Abstract
Accumulated evidence supports that the ubiquitin proteasome pathway (UPP) plays a crucial role in protein metabolism implicated in the regulation of many biological processes such as cell cycle control, DNA damage response, apoptosis, and so on. Therefore, alterations for the ubiquitin proteasome signaling or functional impairments for the ubiquitin proteasome components are involved in the etiology of many diseases, particularly in cancer development. In this minireview, we first give a brief outline for the ubiquitin proteasome pathway, we then discuss with focus for the ubiquitin proteasome pathway in the regulation of cell cycle control and DNA damage response, the relevance for the altered regulation of these signaling pathways in tumorigenesis is also reviewed. We finally assess and summarize the advancement for targeting the ubiquitin proteasome pathway in cancer therapy. A better understanding of the biological functions underlying ubiquitin regulatory mechanisms would provide us a wider prospective on cancer treatment.
Keywords: Ubiquitin proteasome pathway, ubiquitination, cell cycle, DNA damage response, tumorigenesis
Introduction
It is believed that protein biosynthesis and degradation keep a dynamic balance to properly sustain normal cell metabolism. The production of new functional proteins launches the proliferation and differentiation of cells. However, these processes are always accompanied with protein degradation when they accomplish their missions. Thus far, two distinctive proteolytic mechanisms, the lysosome degradation system and the ubiquitin proteasome pathway (UPP) [1], for the intracellular protein degradation have been identified. Lysosomes contain a large variety of hydrolytic enzymes that degrade proteins and other substances taken in by endocytosis [2]. Nevertheless, a majority of cellular proteins are degraded by the proteasome pathway after being tagged with ubiquitin [3]. Therefore, UPP is a common regulatory modification system implicated in the regulation of cell cycle, signal transduction, DNA damage response, apoptosis, and immune response [4].
In this minireview, we will first briefly introduce the ubiquitin proteasome pathway, followed by the discussion with focus for the role of ubiquitination played in cell cycle control and DNA damage response as well as their relevance in tumorigenesis. Finally, we intend to assess and summarize the feasibility of targeting the ubiquitin proteasome pathway as a cancer therapeutic strategy in clinical settings.
The Ubiquitin Proteasome Pathway (UPP)
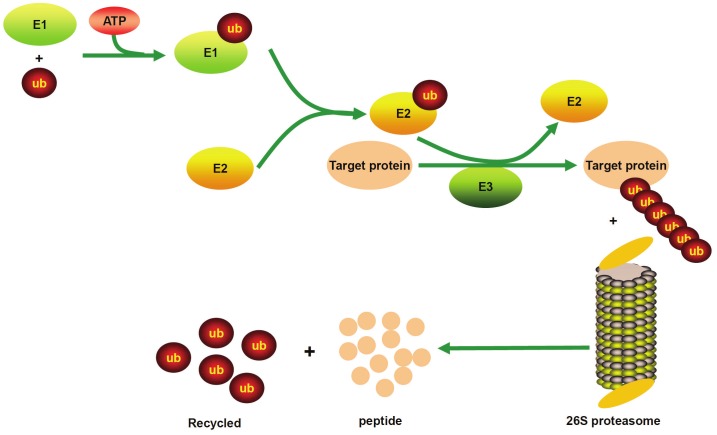
Ubiquitin is a small protein composed of 76 amino acids, and is highly conserved and ubiquitously expressed in the organisms of eukaryotic kingdom [5,6]. The ubiquitin proteasome pathway first processes the attachment of ubiquitin to a target protein which involves three critical enzymes, the ubiquitin activating enzyme or E1 enzyme, the ubiquitin conjugating enzyme or E2 enzyme, and the ubiquitin ligase or E3 ligase [7]. In most cases, ubiquitin tagged target proteins are subjected to degradation, in which the protein complexes are recognized by the 26S proteasome, a large multienzyme complex to mediate protein degradation. This whole process called ubiquitination which includes following three critical steps: firstly, ubiquitin needs to be activated from its precursor by adding to E1 through an ATP-dependent manner, and activated ubiquitin is then transferred to the ubiquitin-conjugating enzyme E2; secondly, E2 interacts with E3 to identify the substrate, and by which ubiquitin is attached to the target protein; and thirdly, the ubiquitin conjugated protein is recognized by the 26S proteasome, and through which, the target protein is degraded to small peptides or amino acids by the proteasome enzymes (Figure 1). Of note, ubiquitin can be released by the deubiquitinating enzymes (DUBs), and therefore, ubiquitin conjugation to target substrates is a reversible process [8].
Figure 1.
The ubiquitin proteasome pathway. Ubiquitin is activated by adding to E1, and E1 transfers ubiquitin to E2, E2 then interacts with E3, leading to the formation of a polyubiquitin chain. Finally, the targeted protein is degraded to small peptides by the 26S proteasome. E1: Ubiquitin activating enzyme; E2: Ubiquitin conjugating enzyme; E3: Ubiquitin ligases.
The Ubiquitin Proteasome Pathway (UPP) in cell cycle control
The cell cycle, or cell-division cycle, is a necessary process for cell to complete proliferation. A normal cell differs from a cancer cell is that normal cell only proliferates in response to stimulatory signals such as the developmental signals or mitogenic signals generated by tissue growth or repair. In contrast, cancer cell proliferation is actually initiated in the absence of those signals, indicating an altered cell cycle control in cancer cells. Control of cell cycle involves three key classes of regulatory molecules, cyclin, cyclin-dependent kinases (CDKs) and cyclin-dependent kinase inhibitors (CKIs) [9]. Ubiquitin-mediated proteolysis of these regulatory molecules is one of the key mechanisms underlying cell cycle control, in which two major ubiquitin ligase complexes are involved to accomplish irreversible cell cycle transitions, one is the Skp1-Cullin-F-box protein (SCF) complex, and the other is the anaphase-promoting complex/cyclosome (APC/C) [10,11].
SCF in cell cycle progression
The SCF complex contains three invariable subunits (SKP1, Cullin and RBX1) and one variable subunit (F-box protein) [12]. The F-box protein recognizes and binds to SKP1, while Cullin acts as a scaffold to link SKP1 with the RBX1 domain. SCF complex functions mainly at the G1 phase to S phase transition [13]. It has been well established that cyclin D, cyclin E and p27 are the key regulators during G1 phase [14]. Cyclin D interacts with CDK4/6, which then inactivates retinoblastoma (Rb) and induces expression of some genes essential for G1-S phase transition. On the other hand, cyclin E binds to CDK2 to promote the expression of cyclin A, which allows cell cycle progression from G1 phase to S phase. Importantly, SKP2, a specific F-box protein within the SCF complex, binds to SKP1 for substrate recognition, which then initiates the ubiquitination and degradation of G1/S cyclins (i.e., cyclin D and cyclin E) and CKIs (e.g., p27), and through which it regulates cell cycle progression [15]. Other than SKP2, FBW7 has also been found to act as an F-box protein for the SCF complex. It was found that the F-box within the FBW7 protein can recruit the SCF complex by directly interacting with SKP1 [16]. Furthermore, a stretch of eight WD40 repeats has also been found within FBW7 with capability for substrate binding [17]. Upon binding to its substrate, FBW7 regulates a network of proteins (e.g., Cyclin E and MYC) with central roles in cell division, cell growth and differentiation [10].
