Abstract
Background. Efavirenz exhibits marked interindividual variability in plasma levels and toxicities. Prior pharmacogenetic studies usually measure exposure via single plasma levels, examine limited numbers of polymorphisms, and rarely model multiple contributors. We analyzed numerous genetic and nongenetic factors impacting short-term and long-term exposure in a large heterogeneous population of human immunodeficiency virus (HIV)–infected women.
Methods. We performed 24-hour intensive pharmacokinetic studies in 111 women receiving efavirenz under actual-use conditions and calculated the area-under-the-concentration-time curve (AUC) to assess short-term exposure; the efavirenz concentration in hair was measured to estimate long-term exposure. A total of 182 single-nucleotide polymorphisms (SNPs) and 45 haplotypes in 9 genes were analyzed in relationship to exposure by use of multivariate models that included a number of nongenetic factors.
Results. Efavirenz AUCs increased 1.26-fold per doubling of the alanine aminotransferase level and 1.23-fold with orange and/or orange juice consumption. Individuals with the CYP2B6 516TT genotype displayed 3.5-fold increases in AUCs and 3.2-fold increases in hair concentrations, compared with individuals with the TG/GG genotype. Another SNP in CYP2B6 (983TT) and a p-glycoprotein haplotype affected AUCs without substantially altering long-term exposure.
Conclusions. This comprehensive pharmacogenomics study showed that individuals with the CYP2B6 516TT genotype displayed >3-fold increases in both short-term and long-term efavirenz exposure, signifying durable effects. Pharmacogenetic testing combined with monitoring of hair levels may improve efavirenz outcomes and reduce toxicities.
Achievement of the full benefits of antiretroviral therapy requires lifelong exposure to adequate but nontoxic levels of these medications. However, significant interindividual variability in drug concentrations occurs, owing to the influence of factors such as treatment adherence, concomitant conditions and medications, illicit substance use, diet, body mass, renal and/or hepatic function, and host genetics. Pharmacogenetic assessment is optimally performed via a comprehensive approach in which genetic traits are assessed in parallel with nongenetic determinants in representative populations. Moreover, since a single plasma drug level does not provide information on long-term exposure, pharmacogenetic studies are best performed using more comprehensive drug exposure measures.
We have previously described methods to measure antiretroviral levels in small samples of hair to assess long-term exposure to treatment [1, 2]. We have shown that protease inhibitor and non-nucleoside reverse transcriptase inhibitor concentrations in hair are consistently powerful independent predictors of virologic outcome [3, 4]. We have also performed intensive pharmacokinetic (PK) studies over 12–24 hours in a large heterogeneous population that we now use to model the contribution of genetic and nongenetic factors to antiretroviral exposure [5, 6]. The area-under-the-concentration-time curve (AUC) calculated from intensive PK sampling data and the concentration in hair samples can be used to measure short-term and long-term exposure, respectively.
Efavirenz is a mainstay of combination antiretroviral therapy, and its dosing in adults is frequently uniform (600 mg daily), despite marked interindividual variability in plasma levels [7, 8]. Although efavirenz is available in a fixed-dose combination administered once daily, rates of discontinuation of this medication in real-world settings, usually due to adverse effects, are high (up to 50%) [9–12]. Central nervous system (CNS) side effects are common and have been linked with high plasma efavirenz levels [13, 14].
Efavirenz is primarily metabolized by the cytochrome p450 (CYP) 2B6 enzyme, and several single nucleotide polymorphisms (SNPs) in the CYP2B6 gene are known to influence plasma levels, responses, and CNS side effects [15–17]. Polymorphisms in other cytochromes have also been linked with variations in efavirenz levels [18], as have SNPs in the multiple drug resistance (MDR1/ABCB1) gene [19]. After SNPs were identified that appeared to influence efavirenz pharmacokinetics, investigators conducted replication studies to examine their associations with efavirenz levels. However, a systematic evaluation of SNPs encompassing the majority of genetic variability in key genes implicated in efavirenz absorption, distribution, metabolism, and excretion (ADME) [20] has not been performed.
The goal of the current study was to examine the contributions of ADME gene polymorphisms to efavirenz exposure when modeled with nongenetic factors. To conduct this study, the association of literature-based SNPs and tagging SNPs selected to capture the majority of genetic variability in CYP2B6 and other implicated ADME genes with efavirenz exposure was examined in a diverse unselected population of HIV-infected women. The effects of these polymorphisms on short-term exposure (evaluated using plasma AUCs) and long-term exposure (evaluated using hair levels) to efavirenz are presented.
METHODS
Study Population and Protocol
The Women's Interagency HIV Study (WIHS) is a large, multicenter cohort study of women with or without HIV infection [5] and is highly representative of HIV-infected women in the United States in terms of factors such as age, race/ethnicity, and socioeconomic characteristics. We previously described the “WIHS Intensive PK Study,” in which 121 participants receiving efavirenz-based therapy underwent 24-hour sampling after a witnessed dose under conditions of routine use at steady-state [6]. The only eligibility criterion for participation in this study was use of an efavirenz-containing antiretroviral regimen.
Laboratory Procedures
Measurement of Short-Term Exposure: Efavirenz AUC Using Intensive Plasma Sampling
Procedures for measuring efavirenz blood levels have been described previously [21]. Plasma was analyzed for efavirenz by standard techniques of liquid chromatography/tandem mass spectrometry (LC/MS/MS) [22]. The absolute recovery of efavirenz from plasma was 99.8%, intraday and interday precision were each <11.7%, and accuracies range from −6.0% to 14.8% [21]. The plasma EFV assay was validated according to the current Food and Drug Administration guidelines for bioanalytical method validation, and all quality control samples were within 15% of their respective nominal value.
