Abstract
Friend of GATA (FOG) proteins regulate GATA factor-activated gene transcription. During vertebrate hematopoiesis, FOG and GATA proteins cooperate to promote erythrocyte and megakaryocyte differentiation. The Drosophila FOG homologue U-shaped (Ush) is expressed similarly in the blood cell anlage during embryogenesis. During hematopoiesis, the acute myeloid leukemia 1 homologue Lozenge and Glial cells missing are required for the production of crystal cells and plasmatocytes, respectively. However, additional factors have been predicted to control crystal cell proliferation. In this report, we show that Ush is expressed in hemocyte precursors and plasmatocytes throughout embryogenesis and larval development, and the GATA factor Serpent is essential for Ush embryonic expression. Furthermore, loss of ush function results in an overproduction of crystal cells, whereas forced expression of Ush reduces this cell population. Murine FOG-1 and FOG-2 also can repress crystal cell production, but a mutant version of FOG-2 lacking a conserved motif that binds the corepressor C-terminal binding protein fails to affect the cell lineage. The GATA factor Pannier (Pnr) is required for eye and heart development in Drosophila. When Ush, FOG-1, FOG-2, or mutant FOG-2 is coexpressed with Pnr during these developmental processes, severe eye and heart phenotypes result, consistent with a conserved negative regulation of Pnr function. These results indicate that the fly and mouse FOG proteins function similarly in three distinct cellular contexts in Drosophila, but may use different mechanisms to regulate genetic events in blood vs. cardial or eye cell lineages.
Members of the GATA family of transcription factors regulate gene expression during the development of a variety of tissues (reviewed in refs. 1 and 2). Functional studies of the GATA N-terminal zinc-finger domain have identified a class of interacting transcriptional coregulators, the Friend of GATA (FOG) family of multitype zinc-finger proteins (3–8). FOG modulation of GATA factor-regulated transcription entails repression or activation of gene expression, depending on the developmental context (3–11). Repression of GATA-1 transcriptional activation by FOG-1 can be mediated through a conserved PXDL amino acid motif, which binds the transcriptional corepressor C-terminal binding protein (CtBP) (11–13). The GATA-FOG regulatory paradigm extends across tissues and taxonomies. In mice, two FOG genes with largely nonoverlapping expression patterns have been identified (6–8). FOG-1 interacts with GATA-1 during hematopoiesis to promote erythrocyte and megakaryocyte differentiation (3, 9, 10). FOG-2 has been shown to repress GATA-4 activation of cardiac promoters in transient transfection assays and is required for cardiac morphogenesis and vascularization (7, 8, 14, 15). In contrast, a single FOG gene has been identified in Xenopus and Drosophila. The expression pattern of Xenopus FOG (xFOG) overlaps that of all six Xenopus GATA factors and, similar to murine FOG-1, appears to regulate erythropoiesis (16). The Drosophila FOG homologue U-shaped (Ush) interacts with the GATA-4 homologue Pannier (Pnr) to regulate heart and sensory bristle development (5, 17).
Ush is also expressed in hemocyte precursors during embryogenesis (17). These cells give rise to two classes of embryonic hemocytes, plasmatocytes and crystal cells (18, 19). Plasmatocytes constitute 90–95% of circulating hemocytes and are macrophage-like cells. Crystal cells, named for their crystalline inclusions, are involved in the defense-related melanization of encapsulated targets (20). During hematopoiesis, the acute myeloid leukemia 1 (AML-1) homologue Lozenge (Lz) and Glial cells missing (Gcm) are required for the production of crystal cells and plasmatocytes, respectively (21). However, additional factors have been predicted to control crystal cell proliferation.