APC/C in mitotic exit
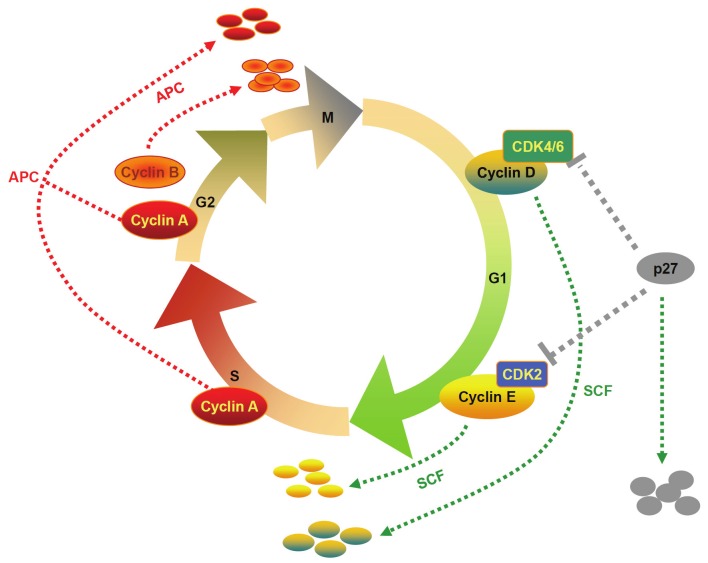
The anaphase-promoting complex, also called cyclosome (APC/C), is implicated in the control of mitotic progression through targeting key mitotic regulators for destruction by the 26S proteasome, which include the anaphase inhibitor securin and the mitotic cyclins A and B [11]. Unlike the SCF complex, APC/C is activated by phosphorylation and stays in active from mid-M phase to the end of G1 phase. APC/C displays different functions depending on its association with two distinctive ancillary proteins, cell division cycle 20 (CDC20) and CDH1 (also called HCT1) [18]. It is believed that CDC20 and CDH1 are primarily responsible for the recognition of substrates by APC/C. CDC20 binds to and activates APC/C during mitosis, which is suppressed until spindle attachment at kinetochores is completed in mitosis [19]. Upon activation, APC/C mediates the degradation of securin and mitotic cyclins during anaphase. In contrast, the activity for CDH1 is regulated by its phosphorylation status. Upon phosphorylation, CDH1 losses its capability to activate APC/C, as conformational changes resulted from phosphorylation prevent CDH1 from binding to the core APC/C subunits. Once activated by CDH1, APC/C initiates the proteolysis of mitotic cyclins throughout the G1 phase [20] (Figure 2).
Figure 2.
The ubiquitin ligase complexes SCF and APC/C in ubiquitin-mediated proteolysis of regulatory molecules essential for cell cycle control. Two major ubiquitin ligase complexes, SCF and APC/C, are responsible for the specific ubiquitination of cell cycle regulators through the proteasome targeted degradation. SCF ligase complexes control the G1 to S transition, while APC/C ligases mediate the onset and exit of mitosis.
The SCF complex and APC/C in tumorigenesis
Given the role of the SCF complex and APC/C played in the control of cell cycle, aberrations in the ubiquitin proteasome pathway would predispose to tumor development [21]. Indeed, accumulated evidence indicates that SKP2 is a proto-oncoprotein. Elevated SKP2 expression has been observed in a variety of human cancers such as in lymphomas [22], prostate cancer [23], melanoma [24], pancreatic cancer [25] and breast carcinomas [26]. In many types of human malignancies, the expressions for SKP2 tend to be higher along with the reduction of the tumor suppressor p27 levels [27]. There is also feasible evidence suggesting that SKP2 mediates the degradation of p27 in G1 phase [10]. Therefore, targeting of SKP2 protein is a booming realm for the treatment of cancer. Similarly, FBW7 recognizes oncoproteins such as cyclin E, MYC, JUN, Notch1 and Notch4 to promote their degradation. A number of cancer related mutations in FBW7 and its substrates have also been identified in a wide range of human cancer tissues [28]. Studies in animals revealed that loss of Fbw7 leads to embryonic lethality due to impaired vascular development, while postnatal studies indicated that targeted deletion of Fbw7 causes chromosomal instability and tumorigenesis [28]. As a result, FBW7 is considered to be a tumor suppressor.
During the mitotic stage of cell division, APC/C is activated by the two highly conserved WD40-repeat proteins, CDC20 and CDH1. CDC20 acts as a co-activator to recruit substrate targets such as securin and mitotic cyclins for destruction, and by which it promotes sister-chromatid separation. CDC20 also functions as a crucial mediator of the spindle checkpoint implicated in the prevention of aneuploidy and genomic instability. In line with these results, CDC20 is found to be overexpressed in some cancers [10], and dysregulation of CDC20-dependent proteolysis is likely to preclude precocious segregation of chromosomes, leading to abnormal chromosome number. Similarly, CDH1 acts as a co-activator to mediate the degradation of mitotic cyclins, non-CDK mitotic kinases and some regulators essential for the formation of pre-replicative complexes. As a result, mutations for CDH1 or its most substrate targets are found in human cancers [10]. Inactivation of CDH1 leads to the accumulation of SKP2 and CDKs associated with the uncontrolled proliferation and genomic instability, leading to tumor development. To date, altered APC/C activity has been found to be implicated in gastric carcinogenesis, colorectal cancer and many other kinds of tumors [29].
The Ubiquitin Proteasome Pathway (UPP) in DNA damage response
Given that double strand DNA breaks could result in dramatic effects on all DNA transactions, DNA damage response (DDR) is thus vital for the maintenance of genomic stability, and its deficits in mammals would lead to various disorders associated with tumor development [30]. Thus far, compelling evidence suggests that pathways relevant to DDR rely on a specialized signal in which ubiquitin-dependent degradation of certain proteins in a programmed manner is essential to ensure the appropriate DNA repair and, as a result, the ubiquitin proteasome pathway plays a pivotal role in the regulation of DNA repair [31] . In this section, we discuss with focus for the impact of p53 ubiquitination and BRCA1 ubiquitin E3 ligase activity on DNA repair response and their relevance in tumorigenesis.
MDM2 mediated p53 ubiquitination in DNA repair and tumorigenesis
Given the role of p53 played in preventing genome mutation, it has been considered as “the guardian of the genome” [32]. Although p53 is subject to a variety of post-translational modifications, ubiquitination of p53 has emerged as a fundamental regulatory mechanism [33]. Studies revealed that p53 can be modified by a number of E3 ubiquitin ligases such as Pirh2, COP1, ARF binding protein and E6AP, while the murine double minute 2 (MDM2) oncoprotein, however, is the most critical negative regulator for p53 activity and the most extensively studied p53 E3 ligase [34].
