Abstract
Background. A genetic bottleneck is known to exist for human immunodeficiency virus (HIV) at the point of sexual transmission. However, the nature of this bottleneck and its effect on viral diversity over time is unclear.
Methods. Interhost and intrahost HIV diversity was analyzed in a stable population in Rakai, Uganda, from 1994 to 2002. HIV-1 envelope sequences from both individuals in initially HIV-discordant relationships in which transmission occurred later were examined using Sanger sequencing of bulk polymerase chain reaction (PCR) products (for 22 couples), clonal analysis (for 3), and next-generation deep sequencing (for 9).
Results. Intrahost viral diversity was significantly higher than changes in interhost diversity (P < .01). The majority of HIV-1–discordant couples examined via bulk PCR (16 of 22 couples), clonal analysis (3 of 3), and next-generation deep sequencing (6 of 9) demonstrated that the viral populations present in the newly infected recipient were more closely related to the donor partner's HIV-1 variants found earlier during infection as compared to those circulating near the estimated time of transmission (P = .03).
Conclusions. These findings suggest that sexual transmission constrains viral diversity at the population level, partially because of the preferential transmission of ancestral as opposed to contemporary strains circulating in the transmitting partner. Future successful vaccine strategies may need to target these transmitted ancestral strains.
(See the editorial commentary by Blish, on pages 1336–8.)
Circulating viruses in individuals with chronic human immunodeficiency virus (HIV) infection manifest significant viral genetic diversity, which increases throughout the course of disease [1, 2]. This increase in viral diversity has been observed over the course of the worldwide AIDS epidemic [3]. However, the process by which this viral diversity changes over time within a stable population experiencing an ongoing HIV epidemic and the effects that new transmissions have on this process are unclear.
In mother-to-child transmission, only a minor subset of the maternal viral population is passed on to the infant [4]. Similar selection of transmitted viruses has been demonstrated in sexual transmission [5, 6]. Following these initial studies, a detailed analysis of acute infections in heterosexuals demonstrated that in the majority of cases only 1 or a limited number of viral strains establish initial infection, after which there is rapid divergence away from these strains as a result of the evolving immune response [7–15]. These findings are supported by analysis of simian immunodeficiency virus (SIV)–infected macaques, in which the initial founder viral strain persisted at a low level after transmission [16, 17]. Taken together, these data suggest that a bottleneck exists at the point of sexual transmission that limits the viral species that can be efficiently transmitted from host to host [18].
In Rakai, Uganda, viral diversity is significantly lower in newly infected individuals than in persons with chronic disease [19]. Furthermore, variants present in newly infected subjects were genetically more closely related to ancestral strains than to viruses circulating in the transmitting partner at the estimated time of transmission [19]. We used data from HIV-discordant couples enrolled in the Rakai Community Cohort Study (RCCS) to explore whether population HIV-1 genetic diversity is constrained because of the bottleneck present during heterosexual transmission.
METHODS
Study Population
The RCCS is a rural, community-based open cohort that, since 1994, has conducted surveillance of persons aged 15–49 years in Rakai district, southwestern Uganda [20]. Data from 1994 and 2002 surveys were used to assess changes in HIV-1 sequence variability over time, prior to the availability of antiretroviral therapy (ART). Individuals who were married or in long-term consensual unions were retrospectively identified in the RCCS, and HIV-discordant couples in which the index infected partners transmitted HIV to their initially HIV-negative partners were identified retrospectively [21]. All subjects provided written consent for their samples to be stored and used for research purposes. The study was approved by institutional review boards in Uganda and at collaborating US institutions. All participants were provided free HIV prevention education and were offered condoms, individual and couples’ HIV testing, and counseling free of cost.
Laboratory Analysis
HIV status was assessed by 2 enzyme immunoassays, and discordant results were confirmed by Western blot (Vitek, bioMérieux, St. Louis, MO). Viral RNA was extracted from all available stored serum samples from HIV-1 antibody–positive subjects from 1994 and 2002, using a QIAmp Viral RNA Mini Kit (Qiagen, Valencia, CA) [22]. The eluted RNA was expanded by reverse-transcription polymerase chain reaction (RT-PCR), using primers for the gag region (forward primer-G00-GACTAGCGGAGGCTAGAAG; reverse primer-G01-AGGGGTCGTTGCCAAAGA) or the env region (forward primer-GP50F1-AAAAATARAACCACTAGGAGTAGCACCCAC; reverse primer-GP41R1-AACGACAAAGGTGAGTATCCCTGCCTAA) of the genome. A subsequent nested PCR was performed in 2 independent reactions to amplify and sequence portions of p24 (HXB2, nucleotides 1249–1704 [forward primer-G60M-CAGCCAAAATTACCCTATAGTRCA; reverse primer-G05M-TGTTGGCTCTGGTCTGYTCT]) and gp41 (HBX2, nucleotides 7858–8260 [forward primer-GP40F1-TCTTAGGAGCAGCAGGAAGCACTATGGG; reverse primer-GP48R2-TCCTACTATCATTATGAATATTTTTATATA]) [23, 24]. Sanger sequences were generated from the nested PCR products directly (ie, from bulk PCR products), and subtypes were assigned as previously described [22, 25]. The resulting sequences were segregated by subtype, and the genetic pairwise distance was analyzed for each region for 1994 and 2002. Changes in genetic pairwise distance means were calculated using a maximum composite likelihood model [22]. Individuals infected with HIV subtype C were removed because of insufficient numbers. Individuals who were HIV positive with sequences available in both 1994 and 2002 were analyzed to determine the change of viral diversity within individuals over the same period. Any individual with a change in virus subtype was removed from the analysis to avoid potential bias due to HIV superinfection or error [26]. The remaining individuals with subtype D (n = 48) or subtype A (n = 19) infections were used to estimate the mean within-individual genetic divergence between 1994 and 2002.