Measurement of Long-Term Exposure: Efavirenz Concentration in Hair
Methods to analyze efavirenz levels in hair have been described elsewhere [1]. A small thatch of hair (approximately 20 strands) is cut as close as possible to the scalp, and the distal portion is labeled. Extraction methods have been described and measurement of efavirenz is performed by LC/MS/MS [1]. This method has been validated for detection of a range of .05–20 nanograms of efavirenz per milligram of hair (ng/mg), with good linearity (R2>.99) and reproducibility (coefficient of variation, <15%).
Blood Collection and Genotyping
Of the 121 participants recruited, genomic DNA could be isolated from cell pellets and successfully genotyped for 111. Genotyping was performed by individuals who were blinded to outcomes, and positive and negative controls were included. Samples were genotyped using a combination of the GoldenGate genotyping platform (Illumina, San Diego, CA) and the TaqMan allelic discrimination assay (Applied Biosystems, Foster City, CA). GoldenGate genotype data were processed according to standard protocols (GenomeStudio, Illumina). Signal-intensity profiles and resulting genotype calls for each SNP were visually inspected by 2 blinded reviewers, with disagreements discussed until consensus was established.
Gene Selection
A literature search identified genes implicated in efavirenz ADME. The custom array was designed to interrogate each candidate gene by using literature-driven SNPs, as well as tagging SNPs. Tagging SNPs were selected to capture neighboring regions in high linkage disequilibrium (LD) across coding and noncoding regions of genes implicated in ADME. The 9 genes studied in each of the 111 participants included adenosine triphosphate–binding cassette protein (ABC) B1 (ABCB1), ABCC2, CYP2B6, CYP2C19, CYP2D6, CYP3A4, CYP3A5, solute carrier-like protein (SCL) A6 (SCL22A6), and uridyl diphosphate glucuronosyltransferase-1 family, polypeptide A1 (UGT1A1).
SNP Selection
Tagging SNPs were required to be common (defined as a minor allele frequency of ≥.05) in public databases (eg, HapMap). To ensure robust analyses, quality control filtering of SNPs was performed, and SNPs with call rates of <93.5% (n = 8) or Hardy-Weinberg P values of < .001 (n = 9) were excluded. SNPs with allele frequencies of <5% (n = 35) were excluded from analysis.
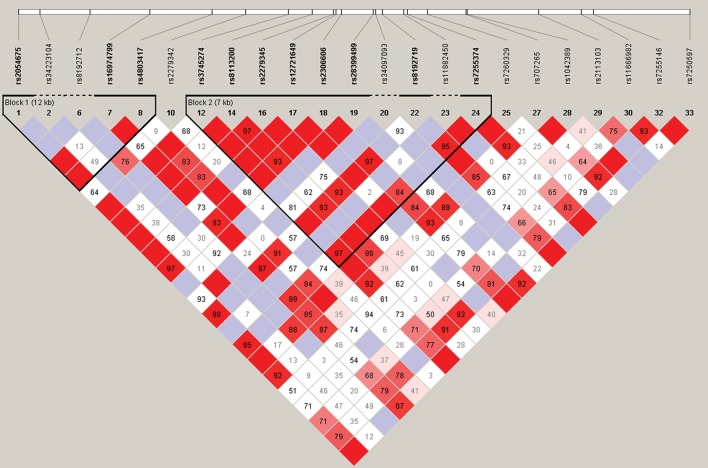
Overall, 182 SNPs among 9 candidate genes (ABCB1: 63 of 70 SNPs; ABCC2: 20 of 28 SNPs; CYP2B6: 23 of 38 SNPs; CYP2C19: 24 of 28 SNPs; CYP2D6: 5 of 7 SNPs; CY3A5/CYP3A4: 21 of 30 SNPs; SCL22A6: 5 of 8 SNPs; and UGT1A1: 22 of 26 SNPs) passed all quality control filters. Figure 1 shows the LD map of the literature-driven and tagging SNPs assessed in CYP2B6.
Figure 1.
Linkage disequilibrium (LD) map of single-nucleotide polymorphisms (SNPs) assessed in CYP2B6 in relationship to efavirenz exposure. An ideogram of CYP2B6 is presented at the top, which represents the physical distance along human chromosome 19 (46 187 595 to 46 230 064; genome build 36.3, contig NT_011109.15). Exons are represented as boxes, with coding regions rendered in gray and untranslated regions rendered in pink; gray lines connecting exons represent introns. Reference sequence (rs) identifiers for each SNP are plotted both in terms of their physical distance (ie, the white bar at the top of the figure) and also equidistantly to render the pairwise LD estimates, which were calculated and visualized with Haploview 4.2. The gene structure for CYP2B6 (ie, reference sequence NM_000767) was rendered with FancyGene 1.4. The correlation statistics (r2 and D’) are provided in the heat map. LD-based haplotype block definition was based on the D’ confidence interval. The haploblock is indicated in a bolded triangle, and its component SNPs are also rendered in bold font, with dashed regions indicating intervening SNPs that are not part of the haploblock. Pairwise D’ values (range, 0–1) were rendered in color, with darker red diamonds representing D’ values approaching 1.0, and progressively lighter red to pink colored diamonds representing progressively smaller D’ values. Grey diamonds represent pairwise D’ values of 1.0 but with log of odds values of <2 (ie, below the significance threshold of 2.0). When the r2 values (range, 0–100) are not equal to 0 or 100, they are provided in a given diamond.
Study Measurements
Outcome Variables
We evaluated factors impacting drug exposure, using 2 methods. First, exposure was assessed by calculation of the AUC divided by dose, in which the plasma AUC was calculated using the trapezoidal rule from 24-hour intensive PK studies, and the dose was the efavirenz dose witnessed at the start of the PK sampling. Second, we determined exposure by measuring efavirenz concentrations in small samples of hair, which is indicative of drug uptake over at least 1 month.
