhydrous (CH3COONa) | Sigma | S2889 |
| Bovine Serum Albumin (BSA) | Lampire Biological Labs | 7500804 |
| Phosphate Buffer Saline (PBS) (10X) | Denville Scientific | CP4390-48 |
| Dylight 549 conjugated NeutrAvidin | Thermo | 22837 |
| Protease Inhibitor Cocktail Tablets | Roche | 4693159001 |
| ChromPure Human IgG, Fc fragment | Jackson Immunoresearch | 009-000-008 |
| ChromPure Human IgG, whole molecule | Jackson Immunoresearch | 009-000-003 |
| ChromPure Mouse IgG, whole molecule | Jackson Immunoresearch | 015-000-003 |
| ChromPure Mouse IgG, Fc fragment | Jackson Immunoresearch | 015-000-008 |
| ChromPure Rabbit IgG, whole molecule | Jackson Immunoresearch | 011-000-003 |
| ChromPure Donkey IgG, whole molecule | Jackson Immunoresearch | 017-000-003 |
| Microarray Scanner | Tecan | LS Reloaded |
Table 2. List of equipments and reagents used in this protocol.
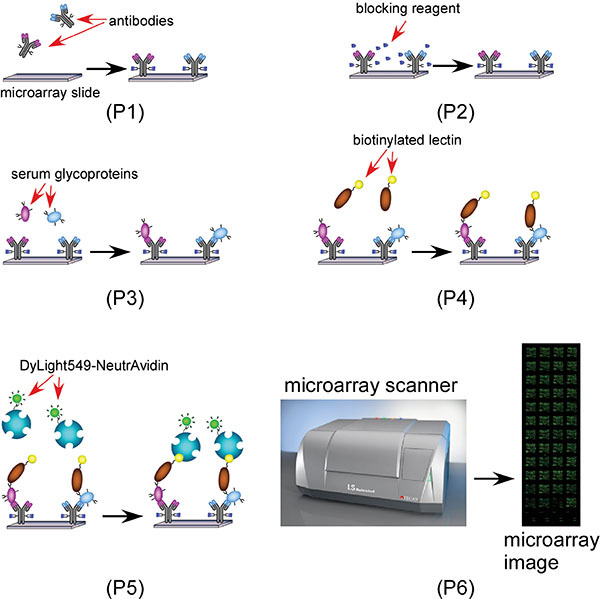 Scheme 1. A scheme showing the lectin antibody microarray based glycan biomarker discovery process. 1 (Step 2 to 4): Block the antibody microarray with the blocker (Glu-hydrazide) and BSA; 2 (Step 5): apply serum samples and capture specific glycoproteins with specific antibodies; 3 (Step 6): apply biotinylated lectin(s); 4 (Step 7): Probe the biotinylated AAL with Dylight 549 labeled NeutrAvidin for microarray imaging.
Scheme 1. A scheme showing the lectin antibody microarray based glycan biomarker discovery process. 1 (Step 2 to 4): Block the antibody microarray with the blocker (Glu-hydrazide) and BSA; 2 (Step 5): apply serum samples and capture specific glycoproteins with specific antibodies; 3 (Step 6): apply biotinylated lectin(s); 4 (Step 7): Probe the biotinylated AAL with Dylight 549 labeled NeutrAvidin for microarray imaging.