Multiple independent PCRs using previously described primers were used to generate full-length envelopes for 3 transmission couples [10, 11]. Products from a minimum of 4 independent PCRs were combined to generate clonal full-length envelope sequences, as previously described [27]. Subtype assignments were generated using the National Center for Biotechnology Information genotyping database and were verified using a phylogenetic analysis of all sequences from each year [22].
Bulk PCR Analysis of Transmission Pairs
Initially, we compared sequences created by Sanger sequencing of bulk PCR products as described above from the viruses circulating in the recipient partner to sequences from viruses in the donor. Viral gp41 sequences from the strains in the donor partner at the time point closest to the estimated time of transmission (ie, donor transmission strains) and at all earlier time points available (ie, donor early strains) were compared to the strains in the previously HIV-negative partner (ie, recipient strains) (Figure 1A). Transmission was assumed to occur halfway between the last HIV-negative and first HIV-positive time point for the recipient partner [28]. Genetic distance between the donor transmission strain and the recipient strain (d0) was compared to the genetic distance between the recipient strain and the donor early strain (d–n) (Figure 1B). In the analysis, we only included sequences from donor and recipient strains that phylogenetically clustered together, to ensure virological linkage among all transmission pairs [29].
Figure 1.
A, A representative initially discordant couple is shown with the time points of positive (+) and negative (−) human immunodeficiency virus (HIV) test results (n/a indicates the individual was not sampled that year), as well as comparisons of the gp41 sequences from the transmitted strain to the donor early (d–3) and the donor transmission (d0) time points. B, The pairwise distance for the 2 time-point comparisons from this representative couple is shown. C, In 16 of 22 couples (72.7%), the donor viral sequence at the earliest time point available had the shortest pairwise distance to the transmitted strain in the recipient.
Clonal Analysis of Transmission Pairs
All full-length envelope sequences were aligned using Clustal X, gap stripped, and further codon aligned manually, using MacClade (version 4.01). Heuristically searched maximum likelihood trees were generated for each transmission pair, using the “best fit” evolutionary model parameters selected by use of Modeltest, version 3.06 [30], and the software package Phylogenetic Analysis Using Parsimony and Other Methods (PAUP 4.02b2a) [31]. All trees were rooted to subtype D sequence D.UG.94.94UG114_U88824 from the Los Alamos database. The most-recent common ancestors (MRCAs) were determined using PAUP.
Next-Generation Sequencing Analysis of Transmission Pairs
gp41 sequences were analyzed from 15 additional couples with at least 2 years between the donor early time point and the estimated time of transmission, using next-generation deep sequencing as previously described, with 1 alteration [29, 32]. Briefly, viral RNA was extracted from 140 µL of serum, and the gp41 region was initially expanded in a manner identical to that of the bulk PCR product, described above. Subsequently, a nested PCR reaction was performed using designed primer sets for gp41 (fourteen 454 bar-coded variations [MID1–MID14]) (Roche, Branford, CT) [29] Successful single-band amplification of gp41 target products was verified by agarose gel electrophoresis. All PCR products were purified by incubating 30 µL of AMPure Beads XP (Agencourt, Beckman Coulter Genomics, Danvers, MA) with 25 µL of PCR product diluted in 25 µL of water. Purified PCR products were quantified using PicoGreen (Invitrogen, Carlsbad, CA), and each template was diluted to 1 × 109 molecules/µL stock. Preparation of templated beads for next-generation sequencing analysis followed the emPCR Method Manual-Lib-L-MV (Roche). Enriched DNA Capture Beads were sequenced on the 454 (Roche) in accordance with the manufacturer's instructions, using a 4-region gasket [29].