Predictor Variables
The 2 exposure outcomes were analyzed in relation to a number of factors that may influence PK. Variables included race, age, hepatitis B/C virus infection status, ovulatory cycle phase or menopausal status, pregnancy status, smoking status, alcohol and substance use, percentage of fat in the typical diet [23], diarrhea, other concurrent symptoms or infections, concomitant medications with potential drug-drug interactions, calculated creatinine clearance rate, body mass index and fat free mass, and results of hepatic function tests [6]. The models examining the hair exposure measure included self-reported adherence to antiretroviral therapy, as estimated by the percentage of prescribed doses consumed over 6 months (using visual analog scales). Level of adherence was categorized as ≤74%, 75%–94%, or ≥95% of prescribed doses.
The variables identified in our previous analyses that independently influenced exposure include age, alanine aminotransferase (ALT) level, albumin level, ideal body weight, orange or orange juice consumption in the preceding 5 days, amenorrhea for ≥12 months, tenofovir use, and race [6]. SNPs were first assessed in relationship to exposure in a bivariate fashion and then added to multivariate models with these nongenetic predictors. For the SNPs, allele and genotype frequencies were determined by gene counting, and Hardy-Weinberg equilibrium was assessed by the χ2 exact test. Measures of LD (ie, D’ and r2) were computed from the participants’ genotypes with Haploview 4.1. LD-based haplotype block definition was based on the D’ confidence interval (CI) [24].
For SNPs in the same haploblock, haplotype analyses were conducted to localize the association signal within each gene and to determine whether haplotypes improved the strength of the association. Haplotypes were constructed using the program PHASE, version 2.1 [25]. To improve the stability of haplotype inference, the haplotype construction procedure was repeated 5 times. Only haplotypes that were inferred with probability estimates of ≥0.85 across the 5 iterations and estimated to occur at a frequency of ≥20% were retained for downstream analyses.
Ancestry informative markers (AIMs) were used to estimate ancestry and as a tool to minimize bias due to population stratification [26]. Homogeneity in ancestry among participants was verified by cluster and principal component (PC) analysis [27]. The number of PCs were sought that distinguished the major racial/ethnic groups in the sample (ie, European, African, and Asian) by visual inspection of scatter plots of orthogonal PCs (ie, PC 1 vs PC2, and PC2 vs PC3). The first 3 PCs were selected to adjust for potential confounding due to population substructure (ie, race/ethnicity) by including them in all multiple regression models. AIMs and their PCs were available for 97 of 111 participant samples.
Statistical Analyses
All analyses were conducted using Stata (version 11.2, College Station, TX) and SAS (version 9.2, SAS Institute, Cary, NC). Descriptive statistics were used to summarize sample characteristics. The AUC and hair outcomes were log transformed for linear regression (modified for hair to account for undetectable levels [28]), but we added 1.0 to the dose-normalized hair level (hair level × 600 mg/dose) before taking logs, to prevent very low levels in hair from becoming large negative values. Four genetic models were assessed for each SNP: unstructured, additive, dominant, and recessive. The genetic model that best fit the data by maximizing the significance of the P value was selected for each SNP. For race/ethnicity, both genetic (AIM-derived PCs) and nongenetic (self-reported) parameters were examined. Since AIMS were only available in a subset of study participants (97 of 111), models with either self-reported race (African American or not) or both self-reported race and AIM PCs were also examined in the same subset and compared with the models in the full sample (n = 111). Although the precision of the estimates was typically less, the models including AIM-derived PCs were essentially the same as those including self-reported race alone.
The first set of models (Supplementary Table 1) show the effect of each individual SNP on log-transformed AUC over dose when combined with nongenetic factors previously shown to influence exposure. For the multivariate models in Tables 1 and 2, genetic predictors were selected in a forward stepwise manner, with adjustment for nongenetic factors, until no remaining genetic candidates met our a priori significance threshold of P < .001. To obtain a more parsimonious final model, we then applied backward elimination to the nongenetic factors until all of the remaining predictors demonstrated P values of <.05. Because efavirenz levels in hair were available for fewer participants, and to allow for a comparison of the findings, we modeled log levels in hair with the same final set of predictors as for log AUC/dose. We then evaluated each remaining genetic factor as a single addition to the hair models.
Table 1.
Factors Associated With Short-Term Efavirenz Exposure Among 111 Subjects From the Women's Interagency HIV Study
| Factor | Fold-Effect on AUC (95% CI) | P | Distribution of Factor in Sample |
|---|---|---|---|
| Oranges or orange juice in preceding 5 d | 1.26 (1.05–1.50) | .012 | Consumed by 76 (68.5%) |
| For every doubling of ALT level | 1.23 (1.11–1.36) | .0001 | Median ALT level, 23 IU/L (range, 8–117 IU/L) |
| CYP2B6 516 G>T (rs3745274) | … | 53 (47.7%), 0 doses | |
| 0 or 1 dose of minor allele (GG, GT) | 1.00 | 1.4 × 10−18 | 44 (39.6%), 1 dose |
| 2 doses of minor allele (TT) | 3.5 (2.7–4.5) | 14 (12.6%), 2 doses | |
| CYP2B6 983 T>C (rs28399499) 0 doses of minor allele (TT) 1 or 2 doses of minor allele (TC, CC) | …1.001.96 (1.54–2.5) | 2.2 × 10−10 | 95 (85.6%), 0 doses15 (13.5%), 1 dose1 (0.9%), 2 doses |
| ABCB1 haplotype (2 SNPs: rs7779562 and rs4148745) | … | 14 (12.6%), 0 doses | |
| 0 doses of the haplotype | 1.00 | 48 (43.2%), 1 dose | |
| 1 or 2 doses of the haplotype | 1.60 (1.24–2.1) | .0004 | 49 (44.1%), 2 doses |
Short-term exposure was evaluated in terms of the AUC per dose.
Abbreviations: ALT, alanine amino transferase; AUC, area under the concentration-time curve; CI, confidence interval.
Table 2.