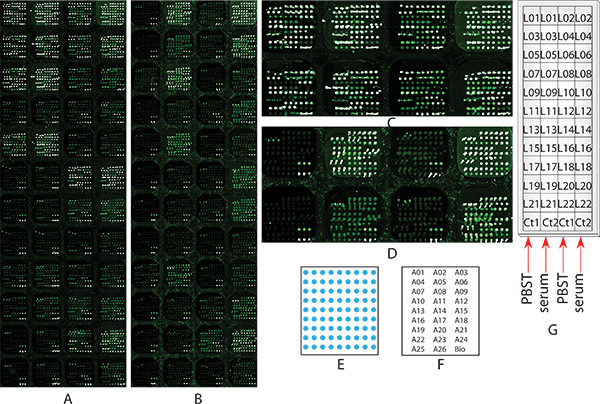 Figure 1. Microarray images of the Sample Experiment 1 glycosylation profiling of multiple serum glycoproteins in HCC patient serum sample by using chemically blocked antibody microarray with multiple lectin detection. Two identical microarray slides, (A) none chemically blocked, or (B) chemically blocked as described in Step 2, both went through all the steps from 2 to 9 for glycosylation profiling, as well as comparison purpose. (A) and (B) are the microarray images scanned at Step 8 in a resolution of 10 micron. (C) the zoom in image of the first two rows of the none chemically blocked microarray slide (A); (D) the zoom in image of the first two rows of the non chemically blocked microarray slide (B)); (E) the diagram of the antibody arrangement within each subarray; (F) array maps: the location of each antibody within the subarray, each antibody name represents 3 spots; (G) Serum sample and lectin location: a diagram shows which subarray each serum sample and lectin was applied onto.
Figure 1. Microarray images of the Sample Experiment 1 glycosylation profiling of multiple serum glycoproteins in HCC patient serum sample by using chemically blocked antibody microarray with multiple lectin detection. Two identical microarray slides, (A) none chemically blocked, or (B) chemically blocked as described in Step 2, both went through all the steps from 2 to 9 for glycosylation profiling, as well as comparison purpose. (A) and (B) are the microarray images scanned at Step 8 in a resolution of 10 micron. (C) the zoom in image of the first two rows of the none chemically blocked microarray slide (A); (D) the zoom in image of the first two rows of the non chemically blocked microarray slide (B)); (E) the diagram of the antibody arrangement within each subarray; (F) array maps: the location of each antibody within the subarray, each antibody name represents 3 spots; (G) Serum sample and lectin location: a diagram shows which subarray each serum sample and lectin was applied onto.
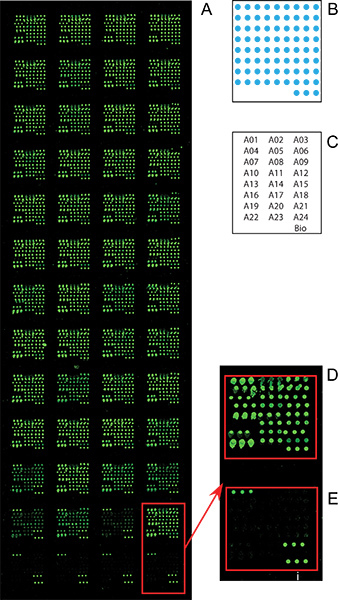 Figure 2. Microarray images of the sample experiment 2 screen for altered fucosylation on specific serum glycoproteins as biomarkers that discriminate liver cirrhosis and HCC patients. The microarray assay was carried out as described in the Sample Experiment 2 section. (A) The whole slide image of the microarray slide from Step 8; (B) the diagram of the antibody arrangement within each subarray; (C) array maps: the location of each antibody within the subarray, each antibody name represents 3 spots; (D) a zoom-in image of a subarray that were incubated with serum sample; (E) a zoom-in image of a subarray that were incubated with PBS control.
Figure 2. Microarray images of the sample experiment 2 screen for altered fucosylation on specific serum glycoproteins as biomarkers that discriminate liver cirrhosis and HCC patients. The microarray assay was carried out as described in the Sample Experiment 2 section. (A) The whole slide image of the microarray slide from Step 8; (B) the diagram of the antibody arrangement within each subarray; (C) array maps: the location of each antibody within the subarray, each antibody name represents 3 spots; (D) a zoom-in image of a subarray that were incubated with serum sample; (E) a zoom-in image of a subarray that were incubated with PBS control.