The resulting next-generation sequencing results were analyzed using the GS Amplicon Variant Analyzer, version 2.5 (Roche). All sequence reads were compared, and similar sequences were combined into a single consensus sequence in accordance with the manufacturer's specifications (Roche). Generated consensus sequences that were within 10 bases from both ends of the amplicon and comprised a cluster of ≥10 individual sequences were determined. In addition, to normalize the number of consensus sequences to the total number of sequence reads acquired for each sample tested, only consensus sequences with sequence read totals of >0.5% of the total sequence read volume for that sample were used in further analysis. Couples in whom the donor and recipient sequences failed to phylogenetically cluster were excluded from the analysis. The sequences for the 2 samples from the donor were grouped separately, and the mean genetic distance between these groups and the recipient consensus sequences was calculated using a maximum composite likelihood model. In addition, the consensus sequences with read totals of >0.5% from each sample within a couple were aligned with a variety of viral sequences from Rakai, rooted on the subtype C population, and were examined using a neighbor-joining phylogenetic tree method to compare the sequence(s) of the populations from either donor time point to the sequence of the recipient population. The transmission category for strains detected in couples was then classified phylogenetically as early (ie, only viral sequences from the donor early strain were highly similar to the recipient strain), preserved (ie, viral sequences from both donor time points were highly similar to those of the recipient strain), or late (ie, only viral sequences from the donor transmission strain were highly similar to those of the recipient strain). All sequences have been submitted to GenBank and are also available on request. Genbank accession numbers: Clonal sequences: JX658572-JX658681 and EU853126-EU853141; Bulk sequences: JX649921-JA649942 and GQ252747,GQ252758, GQ252783, GQ253798, GQ253846, GQ253871, GQ253873, GQ253875-GQ253877, GQ333489, GQ333669, GQ333828, GQ333974, GQ334068, GU017857, GU017864, GU017875, GU017879, GU017882, GU017889, GU017891, GU046796, GU046798, GU046799, GU046800, HM114951, HM114979, HM115186, HM115227, HM115235, HM623170, HM623172-HM623175, HM623177-HM623188; Pyrosequences: JX647046-JX647375.
Statistical Analysis
χ2 tests were used to compare subject characteristics in 1994 and 2002, including age, sex, current marital status, current nonmarital sexual relationship, and number of sex partners during the past year. Differences between the change in diversity observed within individuals and the change in the population as a whole; and between the next-generation sequence reads and the consensus sequence totals were estimated using analysis of variance (ANOVA). The full transmission pair sequence distribution was compared to the null probability of 0.5 with a sample proportions test with continuity correction. SAS, version 9.2 (SAS Institute), was used for all analyses.
RESULTS
Interhost Viral Divergence
Samples from HIV-infected individuals enrolled in the RCCS in 1994 (n = 1072) and 2002 (n = 806) were retrospectively assessed for changes in interhost viral diversity or population viral divergence during this 8-year period [22, 33]. It was found that the 2 populations of HIV-infected participants were significantly different, with the 2002 population composed of a higher percentage of females and of persons reporting nonmarital relationships, condom use, and multiple sex partners in the past year (Supplementary Table 1). These differences reflect the characteristics of HIV-infected individuals in the cohort as a whole during these periods [33].
To examine changes in viral evolution in the population as a whole, direct bulk PCR sequences of p24 and gp41 were examined [22]. Viral diversity significantly increased for both regions in each subtype over the 8-year period (Table 1). Subtype D also diversified at a significantly higher rate than subtype A for both regions (P < .001).
Table 1.
Genetic Distance of All Pairwise Distances for 1994 and 2002, by Genomic Region and Human Immunodeficiency Virus Subtype
| Region, Subtype | Mean Percentage Diversity (95% CI), by Year |
Mean Divergencea (95% CI) | Pb | |
|---|---|---|---|---|
| 1994 | 2002 | |||
| p24, D | 3.97 (0–14.4) | 5.35 (0–13.3) | 1.37 (1.36–1.39) | <.0001 |
| p24, A | 4.91 (.2–14.5) | 6.05 (0–14.4) | 1.15 (1.10–1.19) | <.0001 |
| gp41, D | 6.27 (0–26.6) | 8.57 (0–30.3) | 2.30 (2.28–2.31) | <.0001 |
| gp41, A | 6.81 (.2–19.1) | 8.81 (0–23.1) | 2.00 (1.97–2.04) | <.0001 |
Abbreviation: CI, confidence interval.
a Data were calculated using analysis of variance.
b 1994 and 2002 data were considered statistically significantly different if P ≤ .05.
Intrahost Viral Divergence
HIV-infected individuals with available samples from both 1994 and 2002 were examined to estimate intrahost viral divergence. The mean intrahost divergence in the gp41 region was significantly higher than the mean interhost population divergence for both subtype D and subtype A (Table 2). Although the p24 region trended in a similar fashion, the difference between the mean intrahost divergence and the interhost population divergence was not statistically significant for subtype A and subtype D (Table 2).
Table 2.
Viral Divergence in the Population (Interhost) and Within Infected Individuals (Intrahost) From 1994 and 2002, by Genomic Region and Human Immunodeficiency Virus Subtype
| Region, Subtype | Viral Divergence (95% CI) |
Pa | |
|---|---|---|---|
| Interhost | Intrahost | ||
| p24, D | 1.37 (1.36–1.39) | 1.87 (1.38–2.36) | .18 |
| p24, A | 1.15 (1.10–1.19) | 1.81 (1.11–2.51) | .14 |
| gp41, D | 2.30 (2.29–2.31) | 3.27 (2.61–3.93) | <.01 |
| gp41, A | 2.00 (1.97–2.04) | 2.83 (2.17–3.49) | .05 |
Abbreviation: CI, confidence interval.
a Interhost and intrahost data were considered statistically significantly different if P ≤ .05.