Factors Associated With Long-Term Efavirenz Exposure Among 87 Subjects From the Women's Interagency HIV Study
| Factor | Effect on Hair (95% CI) | P | Distribution of Factor in Sample |
|---|---|---|---|
| Oranges or orange juice in preceding 5 d | 1.19 (.88–1.60) | .27 | Consumed by 57 (65.5%) |
| For every doubling of ALT level | 1.10 (.93–1.30) | .25 | Median ALT, 23 IU/L (range, 8–117 IU/L) |
| CYP2B6 516 G>T (rs3745274) | … | 38 (43.7%), 0 doses | |
| 0 or 1 dose of minor allele (GG, GT) | 1.00 | 1.0 × 10−10 | 36 (41.4%), 1 dose |
| 2 doses of minor allele (TT) | 3.2 (2.2–4.7) | 13 (14.9%), 2 doses | |
| CYP2B6 983 T>C (rs28399499) 0 doses of minor allele (TT) 1 or 2 doses of minor allele (TC,CC) | …1.001.70 (1.09–2.7) | .021 | 77 (88.5%), 0 doses9 (10.3%), 1 dose1 (1.1%), 2 doses |
| ABCB1 haplotype (2 SNPS: rs7779562 and rs4148745) | … | 11 (12.6%), 0 doses | |
| 0 doses of the haplotype | 1.00 | .31 | 38 (43.7%), 1 dose |
| 1 or 2 doses of the haplotype | 1.24 (.82–1.88) | 38 (43.7%), 2 doses | |
| Self-reported adherence≤74%75–94%≥95% | 1.00.94 (.45–1.96)1.10 (.56–2.2) | .88.77 | 4 (4.6%)14 (16.1%)69 (79.3%) |
Long-term exposure was evaluated in terms of the hair level per dose.
Abbreviations: ALT, alanine amino transferase; AUC, area under the concentration-time curve; CI, confidence interval.
RESULTS
Effect of Individual SNPs on Short-Term Efavirenz Exposure
We previously summarized the diverse characteristics of this intensively studied population, including a range of factors that could influence exposure [6]. Of the 111 participants in this study, 8% were white, 13% were Hispanic, 78% were African American, and 1% were of other races/ethnicities. The median age of the participants was 43.1 years (range, 20.6–60.4 years).
The mean efavirenz AUC among these participants was 78 μg × hours/mL; the median AUCs were 54 μg × h/mL (range, 11–519 μg × h/mL). The majority of the participants in this study were taking 600 mg of efavirenz daily, and only 4 of 111 were taking alternative doses.
A total of 230 polymorphisms were selected to interrogate the 9 ADME candidate genes; 48 SNPs failed to meet at least 1 quality control criterion. A total of 182 SNPs and 45 haplotypes were analyzed in relationship to exposure, using linear regression. Supplementary Table 1 shows the relationship between each SNP or haplotype and log-transformed AUC/dose when a single genetic predictor is added to models containing nongenetic predictors. Of the polymorphisms studied in each gene, 1 of 75 in ABCB1, 2 of 23 in ABCC2, 16 of 27 in CYP2B6, and 9 of 32 in UGT1A1 were associated with exposure with a P value of < .01 when combined with nongenetic predictors.
Multivariate Models of Factors Associated With Short-Term Exposure
By using forward selection for the genetic predictors, we generated a final multivariate model of the factors associated with efavirenz AUCs (Table 1). Table 1 also shows the distribution of the SNPs in the sample independently associated with AUC with a P value of <.001 in the final model. Of note, when genetic predictors were included, the only nongenetic predictors that remained independently associated with exposure were increases in the ALT level and consumption of oranges and/or orange juice in the past 5 days. The model explained 53% of the interpatient variability. The effects of genetic factors on exposure were similar (all estimates were within 3%) in the larger models prior to backward elimination, so only the parsimonious models are presented.
Of the SNPs and haplotypes examined, 3 were associated with short-term exposure. Individuals homozygous for the CYP2B6 516T rare allele (“TT” genotype) displayed 3.5-fold (P = 1.4 × 10−18) increases in AUC as compared to carriers of the common “G” allele (“TG” and “GG” genotypes). Individuals homozygous or heterozygous for the CYP2B6 983C common allele (“CC” or “CT” genotype) displayed 1.96-fold (P = 2.2 × 10−10) increases in AUC as compared to carriers of the minor “T” allele (“TT” genotype). A haplotype composed of the common alleles from 2 SNPs (“C” at rs7779562 and “G” at rs4148745) in the transporter ABCB1 was associated with AUC increases of 1.60-fold (P = .0004). Fitting the model for the exposure outcome of Cmax per dose instead of AUC per dose produced similar results. The direction of the association between elevated ALT level and exposure could not be assessed in these cross-sectional analyses. However, repeating the final models without ALT levels did not substantially alter the fixed predictor effects (orange juice consumption and genetic predictors) on exposure.
Multivariate Models of Factors Associated With Long-Term Exposure
Of the 111 participants, long-term exposure measures, as estimated by hair levels, were available for 87. The majority of these 87 participants were taking 600 mg of efavirenz daily; only 3 were receiving alternative doses. The mean concentration of efavirenz in hair was 5.92 ng/mg, and the median concentration was 3.11 ng/mg (range, <.05–41.4). Unlike the intensive PK studies conducted after witnessed dosing, long-term exposure may be affected by adherence, so a self-reported adherence measure was included in these models. Table 2 shows the final multivariate model in relationship to dose-normalized hair levels, along with the distribution of the covariates in the 87 participants. This model explained 34% of the interpatient variability. ALT level and orange juice consumption were not statistically significantly associated with hair levels, although the associations were in the same direction and the estimate for orange juice consumption was reduced only modestly. Self-reported adherence was also not substantially associated with hair concentrations.