Under physiological condition, the cells only maintain low levels of p53, which is controlled by the rapid degradation of p53 via poly-ubiquitination, primarily mediated by the high basal levels of MDM2 [35]. MDM2 acts as the major E3 ubiquitin-protein ligase to interact with p53, and by which it represses p53 transcriptional activity by mediating its ubiquitination and proteasomal degradation [36]. In contrast, p53 undergoes a significant increase in protein stability upon exposing to the DNA damage inducing factors such as stressful insults [35]. It is believed that DNA damage stabilizes p53 in part via the DNA damage signaling pathway that implicates the sensor kinases such as the ataxia telangiectasia mutated (ATM) and ataxia telangiectasia and Rad3-related protein (ATR) kinase, and the effector kinases [37]. The signals generated by these kinases lead to the dissociation of the p53/MDM2 complex along with the activation of p53. Once activated, p53 induces the transcriptional regulation of a variety of genes to arrest cell cycle, a process necessary for DNA damage repair. Nevertheless, when DNA damage is beyond the extent of cellular repair capacity, p53 would then induce apoptosis to prevent the malignant transformation of cells.
In line with its critical role in DNA damage response, mutations in p53 are found in around 50% of human tumors, highlighting the importance of p53 activity in tumor suppression [38]. Particularly, MDM2-mediated p53 ubiquitination has been demonstrated as a classical tumorigenesis pathway [38]. Not surprisingly, overexpression of MDM2 results in the deactivation of p53, which occurs in many types of tumors [39]. Studies in animals further revealed that mouse squamous-cell carcinomas (SCCs) resistant to UV light are related to the p53 defective response caused by MDM2 overexpression along with p53 ubiquitination [40]. Therefore, prevention of MDM2-mediated p53 ubiquitination could be a promising strategy for cancer treatment in many clinical settings.
BRCA1 ubiquitin E3 ligase activity in DNA repair and tumorigenesis
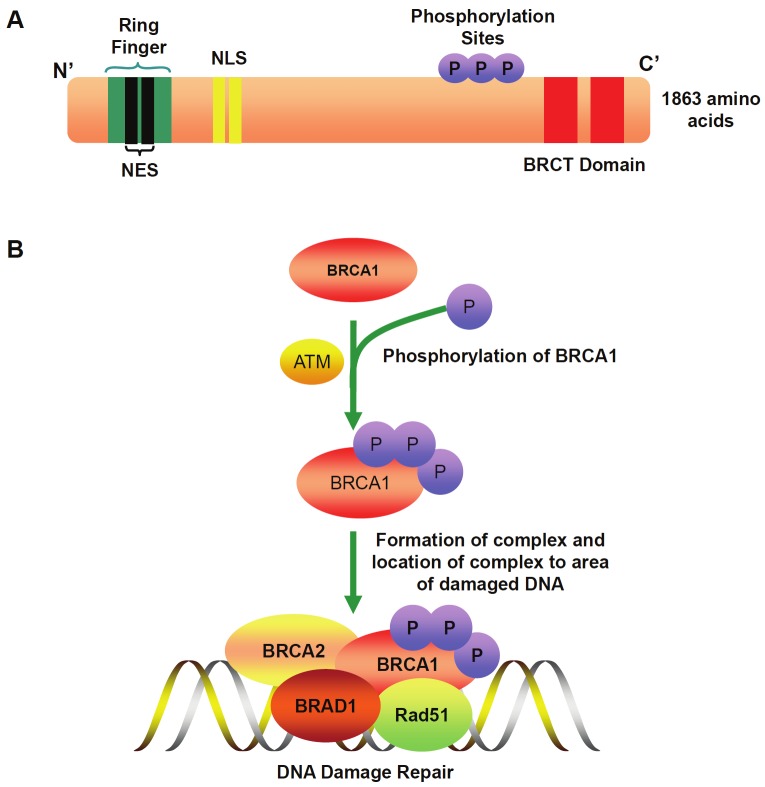
BRCA1 is considered to be a human caretaker gene expressed in the cells of breast and other tissues [41]. It is believed that BRCA1 protects the genome through facilitating the repair of damaged DNA and, in case that the extent of damaged DNA is beyond the cellular capacity to repair, it then shuttles to other cellular compartments to activate cell death pathways to eliminate those cells with persistent DNA lesions. BRCA1 contains several protein-protein interacting domains, including an N-terminal RING domain that possesses E3 ubiquitin ligase activity, and a C-terminal tandem BRCT domain that facilitates phosphoprotein binding (Figure 3A). In response to DNA damage, the original signaling cascade is initiated by the ATM-dependent phosphorylation of BRCA1 [42]. However, BRCA1 needs to form a heterodimer with the BRCA1 associated RING domain protein (BARD1) to display its E3 ubiquitin ligase function [43]. Upon phosphorylation, receptor-associated protein 80 (RAP80) is then acted as a BRCA1-interacting protein to mediate the binding of BRCA1 with ubiquitin [38]. BRCA1 is also required for double strand break recruitment of additional partner proteins, such as BRAC2 and Rad51 [44]. Upon the assembly of a complex with these partners, BRCA1 catalyzes the formation of polyubiquitin chains which is a prerequisite for the recruitment of BRCA1 to the DNA damage site [45] (Figure 3B). At the DNA damaged site, BRCA1 serves to promote high fidelity repair processes including both homologous recombination (HR) and classical nonhomologous DNA end-joining (C-NHEJ), but to suppress mutagenic and errorprone pathways [46].
Figure 3.
A. A diagram shows the sequence domains and motifs in BRCA1. BRCA1 contains an N-terminal RING domain that possesses E3 ubiquitin ligase activity, and a C-terminal tandem BRCT domain that facilitates phosphoprotein binding. BRCA1 possesses 3 phosphorylation sites that initiate the signaling cascade by the ATM-dependent phosphorylation. BRCA1 has two nuclear localization signals (NLS) which import BRCA1 into the nucleus, while two nuclear export sequences (NES) within the RING domain are responsible for the export of BRCA1 from the nucleus into the cytoplasm. B. Signaling cascade for BRCA1-mediated DNA damage repair. In response to DNA damage, ATM phosphorylates BRCA1. Upon phosphorylation BRCA1 forms a heterodimer with BRAD1, which then recruits additional partners such as BRCA2 and Rad5. Upon the assembly of a complex with these partners, BRCA1 is then relocated into the DNA damage site, where it serves to promote high fidelity repair processes.
As mentioned above, BRCA1 is a nuclear-cytoplasmic shuttling protein. It contains two nuclear localization signals (NLS), while two nuclear export sequences (NES) are also found at the N-terminus of the RING finger domain (Figure 3A). NLS imports BRCA1 into the nucleus, where it promotes high fidelity repair of damaged DNA. In contrast, NES exports BRCA1 from the nucleus into the cytoplasm through the chromosome region maintenance 1 (CRM1)/exportin pathway, where it mediates cell apoptosis [47]. It is believed that this subcellular redistribution of BRCA1 plays an important regulatory mechanism in the cellular response to DNA damage [48]. Based on that persistent irradiation dose-dependently induces BRAC-1 nuclear export [49], it is plausible to assume that cytoplasmic BRCA1 shuttling serves as a preventive mechanism for tumorigenesis by eliminating those cells with unrepairable DNA damage.