Viral Diversity in Transmission Pairs
To study more directly what might be anchoring viral evolution in the population, we retrospectively identified HIV-discordant couples with samples available from both partners close to the time of transmission and from the donor prior to the estimated transmission period. We examined viral gp41 sequences from the recipient and the donor viruses at various time points to determine whether variants circulating in the recently infected partner were more closely related to the donor partner's viruses near the time of or prior to the estimated time of transmission (Figure 1). All 6 couples infected with HIV subtype A were excluded from the bulk PCR couple analysis because of an insufficient number of transmission events. Phylogenetic analysis was used to confirm transmission, and 10 unlinked couples or partnerships with possible HIV superinfection were removed. The removal of unlinked couples was planned prior to data analysis; however, removal of couples with possible superinfections was performed after the initial data analysis. For 22 couples, the genetic distance between the gp41 sequences in HIV from the recipient partner with incident infection was calculated relative to both the sequence of donor HIV at the time of transmission and all of the available donor sequences from the years prior to the transmission window. In 16 of these 22 couples (72.7%), the genetic distance between the recipient strain gp41 sequences and the early donor strain sequences was shorter than that between the recipient strain sequences and the donor transmission strain sequences, indicating that the recipient strains were genetically more similar to donor viruses present before rather than near the time of estimated transmission.
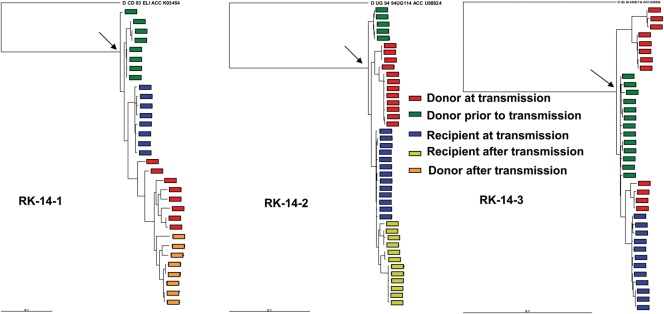
To probe this issue in greater detail, we also examined longitudinally isolated full-length sequences of donor and recipient clonally derived envelopes. Three newly infected subjects were sampled a median of 91 days (range, 17–359 days) after estimated infection. One newly infected subject (RK14-2) was sampled prior to seroconversion and thus was estimated to have had a maximum infection duration of 17 days [34]. All transmitting partners had chronic persistent infection of >2 years duration at the time of estimated transmission. In all 3 couples examined, envelope sequences from virus found in the naive subject early after infection were more closely related to sequences found in virus from the transmitter prior to (median distance = 0.017; range = 0.011–0.026) as opposed to at (median distance = 0.027; range = 0.018–0.055) the time of transmission (Figure 2). In addition, in all 3 couples, recipient virus sequences (median distance = 0.014; range = 0.010–0.023) were closer to sequences of the most recent common ancestor, compared with the donor virus sequences isolated close to the time of estimated transmission (median distance = 0.015; range = 0.015–0.053). If one assumes that each newly infected subject was infected with a single variant, the identity of the transmitting virus was estimated to be the common ancestor for the virus isolated from the subject with incident infection around the time of estimated transmission. In all 3 cases, the transmitted virus was closer to donor viruses circulating earlier (median distance = 0.018; range = 0.011–0.030) as compared to those found around the time of transmission (median distance = 0.029; range = 0.022–0.089).
Figure 2.
Maximum likelihood phylogenetic analysis for multiple full-length envelope sequences from subjects in discordant relationships. Color keys denotes the sequence source and period of sampling. Longitudinal envelopes were collected approximately every 9 months. Arrows point to the most recent common ancestor. The outgroup sequence is labeled at the top of each tree.
Because of the limited sequencing depth provided by bulk PCR and clonal sequence analysis, 15 couples were examined using next-generation deep sequencing of the gp41 region. Of these couples, 3 had at least 1 sample that did not amplify and were subsequently removed. Three additional couples were found to be unlinked or had experienced HIV superinfection and were also removed. Of the remaining 9 couples, the mean total sequence read volumes for all the samples tested was 8588 ± 4055, and the mean number (±SD) of consensus sequences (ie, those with >0.5% of the total read volume) was 12.3 ± 6 (Table 3). There were no significant differences in total read volume or number of consensus sequences between the recipient or donor sample time points.
Table 3.