The major CYP2B6 SNP independently associated with short-term efavirenz exposure was also associated with long-term exposure to efavirenz. Individuals homozygous for the CYP2B6 516T rare allele (“TT” genotype) displayed 3.2-fold increases (P = 1 × 10−10) in efavirenz levels in hair as compared to carriers of the common “G” allele, similar to the 3.5-fold increase seen with AUC. The effects of the CYP2B6 983CC/CT polymorphisms and the ABCB1 haplotype on efavirenz hair levels were in the same direction as their effects on AUC but with smaller estimated effects and larger P values.
We then evaluated each genetic predictor as a single addition to the hair model. One of the genetic factors (an ABCC2 SNP) not selected for in the AUC model met our threshold P value of <.001 when added to factors in Table 2: the unstructured model for this SNP (rs2002042) had an overall P value of 5.0 × 10−5, with those with 1 copy of the rare allele averaging 0.55-fold lower levels in hair than those with no copies (95% CI, .40–.75; P = .0002) and 0.38-fold lower levels than those with 2 copies (95% CI, .24–.61; P = .0001). This pattern did not hold in the corresponding model for AUC.
DISCUSSION
To our knowledge, this is the first report on the association of polymorphisms in CYP2B6 516 with long-term concentrations of efavirenz using hair samples. Since lifelong daily therapy is required for the treatment of HIV infection, studies that identify SNPs that can influence antiretroviral exposure over prolonged periods may guide dose-optimization strategies. Given the extent of treatment discontinuation, toxicities [29], and adherence difficulties in HIV-infected individuals, use of pharmacogenetics to personalize care for these individuals should improve treatment selection, dosing, and outcomes [30]. Indeed, HIV medicine is one of the few fields that routinely uses pharmacogenetic screening to reduce rates of a medication-associated reaction already.
With the availability of new genotyping technology, the paradigm in pharmacogenetics has shifted from examining a small number of genes for a limited number of allelic variants to systematically assessing SNPs in a range of genes relevant for ADME [20]. In a large heterogeneous population of HIV-infected patients, we performed a comprehensive search of 182 SNPs and 45 haplotypes in 9 genes implicated in efavirenz ADME and modeled them with nongenetic traits previously found to influence efavirenz AUC. Host traits were examined in combination with other factors that influence drug metabolism, to identify both genetic and nongenetic contributors to exposure.
Orange juice consumption and 2 additional SNPs (CYP2B6 983C and an ABCB1 haplotype, both of which have been found to influence plasma levels in the literature [15]) were associated with modest elevations in AUC, but less so with hair levels. Estimated effects on hair levels were generally smaller than those on AUCs, possibly reflecting imprecision in our self-reported adherence measure, as well as the need to add 1 before logarithmic transformation [28]. Inhibition of intestinal p-glycoprotein transport or downregulation of enteric CYP3A4 by citrus components in oranges or orange juice could lead to enhanced bioavailability and increased short-term exposure to efavirenz. We identified 1 additional genetic association (rs2002042 in the ABCC2 transporter) that influenced hair levels but not AUCs. The unusual pattern of heterozygotes having lower levels in hair than either type of homozygote must be regarded as preliminary. The most important finding of this study was the consistent effect of the CYP2B6 516T rare allele on tripling efavirenz exposure as measured via AUCs and hair levels.
Efavirenz is prescribed commonly for HIV infection, and a number of studies have examined the relationship between host traits and plasma efavirenz levels [15, 16, 18, 31–44]. Certain CYP2B6 SNPs have consistently been linked to higher plasma efavirenz levels and higher rates of early discontinuation [45, 46]. Individuals homozygous for the CYP2B6 516T rare allele (TT), referred to as “slow metabolizers,” consistently demonstrate higher plasma levels than those in individuals carrying the TG/GG genotypes. However, dose modification of efavirenz on the basis of the presence of SNPs that might transiently increase plasma levels is not routinely performed. Although authors have argued for the synthesis of pharmacogenetic testing with therapeutic drug monitoring to individualize dosing [47], the lack of a “gold standard” to monitor exposure to antiretroviral therapy and the uncertain significance of a SNP's influence on a plasma antiretroviral level remain significant barriers to dose-optimization protocols.
Plasma levels as a measure of exposure have a number of limitations, including insensitivity to day-to-day variation [48], dependence on the accurate reporting of last dose taken, intraindividual variations in diet, medications, and illicit substance use. Plasma levels may not reflect typical patterns of medication use and are subject to “white-coat” effects, in which adherence improves prior to medical visits [49]. Previous studies examining the relationship between genetic traits and efavirenz exposure have used single plasma levels or nonlinear mixed effects modeling of plasma concentrations to calculate population pharmacokinetic parameters as estimates of exposure. Short-term exposure is more robustly estimated by AUCs from intensive PK sampling, which was the measure used in this analysis. Although AUC measurement after witnessed dosing more accurately estimates exposure than single plasma levels, intensive PK sampling is cumbersome and expensive for the routine clinical setting.
We measured long-term drug exposure by monitoring efavirenz concentrations in small samples of hair. Because the concentrations of medications in hair reflect drug uptake from the systemic circulation over weeks to months, they are not subject to bias from the “white-coat” effect or inaccurate recall of the time at which the last dose was taken. Drug levels in hair therefore provide an advantage over plasma concentrations in estimating an average level of exposure over time [50] and provide a more feasible exposure measure in the clinical setting than AUCs. Genetic traits that influence both AUCs and hair concentrations of medications are likely to be durable in their effects.
Given the high rates of efavirenz discontinuation in clinical settings, identifying traits that lead to durable effects on exposure is important. This comprehensive search for SNPs in genes associated with efavirenz ADME demonstrated that individuals homozygous for the CYP2B6 516T (“TT” genotype) had >3-fold increases in short-term and long-term efavirenz exposure. This SNP's effect on exposure over the prolonged duration represented by hair levels is reported for the first time. Hair testing for exposure has feasibility and cost advantages over intensive PK sampling. Polymorphisms that affect exposure may be more important to assess in individuals whose capacity to clear a medication is already limited by nongenetic factors. Therefore, genetic testing coupled with hair measurement may be helpful in optimizing efavirenz dosing in the clinical setting, particularly when other risk factors for high exposure are present.