Considering that Ush is expressed in hemocyte precursors and is structurally and functionally similar to FOG-1 and FOG-2 (7, 8, 11, 12, 17, 22, 23), we wanted to determine its function during hematopoiesis. In this report we show that Ush is expressed in hemocyte precursors and plasmatocytes and functions to limit crystal cell production. Furthermore, this function may be conserved evolutionarily and require CtBP because misexpression of FOG-1 and FOG-2 repressed crystal cell production, whereas a mutant version of FOG-2 lacking the CtBP-binding motif did not. This hematopoietic function of FOG proteins is in contrast to that of Lz and may represent an intersection between the FOG and AML-1 hematopoietic regulatory pathways.
To determine the extent of the functional conservation between the mouse and fly FOG family members, we also coexpressed Ush, FOG-1, FOG-2, or the mutant version of FOG-2 with Pnr during heart and eye development. We observed severe eye and heart phenotypes, consistent with a conserved negative regulation of Pnr transcriptional activity. These results indicate that Ush and the murine FOG proteins function similarly in three distinct cellular contexts during Drosophila development, but may use different mechanisms to regulate genetic events in blood vs. cardial or eye cell lineages.
Materials and Methods
Fly Strains.
y w67c23 served as our wild-type stock. The ush strain used in this study, ush1/SM6, Roi, eve-lacZ, upstream activation sequence (UAS)Ush, and twistGal4 (twiGal4) are described elsewhere (17). The lzGal4 driver (21) was a gift from John Pollock (Carnegie–Mellon University, Pittsburgh). The eyelessGal4 (eyGal4) driver, the srp allele ru1 h1 th1 st1 cu1 srp3 sr1 es ca1/TM3, Sb1 Ser1, the Black cell (Bc) allele Bc1/CyO, and the UASlacZ allele w1118; P{w+mC = UAS-lacZ.NZ}J312 were obtained from the Bloomington Stock Center (Bloomington, IN). lzlacZ embryos and larvae were obtained from mating lzGal4 and UASlacZ strains (21). To generate UASFOG-1, UASFOG-2, and UASΔFOG-2 strains, mouse cDNAs for FOG-1, FOG-2, and ΔFOG-2 were cloned into the P element vector pUAST and injected into y w67c23embryos. Multiple transgenic lines were established by using standard transformation procedures (24) and characterized for each wild-type or mutant sequence. Several lines were tested and showed a moderate to strong dorsal vessel phenotype, similar to that observed with UASUsh (17).
Immunohistochemical Staining of Embryos and Larvae.
Collection, fixation, and immunohistochemical staining of embryos was performed as described previously (24). Dissected larval lymph glands were processed (25) and stained immunohistochemically by using the procedure adapted for embryos. A polyclonal antibody to Drosophila Ush was produced by injecting rabbits with two synthetic peptides: Ush-(231–250) (CSHRIKDTDEAGSDKSGAGG) and Ush-(1174–1191) (VGGHGQQKNKENLQEAAI) (Alpha Diagnostics, San Antonio, TX). A 1:2,000 dilution of the antisera produced an immunostaining pattern similar to that previously reported for ush cRNA detected by in situ hybridization (17). Anti-Srp antibody produced in rabbits was a gift from Mark Brennen (26) and was used at a 1:1,000 dilution. Fluorescent double-antibody labeling of lzlacZ embryos was performed by using anti-Ush and anti-β-galactosidase primary antibodies, followed by Cy3-conjugated donkey anti-rabbit antibody (Jackson ImmunoResearch) and Oregon green 488-conjugated goat anti-mouse antibody (Molecular Probes). Fluorescence was captured and recorded by using an Olympus Fluoview FV500 laser-scanning microscope (Olympus, New Hyde Park, NY).
Gene Expression Analyses in Mutant and Gal4/UAS Embryos.
Embryos obtained from the mating of lzGal4, twiGal4, or eyGal4 and UASUsh, UASFOGS, or UASlzlacZ flies were collected at 29°C, whereas all other embryos were collected at 23°C. To analyze crystal cell production in ush mutant embryos by using the Bc mutation (27), a stock of the genotype ush1 Bc/SM6, Roi, eve-lacZ was generated. A two-generation cross was required to assay lzlacZ expression in ush mutant embryos. In the first generation, ush1/SM6, Roi, eve-lacZ females were crossed to lzGal4 males. The F1 female progeny were either ush1/lzGal4 or ush1 lzGal4/+ and were crossed to ush1/+ ; UASlacZ/+ males, generated by mating ush1/SM6, Roi, eve-lacZ females to UASlacZ males.