Differences in Mean Genetic Distance of Viral Populations Derived From Couples in Which Human Immunodeficiency Virus Was Transmitted, as Determined by Deep Next-Generation Sequencing
| Couple | Recipient |
Donor Early |
Donor Late |
Person-Yearsa | Mean Genetic Distanceb |
Transmission Category |
|||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Reads | Consensus | Reads | Consensus | Reads | Consensus | Early | Late | By Distance | By Phylogeneticsc | ||
| 1 | 10218 | 5 | 19214 | 9 | 4280 | 12 | 6.9 | 2.3 | 0.7 | Late | Late |
| 6 | 6951 | 5 | 15055 | 3 | 9032 | 12 | 2.7 | 0.2 | 0.6 | Early | Preserved |
| 8 | 5649 | 19 | 15191 | 22 | 8042 | 17 | 3.6 | 3.9 | 1.3 | Late | Late |
| 9 | 6871 | 9 | 7529 | 10 | 12396 | 15 | 3.4 | 0.6 | 1.1 | Early | Early |
| 10 | 14306 | 15 | 5257 | 9 | 11205 | 18 | 3.3 | 3.8 | 4.3 | Early | Preserved |
| 11 | 2464 | 6 | 8925 | 27 | 10510 | 12 | 9.8 | 2.2 | 3.1 | Early | Late |
| 12 | 4912 | 10 | 8005 | 18 | 10850 | 17 | 7.6 | 1.9 | 2.3 | Early | Preserved |
| 14 | 8013 | 3 | 5582 | 2 | 2268 | 15 | 5.8 | 2.7 | 1.4 | Late | Late |
| 15 | 7417 | 13 | 5146 | 11 | 6578 | 17 | 3.9 | 2.6 | 3.1 | Early | Preserved |
a Data denote the interval between the donor early and donor late time points.
b Data denote the distance between the recipient consensus sequences and the donor early or late sample, as calculated by a maximum composite likelihood model.
c See Methods for definitions.
In 6 of 9 couples (ie, couples 6, 9–12, and 15), the mean genetic distance between the recipient viral population was shorter to the donor virus population at the earlier time point, compared with the distance to the donor's viral variants at the time of estimated transmission (Table 3). In addition, in 6 couples (ie, 1, 6, 11, 12, 14, and 15), recipient sequences were closer to the MRCA than donor sequences isolated at the time of estimated transmission (Figures 3 and 4). With the assumption that the common ancestor for all the sequences present in the newly infected subject represented the transmitted virus, we calculated the distance from the donor variants prior to and around the time of estimated transmission to this recipient common ancestor. In 5 couples (ie, 6 and 9–12), the estimated transmitted sequence was more closely related to donor variants present prior to transmission as compared to those circulating around the time of transmission.
Figure 3.
Maximum composite likelihood Neighbor-joining phylogenetic trees of gp41 viral consensus sequences (containing >0.5% of total sequence reads) from 3 transmitting couples derived from deep 454 next-generation sequencing of recipient strains (blue), donor early strains (green), and donor transmission strains (red) are shown. The closest or transmitted strain (TS) to the recipient viral population is indicated by an arrow and is either closely related to sequences found in the donor near the time of transmission only (A), in the donor's earlier sample only (B), or both (C). The number of repeated read sequences represented by each consensus sequence is shown at the end of the consensus identifier. Bootstrap values (500 replicates) >80% are shown. Distance is indicated for each individual tree, and samples are grouped with a selection of subtype reference sequences and random sequences from individuals in Rakai (black).
Figure 4.
Maximum likelihood phylogenetic analysis of gp41 viral consensus sequences (containing >0.5% of total sequence reads) from 6 transmitting couple derived from deep 454 next-generation sequencing of the recipient strains (blue), donor early strains (green), and donor transmission strains (red) are shown. The number of repeated read sequences represented by each consensus sequence is shown at the end of the consensus identifier. Distance is indicated for each individual tree.
The donor and recipient strains most closely related to each other were then identified among the phylogenetic trees. In all 3 couples (ie, 1, 8, and 14) in which the genetic distance analyses determined that the recipient strains were more closely related to the donor transmission strain, the phylogenetic analysis confirmed this result (Figure 3A). In the remaining couples, the donor early strains were transmitted, and this was supported by the phylogenetic analysis with 2 distinct patterns (Figure 3B and C). In couple 9, the viral sequence closest to the donor transmitted strain was found only in the donor early sample (Figure 3A). In the remaining 4 couples (ie, 6, 10, 12, and 15), the recipient viral populations were closely related to highly similar or identical viral strains found in the donor at both time points (Figures 3C and 4). In these cases, the newly acquired strains appeared to be residual or archived strains of the earlier virus found in the donor and in all but one instance represented a minority viral population in the donor sample at the time of transmission.
To validate the results of the 3 different sequencing methods, sequences of virus from 3 couples were analyzed by 2 sequencing methods. For couple 6 (RK-14-3), next-generation sequencing and clonal sequencing were used; for couple 9, next-generation sequencing and bulk sequencing were used; and for couple RK-14-1, clonal and bulk sequencing were used. In all 3 cases, results of the different methods agreed that the transmitted strain was more closely related to the viral species found earlier in the donor.
Overall, we identified 22 of 31 couples in which the recipient's viral strain was more closely related to earlier viruses found in the donor. We tested the null hypothesis that the proportion of recipient viral strains would have an equivalent chance (0.5) of being more closely related to the earlier or later donor strains, using a sample proportions test with continuity correction, and found that the observed proportion (0.71; 95% confidence interval, .52–0.85) was significantly different than the null probability (P = .03).