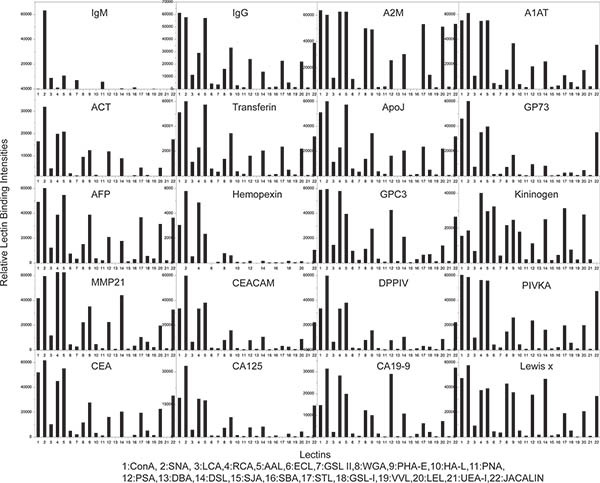 Figure 3. Glycan profiling results from sample experiment 1. Each bar graph represent lectin binding profile (or glycan profiles) of one of the 20 tested protein. Total 22 different lectins were used to analyze the glycan profile of each protein. Please click here to see a larger version of this figure.
Figure 3. Glycan profiling results from sample experiment 1. Each bar graph represent lectin binding profile (or glycan profiles) of one of the 20 tested protein. Total 22 different lectins were used to analyze the glycan profile of each protein. Please click here to see a larger version of this figure.
Discussion
1. Target protein and capture antibody selection
Prior to the antibody microarray assay, some reagents and materials are needed to be considered and prepared. To design an antibody microarray for glycan profiling or glycan biomarker screening, a panel of antibodies specific to glycoprotein candidates should be determined according to the literature or from previous results. These antibodies were usually purchased from different vendors such as R&D Systems etc. IgGs are preferred capture antibodies since our previous tests showed that some IgM and IgE may completely lose their antigen binding affinities after the chemical modification.
2. Design and manufacture of antibody microarray
Manufacture of the antibody microarray is an optional step which needs professional and expensive microarrayer, and well-trained personnel for operation. However, customized antibody microarray manufacture can be readily done from a service provider such as Serome Biosciences Inc. For a robust result, we recommend a non-contact microarrayer, such as the Scienion sciFLEXARRAYER ultra low volume non-contact microarrayer which was used in our antibody microarray manufacture. High binding capacity microarray slides, such as PATH (Gentel Bio Inc. WI) or Slide H (Shott, PA).
3. Select and Prepare Glycan Binding Proteins (GBPs) for glycosylation profiling
GBPs that target different mono- or oligosaccharides can be found in the literature and through a search engine developed by the Haab laboratory 29 and maintained at the Translational Genomics Institute (TGen): https://vai-apps.tgen.org/haab/default.do;jsessionid=83A3F59423A92A49D76D8B4426FAC015?method=search. To select GBPs with high specificity and affinity to the targeted glycan epitopes, select the motif (epitope) from the drop down menu, and click "search." The GBPs specific to this motif (epitope) will be listed according to their logP value from high to low order. Higher logP indicates a stronger binding affinity and specificity to the glycan motif/epitope. Due to the inherent non-specific binding issue from lectin, this method is still not as optimal as the antibody based assay. Thus, we strongly recommend that anti-glycan antibodies are used, such as anti-Lewis x or anti-sialyl Lewis a antibodies, if they are available. Use of multiple GBPs for detection is another strategy to cross check different binding profiles and to obtain reliable binding data.
4. Data analysis
Because this method is used to detect native serum proteins, protein-protein complexes may be captured and detected. Western blot or mass spectrometry is good methods to validate the microarray data. In the meantime, detection of protein levels using the same microarray is another method to learn the details of the glycosylation alteration of the protein, such as whether the changes were due to the total protein level change or just that the glycosylation level increased on each of the proteins.
Disclosures
The authors have nothing to disclose.
Acknowledgments
This work was supported by the Institute for Hepatitis and Virus Research.