Given the role BRCA1 played in the regulation of DNA repair and apoptosis, its importance in cancer development has been highly appreciated. Therefore, BRCA1 and BRCA2 are considered and hereditary breast and ovarian cancer associated genes. Indeed, genetic studies have consistently demonstrated that BRCA1 is implicated in familial breast cancer as manifested by the observation of an increased risk in those subjects carrying certain polymorphisms of this gene [50]. Once BRCA1 itself is mutated or functionally impaired, damaged DNA cannot be repaired properly, which then predisposes to the development of breast cancer [51]. To date, hundreds of variations in the BRCA1 gene have been identified, and studies revealed that abnormalities in the BRCA1 and BRCA2 genes may account for up to 10% of all breast cancers. Although BRCA1 participates in multiple cellular supercomplexes to execute its tasks, but in most of the complexes, however, BRCA1 exists as the RING heterodimer with BRAD1 to provide ubiquitin E3 ligase activity that is required for its tumor suppressor function. Therefore, BRCA1-dependent ubiquitin pathway is closely related to not only familial breast cancers, but also sporadic breast cancers [45]. Nevertheless, a recent study in mice challenged this concept, in which an enzymatically defective Brca1 was introduced into the mouse genome. Unexpectedly, the mutant Brca1 prevented tumor formation to the same degree as that of the wild-type Brca1 [52], suggesting that the E3 ligase activity of BRCA1 is dispensable for tumor suppression. Therefore, additional work is necessary to fully address the impact of BRCA1 E3 ubiquitin ligase activity on tumor development.
Targeting the Ubiquitin-Proteasome Pathway (UUP) in cancer therapy
Along with the significant progress in understanding of the molecular basis for the ubiquitin proteasome pathway in cancer-relevant processes, a substantial of effort has recently been devoted to explore the feasibility by targeting the ubiquitin system for anticancer therapy [53]. In this section, we intend to assess and summarize the studies conducted for the development of therapeutic approaches by targeting several key components of the ubiquitin proteasome pathway.
Inhibition of proteasome in cancer therapy
Proteasome is a multicatalytic enzyme complex (2.5MDa) containing a 20S catalytic core and two 19S regulatory complexes [54]. Given that many proteins targeted by proteasome are involved in the regulation of important processes of carcinogenesis and cancer cell survival, such as cell cycle progression, cell proliferation, differentiation and apoptosis, inhibition of proteasome would lead to cell death or apoptosis [55]. Therefore, a great deal of effort in the past has been devoted for searching proteasome inhibitors for the treatment of cancer [55,56]. Bortezomib was the first inhibitor for the 26S proteasome employed for this purpose [57]. Studies in cells have shown that bortezomib represses NF-κB signaling by blocking IκB degradation [58]. Reduced NF-κB activity is associated with the suppression of pro-inflammatory response genes along with the upregulation of cyclin-dependent kinase inhibitors p21Cip1 and p27Kip1, which then lead to tumor cells undergoing apoptosis [59]. In line with these results, a phase III trial revealed that inclusion of bortezomib to melphalan–prednisone is associated with significant improvement in outcomes in patients with newly diagnosed myeloma who were ineligible for high-dose therapy [60]. These preclinical and clinical studies rendered the approval of bortezomib by FDA for the clinical use, and now bortezomib is considered to be a front-line treatment for newly diagnosed multiple myeloma patients and for patients with relapsed/refractory multiple myeloma and mantle cell lymphoma [53]. Recent studies further demonstrated that other than suppression of NF-κB activity, bortezomib also upregulates NOXA, a proapoptotic protein that promotes tumor cell apoptosis by interacting with the anti-apoptotic proteins of Bcl-2 subfamily Bcl-X(L) and Bcl-2 [61]. Obviously, the clinical success of bortezomib as an anticancer therapy has prompted the development of a new generation of proteasome inhibitors, such as PR-171 (carfilzomib) and NPI-0052 (salinosporamide A) [53]. More recently, two new proteasome inhibitors, CEP-18770 and MLN9708, are underway for clinical trails. More excitingly, significant progress has been made for the development of natural products with potent proteasome-inhibitory activity [55]. Once this type of inhibitors is ready for clinical use, the toxicity to normal cells during the course of anticancer therapy would be significantly minimized.
Inhibitors for MDM2 and E1 enzyme in cancer therapy
Based on the fact that E3 ubiquitin ligases not only catalyze the ubiquitination of a variety of protein substrates, but also determine the specificity of protein substrates, they therefore represent a class of “drugable” targets for pharmaceutical intervention [62]. In line with this assumption, it is believed that a specific E3 ligase inhibitor would manifest higher level selectivity but lower toxicity as compared with that of a particular general proteasome inhibitor [63]. Given that E3 ubiquitin ligases are a large family of proteins, we would limit our focus in this section only to the inhibitors that target MDM2 to stabilize p53 and to reactivate p53 signaling [64].
As aforementioned, p53 has the capability to induce cell cycle arrest and apoptosis. Therefore, approaches aimed to retain the functionality of p53 would be a viable strategy for cancer therapy. A large body of evidence has established that MDM2 acts as a major negative regulator to attenuate p53 activity either by ubiquitination through the proteasome pathway or by masking of its transactivation domain through directly binding to p53, and therefore, librating p53 from MDM2 by antagonizing MDM2 activity would offer a novel approach to stabilize and reactivate p53 pathway [63]. Initially, due to the lack of MDM2 E3 ligase specific inhibitors, the most common approaches employed for this purpose were either to block MDM2 expression, or to disrupt the MDM2-p53 interactions rather than directly targeting its E3 ubiquitin ligase activity [64]. Indeed, studies with MDM2 antisense oligonucleotides have consistently demonstrated that inhibition of MDM2 expression has resulted in p53 stabilization and activation of the p53 pathway in cancer cells and tumor xenografts in nude mice [65-67]. Based on the observation that three residues (i.e., Phe19, Trp23 and Leu26) within the p53 are inserted into a deep hydrophobic pocket on the surface of the MDM2 that are essential for the binding between p53 and MDM2 [68], a substantial of efforts have been invested to screen small molecules that might successfully mimic this interaction. The first group of potent and selective small-molecule MDM2 antagonists were identified from a class of cis-imidazoline compounds and named as nutlins. They bind to the p53 pocket of MDM2 in a manner remarkably similar to the molecular interactions of the crucial amino acid residues from p53 and, as such, nutkines stabilize p53 and activate the p53 pathway, and by which effectively block proliferating cancer cells in G1 and G2 phases [69]. As the research evolves, additional small-molecule MDM2 antagonists have been developed, and the studies for their clinical relevance are underway [70-72].
Along with the development of high throughput technologies, small-molecule inhibitors that specifically target the MDM2 E3 ligase activity have also been developed [64]. For example, a class of compounds named HLI98 was found with potential to inhibit p53 ubiquitination, which then activate p53 signaling and induce cancer cells undergoing apoptosis in a p53-dependent manner [64]. Nevertheless, current assessment revealed that the potency and selectivity of this class of compounds are relatively low, additional modifications for these inhibitors aimed at to increase their potency, E3 ligase specificity and to eliminate p53-independent off-target activities would be essential before their application in clinical settings.