Supplementary Data
Supplementary materials are available at The Journal of Infectious Diseases online (http://jid.oxfordjournals.org/). Supplementary materials consist of data provided by the author that are published to benefit the reader. The posted materials are not copyedited. The contents of all supplementary data are the sole responsibility of the authors. Questions or messages regarding errors should be addressed to the author.
Notes
Acknowledgments. We recognize the late Lewis B. Sheiner, MD, for his many contributions to the field of pharmacokinetics and for the valuable advice he provided during the design of this study. We also thank Claudia Ponath, for her work as the coordinator for the intensive PK studies, and the WIHS participants who contributed to this study.
Data in this manuscript were collected by the WIHS Collaborative Study Group centers (principal investigators): New York City/Bronx Consortium (Kathryn Anastos); Brooklyn, NY (Howard Minkoff); Washington, DC, Metropolitan Consortium (Mary Young); The Connie Wofsy Study Consortium of Northern California (Ruth Greenblatt); Los Angeles County/Southern California Consortium (Alexandra Levine); Chicago Consortium (Mardge Cohen); and Data Coordinating Center (Stephen Gange).
Disclaimer. The contents of this publication are solely the responsibility of the authors and do not necessarily represent the official views of the National Institutes of Health (NIH).
Financial support. This work was supported by the National Institute of Allergy and Infectious Diseases (NIAID; K23 A1067065 to M. G. and R01 AI 65233 to R. M. G.). Additional funding was also provided by the National Center for Research Resources via the University of California San Francisco Clinical and Translational Science Institute (UL1 RR024131) and by the NIH Roadmap for Medical Research (KL2 RR024130 to B. A.).
The WIHS is funded by the NIAID (UO1-AI-35004, UO1-AI-31834, UO1-AI-34994, UO1-AI-34989, UO1-AI-34993, and UO1-AI-42590) and by the Eunice Kennedy Shriver National Institute of Child Health and Human Development (UO1-HD-32632). The study is cofunded by the National Cancer Institute, the National Institute on Drug Abuse, and the National Institute on Deafness and Other Communication Disorders.
Potential conflicts of interest. All authors: No reported conflicts.
All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1.Huang Y, Gandhi M, Greenblatt RM, Gee W, Lin ET, Messenkoff N. Sensitive analysis of anti-HIV drugs, efavirenz, lopinavir and ritonavir, in human hair by liquid chromatography coupled with tandem mass spectrometry. Rapid Commun Mass Spectrom. 2008;22:3401–9. doi: 10.1002/rcm.3750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Huang Y, Yang Q, Yoon K, et al. Microanalysis of the antiretroviral nevirapine in human hair from HIV-infected patients by liquid chromatography-tandem mass spectrometry. Anal Bioanal Chem. 2011;401:1923–33. doi: 10.1007/s00216-011-5278-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gandhi M, Bacchetti P, Ameli N, et al. Atazanavir concentration in hair is the strongest predictor of outcomes on antiretroviral therapy. Clin Infect Dis. 2011;52:1267–75. doi: 10.1093/cid/cir131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gandhi M, Ameli N, Bacchetti P, et al. Protease inhibitor levels in hair strongly predict virologic response to treatment. AIDS. 2009;23:471–8. doi: 10.1097/QAD.0b013e328325a4a9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barkan SE, Melnick SL, Preston-Martin S, et al. The women's interagency HIV study. WIHS collaborative study group. Epidemiology. 1998;9:117–25. [PubMed] [Google Scholar]
- 6.Gandhi M, Benet LZ, Bacchetti P, et al. Nonnucleoside reverse transcriptase inhibitor pharmacokinetics in a large unselected cohort of HIV-infected women. J Acquir Immune Defic Syndr. 2009;50:482–91. doi: 10.1097/qai.0b013e31819c3376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kappelhoff BS, Huitema AD, Yalvac Z, et al. Population pharmacokinetics of efavirenz in an unselected cohort of HIV-1-infected individuals. Clin Pharmacokinet. 2005;44:849–61. doi: 10.2165/00003088-200544080-00006. [DOI] [PubMed] [Google Scholar]
- 8.Molto J, Blanco A, Miranda C, et al. Variability in non-nucleoside reverse transcriptase and protease inhibitors concentrations among HIV-infected adults in routine clinical practice. Br J Clin Pharmacol. 2007;63:715–21. doi: 10.1111/j.1365-2125.2006.02834.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Smith CJ, Sabin CA, Youle MS, et al. Response to efavirenz-containing regimens in previously antiretroviral-naive HIV-positive patients: the role of gender. J Acquir Immune Defic Syndr. 2007;46:62–7. doi: 10.1097/QAI.0b013e31813e5e20. [DOI] [PubMed] [Google Scholar]
- 10.Manfredi R, Calza L, Chiodo F. Efavirenz versus nevirapine in current clinical practice: a prospective, open-label observational study. J Acquir Immune Defic Syndr. 2004;35:492–502. doi: 10.1097/00126334-200404150-00007. [DOI] [PubMed] [Google Scholar]