To analyze the effect of forced FOG expression on the production of crystal cells, lzlacZ (lzGal4/+ ; UASlacZ/+) females were crossed to either UASUsh, UASFOG-1, UASFOG-2, UASFOG-2ΔPIDL, or y w67c23males. Pan-mesodermal expression of FOG transgenes was achieved by crossing twiGal4 virgin females to the same males. eyGal4 virgin females were mated to UASUsh, UASFOG-1, UASFOG-2, UASFOG-2ΔPIDL, or y w67c23males to misexpress the FOG transgenes in the eye imaginal disk, and eye and antennae production was determined in adult males and females.
Results
Ush Expression During Hematopoiesis.
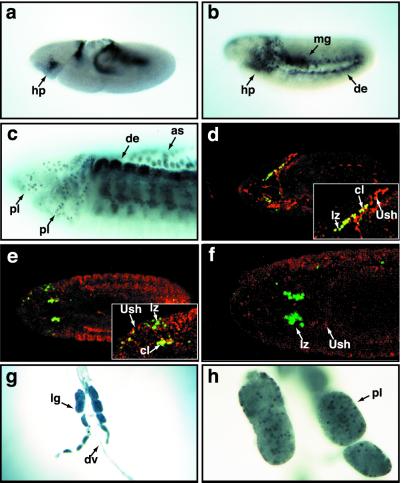
Using an antibody directed against Ush synthetic peptides, we detected Ush protein in an expression pattern similar to that previously reported for the gene transcript. Around embryonic stage 8, both ush RNA and protein can be detected in blood cell precursors (ref. 17; Fig. 1a). By stage 10, Ush-positive hemocyte precursors have spread throughout the lateral and ventral head mesoderm (Fig. 1b). As embryogenesis progressed, we detected Ush in stage 13 plasmatocytes migrating throughout the head mesoderm (Fig. 1c) and down the ventral midline. During the late stages of embryogenesis, Ush continued to be expressed in plasmatocytes circulating throughout the embryonic hemolymph (data not shown).
Figure 1.
Ush is expressed in Drosophila blood cell lineages. (a–c) Immunolocalization of Ush protein in stage 8 (a), stage 10 (b), and stage 13 (c) embryos, lateral views. (d–f) Immunolocalization of β-galactosidase (green) and Ush (red) in lzlacZ stage 10 (d), stage 13 (e), and stage 15 (f) embryos. d and e Insets show colocalization of lz and Ush expression (yellow) in lzlacZ embryos. (g) Larval lymph gland and dorsal vessel (×10). (h) Three of the larval lymph nodes (×40). as, amnioserosa; cl, colocalization; de, dorsal ectoderm; dv, dorsal vessel; hp, hemocyte precursor; lg, lymph gland; lz, lzlacZ; mg, midgut; pl, plasmatocytes.
lz expression in crystal cells is detected first during stage 10 and is maintained in this lineage until the late stages of embryogenesis (21). We used fluorescent antibody staining and confocal microscopy to determine whether Ush and lz were coexpressed in the crystal cell lineage. To detect lz expression in hemocyte precursors and crystal cells, we monitored the expression of a UASlacZ reporter gene driven by lzGal4 (lzlacZ). This reporter is active in hemocyte precursors as early as stage 10 and is expressed in the crystal cell lineage throughout embryogenesis (21). During embryonic stage 10, a number of hemocyte precursors expressed both Ush and lz (Fig. 1d). Later, during stage 13, the number of cells that expressed both lz and Ush decreased (Fig. 1e). Finally, during the late stages of embryogenesis, Ush was not detected in crystal cell lineage, evidenced by its failure to colocalize with the lzlacZ crystal cell marker (Fig. 1f). These results are consistent with a role for ush as a repressor of crystal cell production and suggest that ush expression is down-regulated in hemocyte precursors during crystal cell lineage commitment.