Ubiquitin-activating enzyme E1, which is sitting on the top of the ubiquitination cascade, has also been an attractive target for cancer therapy. Indeed, genetic and chemical inhibition of the E1 enzyme preferentially induced tumor cell death over normal cells along with delayed tumor growth in a mouse model of leukemia [73]. It was noticed that E1 inhibition caused cell death by eliciting endoplasmic reticulum (ER) stress and an unfolded protein response. By screening a commercial library purchased from ChemBridge, Inc., Yang and colleagues characterized the first cell permeable inhibitor of the ubiquitin E1, 4[4-(5-nitro-furan-2-ylmethylene)-3,5-dioxo-pyrazolidin-1-yl]-benzoic acid ethyl ester, which was then named PYR-41 [74]. A subsequent study conducted by Ungermannova and colleagues further characterized another E1 inhibitor, named NSC624206, which prevents p27 ubiquitination by specifically blocking ubiquitin-thioester formation [75]. Together, these data suggest that inhibitors specific for the ubiquitin E1 enzyme could also be an effective alterative approach for the treatment of hematologic malignancies.
Deubiquitination in cancer therapy
Deubiquitination is an important ubiquitinationrelated metabolic pathway that reverses the ubiquitination of target proteins. There is growing research finding suggesting that the aberration of DUBs could result in neoplastic diseases and it also happens frequently [76]. Thus far, around 79 functional DUBs have been characterized in the human proteome, indicating that selective intervention is a reasonable therapeutic objective by either suppression or ablation of oncogene products, or by upregulation of tumor suppressors [77]. For example, deubiquitinating enzyme USP2a is a negative regulator in the MDM2-p53 pathway, and suppression of its activity results in the destabilization of MDM2 and reactivation of wild-type p53 in tumors [78]. Although DUB inhibitors or activators are currently not yet ready for clinical trial, the discovery that CYLD encoded by the cylindromatosis gene is a DUB that removes lysine 63-linked ubiquitin chains from TRAF2, and suppression of its activity blocks NF-κB activation paved the way toward this direction [79]. Indeed, the first proof-of-concept study in humans was actually conducted by Oosterkamp and colleagues based on these results [80]. Therefore, modulation of deubiquitinase activities by either inhibitors or activators could represent a viable therapeutic strategy for cancer treatment [81-83].
Summary and perspectives
Studies in the past several decades conducted both in culture cells and animal models provided convincing evidence supporting that the ubiquitin proteasome pathway regulates diverse signals essential for life processes. As such, its alterations or deficits are predisposed to tumorigenesis. These achievements prompted the development of strategies to target the ubiquitin proteasome pathway for cancer prevention, intervention, and treatment. Given ubiquitination involves a large body of regulatory proteins, intensive research is required for these molecules in which a better understanding of their biological functions would provide us a wider prospective on the treatment of cancer. As the research evolves, it has become clear now that some molecules such as those described in this review, are promising “drugable” targets for cancer therapy. For example, the proteasome inhibitor bortezomib has been successfully employed for the treatment of multiple myeloma and other hemotological malignancies. While these advancements are exciting, unexpected toxicity in bortezomib trials and lack of sustained clinical response toward solid tumors have also been noted. The challenge for future studies would be the development of inhibitors with higher therapeutic potency but minimized toxicity to normal cells, such as those small molecules characterized from natural products.
Acknowledgements
This work was supported by the National Natural Science Foundation of China (81130014), the Chinese Ministry of Science & Technology (2012BAI39B05), and the European Foundation for the Study of Diabetes (EFSD)/ Chinese Diabetes Society (CDS)/Lilly Program for Collaborative Diabetes Research between China and Europe, and the Synergy Award from the Diabetes, Obesity Discovery Institute (DODI) at the Georgia Health Sciences University.
Conflict of interest disclosure
The authors declare no competing financial interests.
References
- 1.Shah SA, Potter MW, Callery MP. Ubiquitin proteasome pathway: implications and advances in cancer therapy. Surg Oncol. 2001;10:43–52. doi: 10.1016/s0960-7404(01)00018-4. [DOI] [PubMed] [Google Scholar]
- 2.Ciechanover A. Intracellular protein degradation: from a vague idea thru the lysosome and the ubiquitin-proteasome system and onto human diseases and drug targeting. Biochim Biophys Acta. 2012;1824:3–13. doi: 10.1016/j.bbapap.2011.03.007. [DOI] [PubMed] [Google Scholar]
- 3.Burger AM, Seth AK. The ubiquitin-mediated protein degradation pathway in cancer: therapeutic implications. Eur J Cancer. 2004;40:2217–2229. doi: 10.1016/j.ejca.2004.07.006. [DOI] [PubMed] [Google Scholar]
- 4.Clague MJ, Urbe S. Ubiquitin: same molecule, different degradation pathways. Cell. 2010;143:682–685. doi: 10.1016/j.cell.2010.11.012. [DOI] [PubMed] [Google Scholar]
- 5.Yang WL, Zhang X, Lin HK. Emerging role of Lys-63 ubiquitination in protein kinase and phosphatase activation and cancer development. Oncogene. 2010;29:4493–4503. doi: 10.1038/onc.2010.190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cook WJ, Jeffrey LC, Xu Y, Chau V. Tertiary structures of class I ubiquitin-conjugating enzymes are highly conserved: crystal structure of yeast Ubc4. Biochemistry. 1993;32:13809–13817. doi: 10.1021/bi00213a009. [DOI] [PubMed] [Google Scholar]
- 7.Pickart CM, Eddins MJ. Ubiquitin: structures, functions, mechanisms. Biochim Biophys Acta. 2004;1695:55–72. doi: 10.1016/j.bbamcr.2004.09.019. [DOI] [PubMed] [Google Scholar]
- 8.Kim JH, Park KC, Chung SS, Bang O, Chung CH. Deubiquitinating enzymes as cellular regulators. J Biochem. 2003;134:9–18. doi: 10.1093/jb/mvg107. [DOI] [PubMed] [Google Scholar]