- 11.Dejesus E, Young B, Morales-Ramirez JO, et al. Simplification of antiretroviral therapy to a single-tablet regimen consisting of efavirenz, emtricitabine, and tenofovir disoproxil fumarate versus unmodified antiretroviral therapy in virologically suppressed HIV-1-infected patients. J Acquir Immune Defic Syndr. 2009;51:163–74. doi: 10.1097/QAI.0b013e3181a572cf. [DOI] [PubMed] [Google Scholar]
- 12.Reekie J, Reiss P, Ledergerber B, et al. A comparison of the long-term durability of nevirapine, efavirenz and lopinavir in routine clinical practice in Europe: a EuroSIDA study. HIV Med. 2011;12:259–68. doi: 10.1111/j.1468-1293.2010.00877.x. [DOI] [PubMed] [Google Scholar]
- 13.Marzolini C, Telenti A, Decosterd L, Greub G, Biollaz J, Buclin T. Efavirenz plasma levels can predict treatment failure and central nervous system side effects in HIV-1-infected patients. AIDS. 2001;15:71–5. doi: 10.1097/00002030-200101050-00011. [DOI] [PubMed] [Google Scholar]
- 14.Gutierrez F, Navarro A, Padilla S, et al. Prediction of neuropsychiatric adverse events associated with long-term efavirenz therapy, using plasma drug level monitoring. Clin Infect Dis. 2005;41:1648–53. doi: 10.1086/497835. [DOI] [PubMed] [Google Scholar]
- 15.Ribaudo HJ, Liu H, Schwab M, et al. Effect of CYP2B6, ABCB1, and CYP3A5 polymorphisms on efavirenz pharmacokinetics and treatment response: an AIDS clinical trials group study. J Infect Dis. 2010;202:717–22. doi: 10.1086/655470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rotger M, Colombo S, Furrer H, et al. Influence of CYP2B6 polymorphism on plasma and intracellular concentrations and toxicity of efavirenz and nevirapine in HIV-infected patients. Pharmacogen and Genom. 2005;15:1–5. doi: 10.1097/01213011-200501000-00001. [DOI] [PubMed] [Google Scholar]
- 17.Haas DW, Ribaudo HJ, Kim RB, et al. Pharmacogenetics of efavirenz and central nervous system side effects: an adult AIDS clinical trials group study. AIDS. 2004;18:2391–400. [PubMed] [Google Scholar]
- 18.di Iulio J, Fayet A, Arab-Alameddine M, et al. In vivo analysis of efavirenz metabolism in individuals with impaired CYP2A6 function. Pharmacogenet Genomics. 2009;19:300–9. doi: 10.1097/FPC.0b013e328328d577. [DOI] [PubMed] [Google Scholar]
- 19.Fellay J, Marzolini C, Meaden E, et al. Response to antiretroviral treatment in HIV-1-infected individuals with allelic variants of the multidrug resistance transporter 1: a pharmacogenetics study. Lancet. 2002;359:30–6. doi: 10.1016/S0140-6736(02)07276-8. [DOI] [PubMed] [Google Scholar]
- 20.Lubomirov R, Csajka C, Telenti A. ADME pathway approach for pharmacogenetic studies of anti-HIV therapy. Pharmacogenomics. 2007;8:623–33. doi: 10.2217/14622416.8.6.623. [DOI] [PubMed] [Google Scholar]
- 21.Egge-Jacobsen W, Unger M, Niemann CU, et al. Automated, fast, and sensitive quantification of drugs in human plasma by LC/LC-MS: quantification of 6 protease inhibitors and 3 nonnucleoside transcriptase inhibitors. Ther Drug Monit. 2004;26:546–62. doi: 10.1097/00007691-200410000-00014. [DOI] [PubMed] [Google Scholar]
- 22.Volosov A, Alexander C, Ting L, Soldin SJ. Simple rapid method for quantification of antiretrovirals by liquid chromatography-tandem mass-spectrometry. Clin Biochem. 2002;35:99–103. doi: 10.1016/s0009-9120(02)00286-2. [DOI] [PubMed] [Google Scholar]
- 23.Block G, Hartman AM, Naughton D. A reduced dietary questionnaire: development and validation. Epidemiology. 1990;1:58–64. doi: 10.1097/00001648-199001000-00013. [DOI] [PubMed] [Google Scholar]
- 24.Gabriel SB, Schaffner SF, Nguyen H, et al. The structure of haplotype blocks in the human genome. Science. 2002;296:2225–95. doi: 10.1126/science.1069424. [DOI] [PubMed] [Google Scholar]
- 25.Stephens M, Smith NJ, Donnelly P. A new statistical method for haplotype reconstruction from population data. Am J Hum Genet. 2001;68:978–89. doi: 10.1086/319501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Halder I, Shriver M, Thomas M, Fernandez JR, Frudakis T. A panel of ancestry informative markers for estimating individual biogeographical ancestry and admixture from four continents: utility and applications. Hum Mutat. 2008;29:648–58. doi: 10.1002/humu.20695. [DOI] [PubMed] [Google Scholar]
- 27.Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006;38:904–9. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 28.Thiébaut R, Jacqmin-Gadda H. Mixed models for longitudinal left-censored repeated measures. Comput Methods Programs Biomed. 2004;74:255–60. doi: 10.1016/j.cmpb.2003.08.004. [DOI] [PubMed] [Google Scholar]
- 29.Keiser O, Fellay J, Opravil M, et al. Adverse events to antiretrovirals in the Swiss HIV Cohort Study: effect on mortality and treatment modification. Antivir Ther. 2007;12:1157–64. [PubMed] [Google Scholar]
- 30.Wang L, McLeod HL, Weinshilboum RM. Genomics and drug response. N Engl J Med. 2011;364:1144–53. doi: 10.1056/NEJMra1010600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Arab-Alameddine M, Di Iulio J, Buclin T, et al. Pharmacogenetics-based population pharmacokinetic analysis of efavirenz in HIV-1-infected individuals. Clin Pharmacol Ther. 2009;85:485–94. doi: 10.1038/clpt.2008.271. [DOI] [PubMed] [Google Scholar]