During larval development, hematopoiesis takes place in the larval lymph glands, which flank the dorsal vessel. Plasmatocytes are specified and develop in the primary and secondary lobes of the gland, whereas crystal cells develop exclusively in the primary lobe (21). We detected Ush in most cells of primary and secondary lobes (Fig. 1 g and h), consistent with expression in the plasmatocyte lineage. The protein was expressed in a differential pattern in the cells of the lymph glands (Fig. 1h), perhaps indicative of down-regulation during hemocyte precursor commitment. This may be analogous to the down-regulation of murine FOG-1 that is required for eosinophil and myeloid lineage differentiation (3, 28).
Srp Is Required for Ush Hematopoietic Expression.
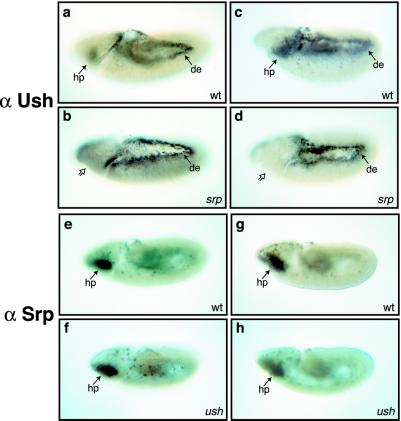
Srp function is required for hemocyte development and for differentiation of plasmatocytes and crystal cells (21, 29, 30). Furthermore, studies using amorphic alleles of srp indicate that it is required for hemocyte precursor specification (29). Srp is expressed first in the hemocyte precursors during embryonic stage 5, and, similar to Ush, its expression is maintained in plasmatocytes throughout embryogenesis (21, 30). To determine whether an epistatic relationship exists between srp and ush, we assayed Ush expression in srp mutant embryos and Srp expression in ush mutant embryos (Fig. 2). We used the hypomorphic allele srp3, which results in the production of hemocyte precursors, even with the reduction of Srp function (30). In srp embryos, Ush was not detected in hemocyte precursors, plasmatocytes, or midgut (Fig. 2 b and d), unlike the wild-type expression pattern (Fig. 2 a and c). In contrast, Srp was observed in hemocyte precursors and plasmatocytes in both wild-type (Fig. 2 e and g) and ush mutant embryos (Fig. 2 f and h). This result suggests ush resides downstream of srp in the hematopoiesis hierarchy and ush expression requires Srp function. Furthermore, ush is not required for the specification of hemocyte precursors or plasmatocytes, because these Srp-positive cells were detected in ush mutant embryos. Finally, wild-type levels of ush were present in the dorsal ectoderm of srp mutant embryos (Fig. 2 b and d), indicating that dynamic ush expression is under the control of multiple regulators during embryogenesis.
Figure 2.
The epistatic relationship between Ush and Srp. Ush is detected in the hemocyte precursors (hp) of wild-type (a and c) but not srp mutant (b and d) embryos, whereas expression remains unaltered in the dorsal ectoderm (de) of both genotypes. In contrast, the expression of Srp (e–h) is unaltered in ush mutant embryos (f and h), compared with wild-type embryos (e and g). Lateral views of stage 9 (a, b, e, and f) and stage 10 (c, d, g, and h) embryos. Solid arrows indicate tissues in which protein expression is detected. Open arrows indicate tissues in which Ush is not detected.
Ush Functions to Prevent the Overproduction of Crystal Cells.