DISCUSSION
We examined HIV transmission in a heterosexual population and observed that viral diversity increased from 1994 to 2002 in the HIV-infected population as a whole. This occurred at a significantly slower rate than viral divergence within individuals in the same population who survived over the 8-year period. We hypothesized that this anchoring of the interhost viral population may be due to a selection of distinct viral strains during sexual transmission. To address this hypothesis, we examined viral strains in transmitting couples using different methods and found that in a significant proportion of the couples tested the viruses circulating in the newly infected subject were more closely related to viruses present in the donor partner prior to as opposed to around the time of estimated transmission.
The increased viral diversity seen in both regions of the viral genome supports findings from other settings and suggests that HIV diversity is increasing in Africa [1, 3, 35]. It has been speculated that this difference between intrahost and interhost divergence is most likely due to the evolving selective pressure of the ongoing immune response within an individual [1, 36]. Others have suggested that the transmission bottleneck in HIV is one of opportunity, rather than an inherent transmission profile passed from host to host [1, 37]. However, our analysis of discordant couples suggests a possible selection bias for transmission of viral species found earlier in the infection of the donor partner. A similar selection process is seen in mother-to-child transmission [4]. This project expands on our previous findings that transmitted strains were genetically closer to a calculated common ancestral strain in Rakai by demonstrating that the transmitted strains are found in the donor sample at earlier time points and that this selection of sexually transmitted strains of HIV reduces viral diversification at the population level [19]. Our data support the previous suggestion by Zhu et al that viral diversity is essentially reset with each individual transmission event [5].
The anchoring of the viral diversity was only statistically significant in the gp41 region, although results in the p24 region trended in a similar fashion (Table 2). This is most likely due to the lower diversity rate seen in the nonenvelope portions of the viral genome [35]. One limitation of this work is the relatively long time frame between sampling, which limits the ability to narrow the transmission window in these couples.
Three couples were examined via clonal analysis, and in all 3 the sequences in the donor strains from the earlier time point, as opposed to sequences in strains circulating around the estimated time of transmission, were more closely related to the variants found in the newly infected subject and the estimated transmitted variants. The next-generation sequencing data further supported these findings [19]. Our results support recent findings in the SIV macaque model, which postulate that transmitted strains can be sequestered in a long-lived reservoir during the early stage of infection, persist at a low level in the serum, and are potentially preferentially selected for transmission [38].
It can be argued that viruses present in the donor early in infection are not preferentially acquired by the recipient and that, instead, the transmitted variants revert toward the ancestral state in the newly infected individual because there is minimal immune pressure early after infection [39]. However, in 1 subject (RK14-2) sampled prior to seroconversion, viruses found after newly acquired infection were more closely related to the MRCA and donor sequences of viruses present years prior to transmission. This suggests that viruses more closely related to the ancestral strains are present even prior to the generation of a humoral response. Our conclusion, however, is based on findings for 1 subject. Further longitudinal analysis of couples in which the recipient is sampled during the acute stage of disease will be needed to determine whether reversion after acquisition or selection from donor variants accounts for the ancestral genotypes found in newly infected subjects.
It has been shown that transmitted and early strains of HIV manifest distinct phenotypes, such as unique glycosylation patterns and binding potential [12, 13, 40]. In light of these previous studies and our current data, we propose that previously sexually transmitted viral strains may have a distinct advantage in transmission efficiency and are sustained or archived, allowing for subsequent transmission events. Furthermore, these findings suggest that future studies may need to identify these HIV strains that are preferentially transmitted and to examine them for molecular determinants important for transmission, as well as include them as targets for future vaccines.
Supplementary Data
Supplementary materials are available at The Journal of Infectious Diseases online (http://jid.oxfordjournals.org/). Supplementary materials consist of data provided by the author that are published to benefit the reader. The posted materials are not copyedited. The contents of all supplementary data are the sole responsibility of the authors. Questions or messages regarding errors should be addressed to the author.
Notes
Acknowledgments. We thank all participants in the Rakai Community Cohort Study; all staff at the Rakai Health Sciences Program; Susanna Lamers, for her technical expertise; and Dr Stuart Ray, for helpful discussions.
Financial support. This work was supported in part by funding from the Division of Intramural Research, National Institutes of Allergy and Infectious Diseases (NIAID), National Institutes of Health; the NIAID (grants R01 A134826, R01 A134265 and R01 AI077473 to M. S.); the Eunice Kennedy Shriver National Institute of Child Health & Human Development (grant 5P30HD06826); the World Bank STI Project, Uganda; the Henry M. Jackson Foundation; the Fogarty Foundation (grant 5D43TW00010); and the Bill and Melinda Gates Institute for Population and Reproductive Health at the Bloomberg School of Public Health, Johns Hopkins University.
Potential conflicts of interest. All authors: No reported conflicts.