References
- Fang M. The ER UDPase ENTPD5 promotes protein N-glycosylation, the Warburg effect, and proliferation in the PTEN pathway. Cell. 2010;143:711–724. doi: 10.1016/j.cell.2010.10.010. [DOI] [PubMed] [Google Scholar]
- Marino K, Bones J, Kattla JJ, Rudd PM. A systematic approach to protein glycosylation analysis: a path through the maze. Nat. Chem. Biol. 2010;6:713–723. doi: 10.1038/nchembio.437. [DOI] [PubMed] [Google Scholar]
- Shental-Bechor D, Levy Y. Effect of glycosylation on protein folding: a close look at thermodynamic stabilization. Proc. Natl. Acad. Sci. U. S. A. 2008;105:8256–8261. doi: 10.1073/pnas.0801340105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hossler P, Khattak SF, Li ZJ. Optimal and consistent protein glycosylation in mammalian cell culture. Glycobiology. 2009;19:936–949. doi: 10.1093/glycob/cwp079. [DOI] [PubMed] [Google Scholar]
- Nothaft H, Szymanski CM. Protein glycosylation in bacteria: sweeter than ever. Nat. Rev. Microbiol. 2011;8:765–778. doi: 10.1038/nrmicro2383. [DOI] [PubMed] [Google Scholar]
- Sola RJ, Griebenow K. Effects of glycosylation on the stability of protein pharmaceuticals. J. Pharm. Sci. 2009;98:1223–1245. doi: 10.1002/jps.21504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C, Lubman DM. Analysis of serum protein glycosylation with antibody-lectin microarray for high-throughput biomarker screening. Methods Mol. Biol. 2011;723:15–28. doi: 10.1007/978-1-61779-043-0_2. [DOI] [PubMed] [Google Scholar]
- Dwek MV, Jenks A, Leathem AJ. A sensitive assay to measure biomarker glycosylation demonstrates increased fucosylation of prostate specific antigen (PSA) in patients with prostate cancer compared with benign prostatic hyperplasia. Clin. Chim. Acta. 2010;411:1935–1939. doi: 10.1016/j.cca.2010.08.009. [DOI] [PubMed] [Google Scholar]
- Drake PM. Sweetening the pot: adding glycosylation to the biomarker discovery equation. Clin. Chem. 2010;56:223–236. doi: 10.1373/clinchem.2009.136333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y-P, Park S, Oh E, Oh Y-H, Kim H-S. On-chip detection of protein glycosylation based on energy transfer between nanoparticles. Biosensors & Bioelectronics. 2009;24:1189–1194. doi: 10.1016/j.bios.2008.07.012. [DOI] [PubMed] [Google Scholar]
- Boland M, Rudd PM. Disease related glycosylation changes and biomarker discovery: challenges and possibilities in an emerging field. Editorial. Dis. Markers. 2008;25:189–192. doi: 10.1155/2008/529423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norton PA. N-linked glycosylation of the liver cancer biomarker GP73. J. Cell Biochem. 2008;104:136–149. doi: 10.1002/jcb.21610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa T. Glycomic analysis of alpha-fetoprotein L3 in hepatoma cell lines and hepatocellular carcinoma patients. J. Proteome Res. 2008;7:2222–2233. doi: 10.1021/pr700841q. [DOI] [PubMed] [Google Scholar]
- Durazo FA. Des-gamma-carboxyprothrombin, alpha-fetoprotein and AFP-L3 in patients with chronic hepatitis, cirrhosis and hepatocellular carcinoma. J. Gastroenterol Hepatol. 2008;23:1541–1548. doi: 10.1111/j.1440-1746.2008.05395.x. [DOI] [PubMed] [Google Scholar]
- Kobayashi M. Fucosylated fraction of alpha-fetoprotein, L3, as a useful prognostic factor in patients with hepatocellular carcinoma with special reference to low concentrations of serum alpha-fetoprotein. Hepatol. Res. 2007;37:914–922. doi: 10.1111/j.1872-034X.2007.00147.x. [DOI] [PubMed] [Google Scholar]