- 9.Park MT, Lee SJ. Cell cycle and cancer. J Biochem Mol Biol. 2003;36:60–65. doi: 10.5483/bmbrep.2003.36.1.060. [DOI] [PubMed] [Google Scholar]
- 10.Nakayama KI, Nakayama K. Ubiquitin ligases: cell-cycle control and cancer. Nat Rev Cancer. 2006;6:369–381. doi: 10.1038/nrc1881. [DOI] [PubMed] [Google Scholar]
- 11.Mocciaro A, Rape M. Emerging regulatory mechanisms in ubiquitin-dependent cell cycle control. J Cell Sci. 2012;125:255–263. doi: 10.1242/jcs.091199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Skaar JR, Pagano M. Control of cell growth by the SCF and APC/C ubiquitin ligases. Curr Opin Cell Biol. 2009;21:816–824. doi: 10.1016/j.ceb.2009.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kurland JF, Tansey WP. Crashing waves of destruction: the cell cycle and APC(Cdh1) regulation of SCF(Skp2) Cancer Cell. 2004;5:305–306. doi: 10.1016/s1535-6108(04)00091-1. [DOI] [PubMed] [Google Scholar]
- 14.Csikasz-Nagy A, Palmisano A, Zamborszky J. Molecular network dynamics of cell cycle control: transitions to start and finish. Methods Mol Biol. 2011;761:277–291. doi: 10.1007/978-1-61779-182-6_19. [DOI] [PubMed] [Google Scholar]
- 15.Egozi D, Shapira M, Paor G, Ben-Izhak O, Skorecki K, Hershko DD. Regulation of the cell cycle inhibitor p27 and its ubiquitin ligase Skp2 in differentiation of human embryonic stem cells. FASEB J. 2007;21:2807–2817. doi: 10.1096/fj.06-7758com. [DOI] [PubMed] [Google Scholar]
- 16.Bai C, Sen P, Hofmann K, Ma L, Goebl M, Harper JW, Elledge SJ. SKP1 connects cell cycle regulators to the ubiquitin proteolysis machinery through a novel motif, the F-box. Cell. 1996;86:263–274. doi: 10.1016/s0092-8674(00)80098-7. [DOI] [PubMed] [Google Scholar]
- 17.Hao B, Oehlmann S, Sowa ME, Harper JW, Pavletich NP. Structure of a Fbw7-Skp1-cyclin E complex: multisite-phosphorylated substrate recognition by SCF ubiquitin ligases. Mol Cell. 2007;26:131–143. doi: 10.1016/j.molcel.2007.02.022. [DOI] [PubMed] [Google Scholar]
- 18.Meyer HJ, Rape M. Processive ubiquitin chain formation by the anaphase-promoting complex. Semin Cell Dev Biol. 2011;22:544–550. doi: 10.1016/j.semcdb.2011.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Foe IT, Foster SA, Cheung SK, DeLuca SZ, Morgan DO, Toczyski DP. Ubiquitination of Cdc20 by the APC occurs through an intramolecular mechanism. Curr Biol. 2011;21:1870–1877. doi: 10.1016/j.cub.2011.09.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Toda K, Naito K, Mase S, Ueno M, Uritani M, Yamamoto A, Ushimaru T. APC/C-Cdh1-dependent anaphase and telophase progression during mitotic slippage. Cell Div. 2012;7:4. doi: 10.1186/1747-1028-7-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yamasaki L, Pagano M. Cell cycle, proteolysis and cancer. Curr Opin Cell Biol. 2004;16:623–628. doi: 10.1016/j.ceb.2004.08.005. [DOI] [PubMed] [Google Scholar]
- 22.Abdou AG, Asaad NY, Abd El-Wahed MM, Samaka RM, Allah MS. The prognostic value of Skp2 expression in Egyptian diffuse large B-cell lymphoma. Appl Immunohistochem Mol Morphol. 2012;20:47–55. doi: 10.1097/PAI.0b013e318219a19f. [DOI] [PubMed] [Google Scholar]
- 23.Wang Z, Gao D, Fukushima H, Inuzuka H, Liu P, Wan L, Sarkar FH, Wei W. Skp2: a novel potential therapeutic target for prostate cancer. Biochim Biophys Acta. 2012;1825:11–17. doi: 10.1016/j.bbcan.2011.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen G, Cheng Y, Zhang Z, Martinka M, Li G. Cytoplasmic Skp2 expression is increased in human melanoma and correlated with patient survival. PLoS One. 2011;6:e17578. doi: 10.1371/journal.pone.0017578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Einama T, Kagata Y, Tsuda H, Morita D, Ogata S, Ueda S, Takigawa T, Kawarabayashi N, Fukatsu K, Sugiura Y, Matsubara O, Hatsuse K. High-level Skp2 expression in pancreatic ductal adenocarcinoma: correlation with the extent of lymph node metastasis, higher histological grade, and poorer patient outcome. Pancreas. 2006;32:376–381. doi: 10.1097/01.mpa.0000220862.78248.c4. [DOI] [PubMed] [Google Scholar]
- 26.Traub F, Mengel M, Luck HJ, Kreipe HH, von Wasielewski R. Prognostic impact of Skp2 and p27 in human breast cancer. Breast Cancer Res Treat. 2006;99:185–191. doi: 10.1007/s10549-006-9202-3. [DOI] [PubMed] [Google Scholar]
- 27.Shapira M, Ben-Izhak O, Slotky M, Goldin O, Lahav-Baratz S, Hershko DD. Expression of the ubiquitin ligase subunit cyclin kinase subunit 1 and its relationship to S-phase kinase protein 2 and p27Kip1 in prostate cancer. J Urol. 2006;176:2285–2289. doi: 10.1016/j.juro.2006.07.051. [DOI] [PubMed] [Google Scholar]
- 28.Welcker M, Clurman BE. FBW7 ubiquitin ligase: a tumour suppressor at the crossroads of cell division, growth and differentiation. Nat Rev Cancer. 2008;8:83–93. doi: 10.1038/nrc2290. [DOI] [PubMed] [Google Scholar]
- 29.Smolders L, Teodoro JG. Targeting the anaphase promoting complex: common pathways for viral infection and cancer therapy. Expert Opin Ther Targets. 2011;15:767–780. doi: 10.1517/14728222.2011.558008. [DOI] [PubMed] [Google Scholar]
- 30.Hoeijmakers JH. DNA damage, aging, and cancer. N Engl J Med. 2009;361:1475–1485. doi: 10.1056/NEJMra0804615. [DOI] [PubMed] [Google Scholar]
- 31.Feng L, Chen J. The E3 ligase RNF8 regulates KU80 removal and NHEJ repair. Nat Struct Mol Biol. 2012;19:201–206. doi: 10.1038/nsmb.2211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lane DP. Cancer. p53, guardian of the genome. Nature. 1992;358:15–16. doi: 10.1038/358015a0. [DOI] [PubMed] [Google Scholar]
- 33.Brooks CL, Gu W. Ubiquitination, phosphorylation and acetylation: the molecular basis for p53 regulation. Curr Opin Cell Biol. 2003;15:164–171. doi: 10.1016/s0955-0674(03)00003-6. [DOI] [PubMed] [Google Scholar]
- 34.Coutts AS, Adams CJ, La Thangue NB. p53 ubiquitination by Mdm2: a never ending tail? DNA Repair (Amst) 2009;8:483–490. doi: 10.1016/j.dnarep.2009.01.008. [DOI] [PubMed] [Google Scholar]
- 35.Kubbutat MH, Jones SN, Vousden KH. Regulation of p53 stability by Mdm2. Nature. 1997;387:299–303. doi: 10.1038/387299a0. [DOI] [PubMed] [Google Scholar]