All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1.Lemey P, Rambaut A, Pybus OG. HIV evolutionary dynamics within and among hosts. AIDS Rev. 2006;8:125–40. [PubMed] [Google Scholar]
- 2.Taylor BS, Sobieszczyk ME, McCutchan FE, Hammer SM. The challenge of HIV-1 subtype diversity. N Engl J Med. 2008;358:1590–602. doi: 10.1056/NEJMra0706737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kuiken CL, Zwart G, Baan E, Coutinho RA, van den Hoek JA, Goudsmit J. Increasing antigenic and genetic diversity of the V3 variable domain of the human immunodeficiency virus envelope protein in the course of the AIDS epidemic. Proc Natl Acad Sci U S A. 1993;90:9061–5. doi: 10.1073/pnas.90.19.9061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wolinsky SM, Wike CM, Korber BT, et al. Selective transmission of human immunodeficiency virus type-1 variants from mothers to infants. Science. 1992;255:1134–7. doi: 10.1126/science.1546316. [DOI] [PubMed] [Google Scholar]
- 5.Zhu T, Mo H, Wang N, et al. Genotypic and phenotypic characterization of HIV-1 patients with primary infection. Science. 1993;261:1179–81. doi: 10.1126/science.8356453. [DOI] [PubMed] [Google Scholar]
- 6.Poss M, Martin HL, Kreiss JK, et al. Diversity in virus populations from genital secretions and peripheral blood from women recently infected with human immunodeficiency virus type 1. J Virol. 1995;69:8118–22. doi: 10.1128/jvi.69.12.8118-8122.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Goonetilleke N, Liu MK, Salazar-Gonzalez JF, et al. The first T cell response to transmitted/founder virus contributes to the control of acute viremia in HIV-1 infection. J Exp Med. 2009;206:1253–72. doi: 10.1084/jem.20090365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lee HY, Giorgi EE, Keele BF, et al. Modeling sequence evolution in acute HIV-1 infection. J Theor Biol. 2009;261:341–60. doi: 10.1016/j.jtbi.2009.07.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rong R, Li B, Lynch RM, et al. Escape from autologous neutralizing antibodies in acute/early subtype C HIV-1 infection requires multiple pathways. PLoS Pathog. 2009;5:e1000594. doi: 10.1371/journal.ppat.1000594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Salazar-Gonzalez JF, Salazar MG, Keele BF, et al. Genetic identity, biological phenotype, and evolutionary pathways of transmitted/founder viruses in acute and early HIV-1 infection. J Exp Med. 2009;206:1273–89. doi: 10.1084/jem.20090378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Keele BF, Giorgi EE, Salazar-Gonzalez JF, et al. Identification and characterization of transmitted and early founder virus envelopes in primary HIV-1 infection. Proc Natl Acad Sci U S A. 2008;105:7552–7. doi: 10.1073/pnas.0802203105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Derdeyn CA, Decker JM, Bibollet-Ruche F, et al. Envelope-constrained neutralization-sensitive HIV-1 after heterosexual transmission. Science. 2004;303:2019–22. doi: 10.1126/science.1093137. [DOI] [PubMed] [Google Scholar]
- 13.Chohan B, Lang D, Sagar M, et al. Selection for human immunodeficiency virus type 1 envelope glycosylation variants with shorter V1-V2 loop sequences occurs during transmission of certain genetic subtypes and may impact viral RNA levels. J Virol. 2005;79:6528–31. doi: 10.1128/JVI.79.10.6528-6531.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Long EM, Martin HL, Jr, Kreiss JK, et al. Gender differences in HIV-1 diversity at time of infection. Nat Med. 2000;6:71–5. doi: 10.1038/71563. [DOI] [PubMed] [Google Scholar]
- 15.Sagar M, Lavreys L, Baeten JM, et al. Identification of modifiable factors that affect the genetic diversity of the transmitted HIV-1 population. AIDS. 2004;18:615–9. doi: 10.1097/00002030-200403050-00005. [DOI] [PubMed] [Google Scholar]
- 16.Love TM, Thurston SW, Keefer MC, Dewhurst S, Lee HY. Mathematical modeling of ultradeep sequencing data reveals that acute CD8+ T-lymphocyte responses exert strong selective pressure in simian immunodeficiency virus-infected macaques but still fail to clear founder epitope sequences. J Virol. 2010;84:5802–14. doi: 10.1128/JVI.00117-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bar KJ, Li H, Chamberland A, et al. Wide variation in the multiplicity of HIV-1 infection among injection drug users. J Virol. 2010;84:6241–7. doi: 10.1128/JVI.00077-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ray SC, Quinn TC. Sex and the genetic diversity of HIV-1. Nat Med. 2000;6:23–5. doi: 10.1038/71487. [DOI] [PubMed] [Google Scholar]
- 19.Sagar M, Laeyendecker O, Lee S, et al. Selection of HIV variants with signature genotypic characteristics during heterosexual transmission. J Infect Dis. 2009;199:580–9. doi: 10.1086/596557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wawer MJ, Gray RH, Sewankambo NK, et al. A randomized, community trial of intensive sexually transmitted disease control for AIDS prevention, Rakai, Uganda. AIDS. 1998;12:1211–25. doi: 10.1097/00002030-199810000-00014. [DOI] [PubMed] [Google Scholar]