- Maisey NR. CA19-9 as a prognostic factor in inoperable pancreatic cancer: the implication for clinical trials. Br. J. Cancer. 2005;93:740–743. doi: 10.1038/sj.bjc.6602760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talar-Wojnarowska R. Clinical value of serum neopterin, tissue polypeptide-specific antigen and CA19-9 levels in differential diagnosis between pancreatic cancer and chronic pancreatitis. Pancreatology. 2010;10:689–694. doi: 10.1159/000320693. [DOI] [PubMed] [Google Scholar]
- Chen S. Multiplexed analysis of glycan variation on native proteins captured by antibody microarrays. Nat. Methods. 2007;4:437–444. doi: 10.1038/nmeth1035. [DOI] [PubMed] [Google Scholar]
- Shao C. Antibody microarray analysis of serum glycans in esophageal spuamous cell carcinoma cases and controls. Proteomics Clinical Applications. 2009;3:923–931. doi: 10.1002/prca.200800245. [DOI] [PubMed] [Google Scholar]
- Chen S, Haab BB. Analysis of glycans on serum proteins using antibody microarrays. Methods Mol. Biol. 2009;520:39–58. doi: 10.1007/978-1-60327-811-9_4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yue T. The Prevalence and Nature of Glycan Alterations on Specific Proteins in Pancreatic Cancer Patients Revealed Using Antibody-Lectin Sandwich Arrays. Molecular & Cellular Proteomics. 2009;8:1697–1707. doi: 10.1074/mcp.M900135-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf-Yadlin A, Sevecka M, MacBeath G. Dissecting protein function and signaling using protein microarrays. Current Opinion in Chemical Biology. 2009;13:398–405. doi: 10.1016/j.cbpa.2009.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richard E. Proteomics as Applied to Inherited Metabolic Diseases. Current Proteomics. 2009;6:140–153. [Google Scholar]
- Nolen B, Winans M, Marrangoni A, Lokshin A. Aberrant tumor-associated antigen autoantibody profiles in healthy controls detected by multiplex bead-based immunoassay. Journal of Immunological Methods. 2009;344:116–120. doi: 10.1016/j.jim.2009.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuno A. Focused Differential Glycan Analysis with the Platform Antibody-assisted Lectin Profiling for Glycan-related Biomarker Verification. Molecular & Cellular Proteomics. 2009;8:99–108. doi: 10.1074/mcp.M800308-MCP200. [DOI] [PubMed] [Google Scholar]
- Hsu K-L, Mahal LK. Sweet tasting chips: microarray-based analysis of glycans. Current Opinion in Chemical Biology. 2009;13:427–432. doi: 10.1016/j.cbpa.2009.07.013. [DOI] [PubMed] [Google Scholar]
- Borrebaeck CAK, Wingren C. High-throughput proteomics using antibody microarrays: an update. Expert Review of Molecular Diagnostics. 2007;7:673–686. doi: 10.1586/14737159.7.5.673. [DOI] [PubMed] [Google Scholar]
- Sanchez-Carbayo M. Antibody array-based technologies for cancer protein profiling and functional proteomic analyses using serum and tissue specimens. Tumor Biology. 2010;31:103–112. doi: 10.1007/s13277-009-0014-z. [DOI] [PubMed] [Google Scholar]
- Porter A. A motif-based analysis of glycan array data to determine the specificities of glycan-binding proteins. Glycobiology. 2010;20:369–380. doi: 10.1093/glycob/cwp187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maupin KA. Glycogene Expression Alterations Associated with Pancreatic Cancer Epithelial-Mesenchymal Transition in Complementary Model Systems. Plos One. 2010;5 doi: 10.1371/journal.pone.0013002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sevecka M, Wolf-Yadlin A, MacBeath G. Lysate Microarrays Enable High-throughput, Quantitative Investigations of Cellular Signaling. Molecular & Cellular Proteomics. 2011;10 doi: 10.1074/mcp.M110.005363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M. Novel fucosylated biomarkers for the early detection of hepatocellular carcinoma. Cancer Epidemiol. Biomarkers Prev. 2009;18:1914–1921. doi: 10.1158/1055-9965.EPI-08-0980. [DOI] [PMC free article] [PubMed] [Google Scholar]