- 36.Yang Y, Li CC, Weissman AM. Regulating the p53 system through ubiquitination. Oncogene. 2004;23:2096–2106. doi: 10.1038/sj.onc.1207411. [DOI] [PubMed] [Google Scholar]
- 37.Lakin ND, Jackson SP. Regulation of p53 in response to DNA damage. Oncogene. 1999;18:7644–7655. doi: 10.1038/sj.onc.1203015. [DOI] [PubMed] [Google Scholar]
- 38.Soussi T, Wiman KG. Shaping genetic alterations in human cancer: the p53 mutation paradigm. Cancer Cell. 2007;12:303–312. doi: 10.1016/j.ccr.2007.10.001. [DOI] [PubMed] [Google Scholar]
- 39.Chene P. Inhibiting the p53-MDM2 interaction: an important target for cancer therapy. Nat Rev Cancer. 2003;3:102–109. doi: 10.1038/nrc991. [DOI] [PubMed] [Google Scholar]
- 40.Knights CD, Liu Y, Appella E, Kulesz-Martin M. Defective p53 post-translational modification required for wild type p53 inactivation in malignant epithelial cells with mdm2 gene amplification. J Biol Chem. 2003;278:52890–52900. doi: 10.1074/jbc.M300279200. [DOI] [PubMed] [Google Scholar]
- 41.Deng CX, Scott F. Role of the tumor suppressor gene Brca1 in genetic stability and mammary gland tumor formation. Oncogene. 2000;19:1059–1064. doi: 10.1038/sj.onc.1203269. [DOI] [PubMed] [Google Scholar]
- 42.Cortez D, Wang Y, Qin J, Elledge SJ. Requirement of ATM-dependent phosphorylation of brca1 in the DNA damage response to double-strand breaks. Science. 1999;286:1162–1166. doi: 10.1126/science.286.5442.1162. [DOI] [PubMed] [Google Scholar]
- 43.Mallery DL, Vandenberg CJ, Hiom K. Activation of the E3 ligase function of the BRCA1/BARD1 complex by polyubiquitin chains. EMBO J. 2002;21:6755–6762. doi: 10.1093/emboj/cdf691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Greenberg RA, Sobhian B, Pathania S, Cantor SB, Nakatani Y, Livingston DM. Multifactorial contributions to an acute DNA damage response by BRCA1/BARD1-containing complexes. Genes Dev. 2006;20:34–46. doi: 10.1101/gad.1381306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wu W, Koike A, Takeshita T, Ohta T. The ubiquitin E3 ligase activity of BRCA1 and its biological functions. Cell Div. 2008;3:1. doi: 10.1186/1747-1028-3-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhang J, Powell SN. The role of the BRCA1 tumor suppressor in DNA double-strand break repair. Mol Cancer Res. 2005;3:531–539. doi: 10.1158/1541-7786.MCR-05-0192. [DOI] [PubMed] [Google Scholar]
- 47.Yang ES, Xia F. BRCA1 16 years later: DNA damage-induced BRCA1 shuttling. FEBS J. 2010;277:3079–3085. doi: 10.1111/j.1742-4658.2010.07734.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jiang J, Yang ES, Jiang G, Nowsheen S, Wang H, Wang T, Wang Y, Billheimer D, Chakravarthy AB, Brown M, Haffty B, Xia F. p53-dependent BRCA1 nuclear export controls cellular susceptibility to DNA damage. Cancer Res. 2011;71:5546–5557. doi: 10.1158/0008-5472.CAN-10-3423. [DOI] [PubMed] [Google Scholar]
- 49.Feng Z, Kachnic L, Zhang J, Powell SN, Xia F. DNA damage induces p53-dependent BRCA1 nuclear export. J Biol Chem. 2004;279:28574–28584. doi: 10.1074/jbc.M404137200. [DOI] [PubMed] [Google Scholar]
- 50.Ahmed M, Lalloo F, Evans DG. Update on genetic predisposition to breast cancer. Expert Rev Anticancer Ther. 2009;9:1103–1113. doi: 10.1586/era.09.38. [DOI] [PubMed] [Google Scholar]
- 51.Friedenson B. The BRCA1/2 pathway prevents hematologic cancers in addition to breast and ovarian cancers. BMC Cancer. 2007;7:152. doi: 10.1186/1471-2407-7-152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shakya R, Reid LJ, Reczek CR, Cole F, Egli D, Lin CS, deRooij DG, Hirsch S, Ravi K, Hicks JB, Szabolcs M, Jasin M, Baer R, Ludwig T. BRCA1 tumor suppression depends on BRCT phosphoprotein binding, but not its E3 ligase activity. Science. 2011;334:525–528. doi: 10.1126/science.1209909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hoeller D, Dikic I. Targeting the ubiquitin system in cancer therapy. Nature. 2009;458:438–444. doi: 10.1038/nature07960. [DOI] [PubMed] [Google Scholar]
- 54.Voges D, Zwickl P, Baumeister W. The 26S proteasome: a molecular machine designed for controlled proteolysis. Annu Rev Biochem. 1999;68:1015–1068. doi: 10.1146/annurev.biochem.68.1.1015. [DOI] [PubMed] [Google Scholar]
- 55.Frezza M, Schmitt S, Dou QP. Targeting the ubiquitin-proteasome pathway: an emerging concept in cancer therapy. Curr Top Med Chem. 2011;11:2888–2905. doi: 10.2174/156802611798281311. [DOI] [PubMed] [Google Scholar]
- 56.Lecker SH, Goldberg AL, Mitch WE. Protein degradation by the ubiquitin-proteasome pathway in normal and disease states. J Am Soc Nephrol. 2006;17:1807–1819. doi: 10.1681/ASN.2006010083. [DOI] [PubMed] [Google Scholar]
- 57.Adams J. Development of the proteasome inhibitor PS-341. Oncologist. 2002;7:9–16. doi: 10.1634/theoncologist.7-1-9. [DOI] [PubMed] [Google Scholar]
- 58.Adams J. The development of proteasome inhibitors as anticancer drugs. Cancer Cell. 2004;5:417–421. doi: 10.1016/s1535-6108(04)00120-5. [DOI] [PubMed] [Google Scholar]
- 59.Ling YH, Liebes L, Jiang JD, Holland JF, Elliott PJ, Adams J, Muggia FM, Perez-Soler R. Mechanisms of proteasome inhibitor PS-341-induced G(2)-M-phase arrest and apoptosis in human non-small cell lung cancer cell lines. Clin Cancer Res. 2003;9:1145–1154. [PubMed] [Google Scholar]
- 60.San Miguel JF, Schlag R, Khuageva NK, Dimopoulos MA, Shpilberg O, Kropff M, Spicka I, Petrucci MT, Palumbo A, Samoilova OS, Dmoszynska A, Abdulkadyrov KM, Schots R, Jiang B, Mateos MV, Anderson KC, Esseltine DL, Liu K, Cakana A, van de Velde H, Richardson PG. Bortezomib plus melphalan and prednisone for initial treatment of multiple myeloma. N Engl J Med. 2008;359:906–917. doi: 10.1056/NEJMoa0801479. [DOI] [PubMed] [Google Scholar]
- 61.Chen D, Frezza M, Schmitt S, Kanwar J, Dou QP. Bortezomib as the first proteasome inhibitor anticancer drug: current status and future perspectives. Curr Cancer Drug Targets. 2011;11:239–253. doi: 10.2174/156800911794519752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Pray TR, Parlati F, Huang J, Wong BR, Payan DG, Bennett MK, Issakani SD, Molineaux S, Demo SD. Cell cycle regulatory E3 ubiquitin ligases as anticancer targets. Drug Resist Updat. 2002;5:249–258. doi: 10.1016/s1368-7646(02)00121-8. [DOI] [PubMed] [Google Scholar]