- 21.Hollingsworth TD, Laeyendecker O, Shirreff G, et al. HIV-1 transmitting couples have similar viral load set-points in Rakai, Uganda. PLoS Pathog. 2010;6:e1000876. doi: 10.1371/journal.ppat.1000876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Collinson-Streng AN, Redd AD, Sewankambo NK, et al. Geographic HIV type 1 subtype distribution in Rakai district, Uganda. AIDS Res Hum Retroviruses. 2009;25:1045–8. doi: 10.1089/aid.2009.0127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yang C, Dash BC, Simon F, et al. Detection of diverse variants of human immunodeficiency virus-1 groups M, N, and O and simian immunodeficiency viruses from chimpanzees by using generic pol and env primer pairs. J Infect Dis. 2000;181:1791–5. doi: 10.1086/315439. [DOI] [PubMed] [Google Scholar]
- 24.Yang C, Dash B, Hanna SL, et al. Predominance of HIV type 1 subtype G among commercial sex workers from Kinshasa, Democratic Republic of Congo. AIDS Res Hum Retroviruses. 2001;17:361–5. doi: 10.1089/08892220150503726. [DOI] [PubMed] [Google Scholar]
- 25.Conroy SA, Laeyendecker O, Redd AD, et al. Changes in the distribution of HIV type 1 subtypes D and A in Rakai District, Uganda between 1994 and 2002. AIDS Res Hum Retroviruses. 2010;26:1087–91. doi: 10.1089/aid.2010.0054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Piantadosi A, Chohan B, Chohan V, McClelland RS, Overbaugh J. Chronic HIV-1 infection frequently fails to protect against superinfection. PLoS Pathog. 2007;3:e177. doi: 10.1371/journal.ppat.0030177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Etemad B, Fellows A, Kwambana B, et al. Human immunodeficiency virus type 1 V1-to-V5 envelope variants from the chronic phase of infection use CCR5 and fuse more efficiently than those from early after infection. J Virol. 2009;83:9694–708. doi: 10.1128/JVI.00925-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kiwanuka N, Laeyendecker O, Gray RH, et al. HIV-1 subtypes and differences in heterosexual transmission of HIV among HIV-1 discordant couples in Rakai, Uganda. AIDS. 2009;23:2479–84. doi: 10.1097/QAD.0b013e328330cc08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Redd AD, Collinson-Streng A, Martens C, et al. Identification of HIV superinfection in seroconcordant couples in Rakai, Uganda using next generation deep sequencing. J Clin Microbiol. 2011;49:2859–67. doi: 10.1128/JCM.00804-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Posada D, Crandall KA. MODELTEST: testing the model of DNA substitution. Bioinformatics. 1998;14:817–8. doi: 10.1093/bioinformatics/14.9.817. [DOI] [PubMed] [Google Scholar]
- 31.Swofford DL. Phylogenetic analysis using parsimony. 4th ed. Sunderland, MA: Sinauer Associates; 1998. [Google Scholar]
- 32.Eshleman SH, Hudelson SE, Redd AD, et al. Analysis of genetic linkage of HIV from couples enrolled in the HIV Prevention Trials Network 052 trial. J Infect Dis. 2011;204:1918–26. doi: 10.1093/infdis/jir651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Conroy SA, Laeyendecker O, Redd AD, et al. Changes in the distribution of HIV-1 subtypes D and A in Rakai District, Uganda between 1994 and 2002. AIDS Res Hum Retroviruses. 2010;26:1087–91. doi: 10.1089/aid.2010.0054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cohen MS, Gay CL, Busch MP, Hecht FM. The detection of acute HIV infection. J Infect Dis. 2010;202(Suppl 2):S270–7. doi: 10.1086/655651. [DOI] [PubMed] [Google Scholar]
- 35.Abecasis AB, Vandamme AM, Lemey P. Quantifying differences in the tempo of human immunodeficiency virus type 1 subtype evolution. J Virol. 2009;83:12917–24. doi: 10.1128/JVI.01022-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rambaut A, Posada D, Crandall KA, Holmes EC. The causes and consequences of HIV evolution. Nat Rev Genet. 2004;5:52–61. doi: 10.1038/nrg1246. [DOI] [PubMed] [Google Scholar]
- 37.Hedskog C, Mild M, Albert J. Transmission of the X4 phenotype of HIV-1: is there evidence against the “random transmission” hypothesis? J Infect Dis. 2012;205:163–5. doi: 10.1093/infdis/jir719. [DOI] [PubMed] [Google Scholar]
- 38.Whitney JB, Hraber PT, Luedemann C, et al. Genital tract sequestration of SIV following acute infection. PLoS Pathog. 2011;7:e1001293. doi: 10.1371/journal.ppat.1001293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wood N, Bhattacharya T, Keele BF, et al. HIV evolution in early infection: selection pressures, patterns of insertion and deletion, and the impact of APOBEC. PLoS Pathog. 2009;5:e1000414. doi: 10.1371/journal.ppat.1000414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nawaz F, Cicala C, Van Ryk D, et al. The genotype of early-transmitting HIV gp120s promotes alpha (4) beta(7)-reactivity, revealing alpha (4) beta(7) +/CD4+ T cells as key targets in mucosal transmission. PLoS Pathog. 2010;7:e1001301. doi: 10.1371/journal.ppat.1001301. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.