- 32.Haas DW, Gebretsadik T, Mayo G, et al. Associations between CYP2B6 polymorphisms and pharmacokinetics after a single dose of nevirapine or efavirenz in African Americans. J Infect Dis. 2009;199:872–80. doi: 10.1086/597125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cabrera SE, Santos D, Valverde MP, et al. Influence of the cytochrome P450 2B6 genotype on population pharmacokinetics of efavirenz in human immunodeficiency virus patients. Antimicrob Agents Chemother. 2009;53:2791–8. doi: 10.1128/AAC.01537-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ribaudo HJ, Haas D, Tierney C, et al. Pharmacogenetics of plasma efavirenz exposure after treatment discontinuation: an adults AIDS clinical trials group study. Clin Infect Dis. 2006;42:401–7. doi: 10.1086/499364. [DOI] [PubMed] [Google Scholar]
- 35.Haas D, Ribaudo HJ, Kim RB, et al. Pharmacogenetics of efavirenz and central nervous system side effects: an adult AIDS clinical trials group study. AIDS. 2004;18:2391–400. [PubMed] [Google Scholar]
- 36.Haas D, Smeaton L, Shafer R, et al. Pharmacogenetics of long-term responses to antiretroviral regimens containing efavirenz and/or nelfinavir: an adult AIDS clinical trails group study. J Infect Dis. 2005;192:1931–42. doi: 10.1086/497610. [DOI] [PubMed] [Google Scholar]
- 37.Burger D, van der Heiden I, la Porte C, et al. Interpatient variability in the pharmacokinetics of the HIV non-nucleoside reverse transcriptase inhibitor efavirenz: the effect of gender, race, and CYP2B6 polymorphism. Br J Clin Pharmacol. 2006;61:148–54. doi: 10.1111/j.1365-2125.2005.02536.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sanchez A, Cabrera S, Santos D, et al. Population pharmacokinetic/pharmacogenetic model for optimization of efavirenz therapy in Caucasian HIV-infected patients. Antimicrob Agents Chemother. 2011;55:5314–24. doi: 10.1128/AAC.00194-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Maimbo M, Kiyotani K, Mushiroda T, Masimirembwa C, Nakamura Y. CYP2B6 genotype is a strong predictor of systemic exposure to efavirenz in HIV-infected Zimbabweans. Eur J Clin Pharmacol. 2012;68:267–71. doi: 10.1007/s00228-011-1118-0. [DOI] [PubMed] [Google Scholar]
- 40.Gupta SK, Rosenkranz SL, Cramer YS, et al. The pharmacokinetics and pharmacogenomics of efavirenz and lopinavir/ritonavir in HIV-infected persons requiring hemodialysis. AIDS. 2008;22:1919–27. doi: 10.1097/QAD.0b013e32830e011f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lindfelt T, O'Brien J, Song JC, Patel R, Winslow DL. Efavirenz plasma concentrations and cytochrome 2B6 polymorphisms. Ann Pharmacother. 2010;44:1572–8. doi: 10.1345/aph.1P141. [DOI] [PubMed] [Google Scholar]
- 42.Rodriguez-Novoa S, Barreiro P, Rendon A, Jimenez-Nacher I, Gonzalez-Lahoz J, Soriano V. Influence of 516G>T polymorphisms at the gene encoding the CYP450–2B6 isoenzyme on efavirenz plasma concentrations in HIV-infected subjects. Clin Infect Dis. 2005;40:1358–61. doi: 10.1086/429327. [DOI] [PubMed] [Google Scholar]
- 43.Tsuchiya K, Gatanaga H, Tachikawa N, et al. Homozygous CYP2B6 *6 (Q172H and K262R) correlates with high plasma efavirenz concentrations in HIV-1 patients treated with standard efavirenz-containing regimens. Biochem Biophys Res Commun. 2004;319:1322–6. doi: 10.1016/j.bbrc.2004.05.116. [DOI] [PubMed] [Google Scholar]
- 44.Maimbo M, Kiyotani K, Mushiroda T, Masimirembwa C, Nakamura Y. CYP2B6 genotype is a strong predictor of systemic exposure to efavirenz in HIV-infected Zimbabweans. Eur J Clin Pharmacol. 2012;68:267–71. doi: 10.1007/s00228-011-1118-0. [DOI] [PubMed] [Google Scholar]
- 45.Wyen C, Hendra H, Siccardi M, et al. Cytochrome P450 2B6 (CYP2B6) and constitutive androstane receptor (CAR) polymorphisms are associated with early discontinuation of efavirenz-containing regimens. J Antimicrob Chemother. 2011;66:2092–8. doi: 10.1093/jac/dkr272. [DOI] [PubMed] [Google Scholar]
- 46.Lubomirov R, Colombo S, di Iulio J, et al. Association of pharmacogenetic markers with premature discontinuation of first-line anti-HIV therapy: an observational cohort study. J Infect Dis. 2011;203:246–57. doi: 10.1093/infdis/jiq043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rotger M, Telenti A. Optimizing efavirenz treatment: CYP2B6 genotyping or therapeutic drug monitoring? Eur J Clin Pharmacol. 2008;64:335–6. doi: 10.1007/s00228-007-0440-z. [DOI] [PubMed] [Google Scholar]
- 48.Nettles RE, Kieffer TL, Parsons T, et al. Marked intraindividual variability in antiretroviral concentrations may limit the utility of therapeutic drug monitoring. Clin Infect Dis. 2006;42:1189–96. doi: 10.1086/501458. [DOI] [PubMed] [Google Scholar]
- 49.Podsadecki TJ, Vrijens BC, Tousset EP, Rode RA, Hanna GJ. White coat compliance limits the reliability of therapeutic drug monitoring in HIV-1-infected patients. HIV Clin Trials. 2008;9:238–46. doi: 10.1310/hct0904-238. [DOI] [PubMed] [Google Scholar]
- 50.Beumer JH, Bosman IJ, Maes RA. Hair as a biological specimen for therapeutic drug monitoring. Int J Clin Pract. 2001;55:353–7. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.