- 63.Di J, Zhang Y, Zheng J. Reactivation of p53 by inhibiting Mdm2 E3 ligase: a novel antitumor approach. Curr Cancer Drug Targets. 2011;11:987–994. doi: 10.2174/156800911797264789. [DOI] [PubMed] [Google Scholar]
- 64.Yang Y, Ludwig RL, Jensen JP, Pierre SA, Medaglia MV, Davydov IV, Safiran YJ, Oberoi P, Kenten JH, Phillips AC, Weissman AM, Vousden KH. Small molecule inhibitors of HDM2 ubiquitin ligase activity stabilize and activate p53 in cells. Cancer Cell. 2005;7:547–559. doi: 10.1016/j.ccr.2005.04.029. [DOI] [PubMed] [Google Scholar]
- 65.Chen L, Agrawal S, Zhou W, Zhang R, Chen J. Synergistic activation of p53 by inhibition of MDM2 expression and DNA damage. Proc Natl Acad Sci U S A. 1998;95:195–200. doi: 10.1073/pnas.95.1.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Tortora G, Caputo R, Damiano V, Bianco R, Chen J, Agrawal S, Bianco AR, Ciardiello F. A novel MDM2 anti-sense oligonucleotide has anti-tumor activity and potentiates cytotoxic drugs acting by different mechanisms in human colon cancer. Int J Cancer. 2000;88:804–809. doi: 10.1002/1097-0215(20001201)88:5<804::aid-ijc19>3.0.co;2-z. [DOI] [PubMed] [Google Scholar]
- 67.Zhang R, Wang H, Agrawal S. Novel antisense anti-MDM2 mixed-backbone oligonucleotides: proof of principle, in vitro and in vivo activities, and mechanisms. Curr Cancer Drug Targets. 2005;5:43–49. doi: 10.2174/1568009053332663. [DOI] [PubMed] [Google Scholar]
- 68.Kussie PH, Gorina S, Marechal V, Elenbaas B, Moreau J, Levine AJ, Pavletich NP. Structure of the MDM2 oncoprotein bound to the p53 tumor suppressor transactivation domain. Science. 1996;274:948–953. doi: 10.1126/science.274.5289.948. [DOI] [PubMed] [Google Scholar]
- 69.Shinohara T, Uesugi M. In-vivo activation of the p53 pathway by small-molecule antagonists of MDM2. Tanpakushitsu Kakusan Koso. 2007;52:1816–1817. [PubMed] [Google Scholar]
- 70.Grasberger BL, Lu T, Schubert C, Parks DJ, Carver TE, Koblish HK, Cummings MD, LaFrance LV, Milkiewicz KL, Calvo RR, Maguire D, Lattanze J, Franks CF, Zhao S, Ramachandren K, Bylebyl GR, Zhang M, Manthey CL, Petrella EC, Pantoliano MW, Deckman IC, Spurlino JC, Maroney AC, Tomczuk BE, Molloy CJ, Bone RF. Discovery and cocrystal structure of benzodiazepine-dione HDM2 antagonists that activate p53 in cells. J Med Chem. 2005;48:909–912. doi: 10.1021/jm049137g. [DOI] [PubMed] [Google Scholar]
- 71.Issaeva N, Bozko P, Enge M, Protopopova M, Verhoef LG, Masucci M, Pramanik A, Selivanova G. Small molecule RITA binds to p53, blocks p53-HDM-2 interaction and activates p53 function in tumors. Nat Med. 2004;10:1321–1328. doi: 10.1038/nm1146. [DOI] [PubMed] [Google Scholar]
- 72.Ding K, Lu Y, Nikolovska-Coleska Z, Qiu S, Ding Y, Gao W, Stuckey J, Krajewski K, Roller PP, Tomita Y, Parrish DA, Deschamps JR, Wang S. Structure-based design of potent non-peptide MDM2 inhibitors. J Am Chem Soc. 2005;127:10130–10131. doi: 10.1021/ja051147z. [DOI] [PubMed] [Google Scholar]
- 73.Xu GW, Ali M, Wood TE, Wong D, Maclean N, Wang X, Gronda M, Skrtic M, Li X, Hurren R, Mao X, Venkatesan M, Beheshti Zavareh R, Ketela T, Reed JC, Rose D, Moffat J, Batey RA, Dhe-Paganon S, Schimmer AD. The ubiquitin-activating enzyme E1 as a therapeutic target for the treatment of leukemia and multiple myeloma. Blood. 2010;115:2251–2259. doi: 10.1182/blood-2009-07-231191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Yang Y, Kitagaki J, Dai RM, Tsai YC, Lorick KL, Ludwig RL, Pierre SA, Jensen JP, Davydov IV, Oberoi P, Li CC, Kenten JH, Beutler JA, Vousden KH, Weissman AM. Inhibitors of ubiquitin-activating enzyme (E1), a new class of potential cancer therapeutics. Cancer Res. 2007;67:9472–9481. doi: 10.1158/0008-5472.CAN-07-0568. [DOI] [PubMed] [Google Scholar]
- 75.Ungermannova D, Parker SJ, Nasveschuk CG, Chapnick DA, Phillips AJ, Kuchta RD, Liu X. Identification and mechanistic studies of a novel ubiquitin E1 inhibitor. J Biomol Screen. 2012;17:421–434. doi: 10.1177/1087057111433843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Luise C, Capra M, Donzelli M, Mazzarol G, Jodice MG, Nuciforo P, Viale G, Di Fiore PP, Confalonieri S. An atlas of altered expression of deubiquitinating enzymes in human cancer. PLoS One. 2011;6:e15891. doi: 10.1371/journal.pone.0015891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Nicholson B, Marblestone JG, Butt TR, Mattern MR. Deubiquitinating enzymes as novel anticancer targets. Future Oncol. 2007;3:191–199. doi: 10.2217/14796694.3.2.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Allende-Vega N, Sparks A, Lane DP, Saville MK. MdmX is a substrate for the deubiquitinating enzyme USP2a. Oncogene. 2010;29:432–441. doi: 10.1038/onc.2009.330. [DOI] [PubMed] [Google Scholar]
- 79.Brummelkamp TR, Nijman SM, Dirac AM, Bernards R. Loss of the cylindromatosis tumour suppressor inhibits apoptosis by activating NF-kappaB. Nature. 2003;424:797–801. doi: 10.1038/nature01811. [DOI] [PubMed] [Google Scholar]
- 80.Oosterkamp HM, Neering H, Nijman SM, Dirac AM, Mooi WJ, Bernards R, Brummelkamp TR. An evaluation of the efficacy of topical application of salicylic acid for the treatment of familial cylindromatosis. Br J Dermatol. 2006;155:182–185. doi: 10.1111/j.1365-2133.2006.07224.x. [DOI] [PubMed] [Google Scholar]
- 81.Li Z, Wang D, Messing EM, Wu G. VHL protein-interacting deubiquitinating enzyme 2 deubiquitinates and stabilizes HIF-1alpha. EMBO Rep. 2005;6:373–378. doi: 10.1038/sj.embor.7400377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Lee BH, Lee MJ, Park S, Oh DC, Elsasser S, Chen PC, Gartner C, Dimova N, Hanna J, Gygi SP, Wilson SM, King RW, Finley D. Enhancement of proteasome activity by a small-molecule inhibitor of USP14. Nature. 2010;467:179–184. doi: 10.1038/nature09299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Stevenson LF, Sparks A, Allende-Vega N, Xirodimas DP, Lane DP, Saville MK. The deubiquitinating enzyme USP2a regulates the p53 pathway by targeting Mdm2. EMBO J. 2007;26:976–986. doi: 10.1038/sj.emboj.7601567. [DOI] [PMC free article] [PubMed] [Google Scholar]