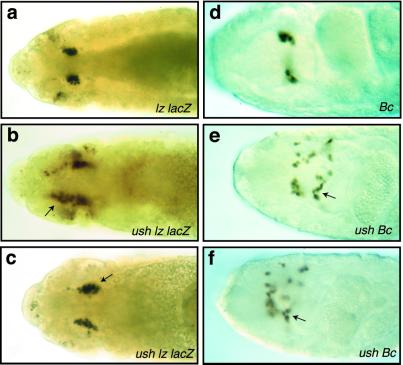
Previous studies have shown that ush functions to prevent the overproduction of sensory bristles, cardial cells, and pericardial cells (5, 17). These observations, together with our findings that ush appeared to be down-regulated during crystal cell lineage commitment (Fig. 1), suggested that Ush may act to limit crystal cell production. To test this hypothesis, we assayed for increased numbers of crystal cells in ush mutant embryos. Crystal cells are localized in a bilateral cluster of cells within the head mesoderm and require lz expression from embryonic stage 10 through 14 for their development (21). The lzlacZ genotype served as a crystal cell marker. Expression of lzlacZ was assayed in stage 13 embryos because during this stage the germ-band retraction phenotype can be used to distinguish ush mutant from wild-type embryos. Homozygous ush embryos showed an increase in the number of lzlacZ-expressing cells (Fig. 3 b and c) compared with the wild-type control (Fig. 3a). We confirmed these results by using embryos harboring the Bc mutation, which renders crystal cells visible in late-stage homozygous embryos (27). Crystal cell production in ush Bc embryos was compared with the Bc parental strain, which has the wild-type ush allele. Again, homozygous ush Bc embryos had an increase in the number of crystal cells (Fig. 3 e and f) compared with the Bc embryos from the parental strain (Fig. 3d). Because the number of crystal cells in wild-type embryo populations can vary more than 2-fold, we sampled 20 wild-type and 20 ush embryos and showed a 30% overall increase in the number of crystal cells by using either the lzlacZ or Bc marker. These results indicate that Ush functions to repress crystal cell production during hematopoiesis.
Figure 3.
ush gene functions to limit crystal cell production. (a–c) Dorsal view of stage 13 embryos stained for β-galactosidase protein to assay lzlacZ reporter gene expression. (a) lzlacZ/+ embryo. (b and c) ush lzlacZ/+ embryos. (d) Dorsal view of stage 16 homozygous Bc embryo. (e and f) Dorsal view of stage 16 homozygous ush Bc embryos. Arrows point to the increased numbers of crystal cells in ush homozygous mutant embryos.
Misexpression of FOG Transgenes Represses Crystal Cell Production.
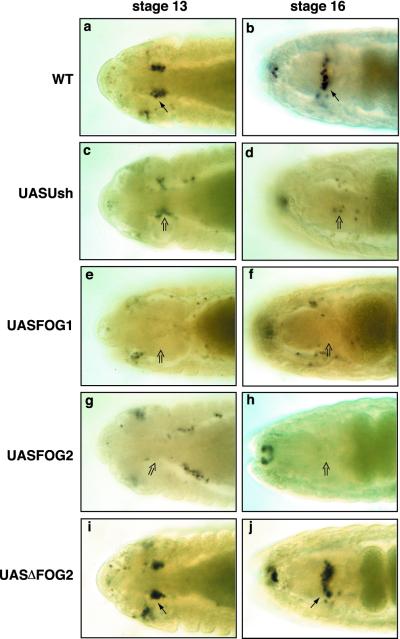
To demonstrate further that Ush repressed crystal cell production, we expressed Ush in crystal cells by using the Gal4/UAS binary system (31). The lzGal4 driver was used to express UASUsh in crystal cells, and their production was monitored by using the lzlacZ marker. Embryos with forced expression of Ush in crystal cells had a significant reduction in the number of these cells. Compared with similarly staged wild-type controls (Fig. 4 a and b), UASUsh stage 13 and 16 embryos (Fig. 4 c and d) had a 30% and 85% reduction in number of crystal cells, respectively. A sample of 40 stage 13–16 UASUsh embryos averaged a 30% reduction in the number of crystal cells compared with wild-type controls. The phenotype of individual embryos within this population ranged from being completely devoid of crystal cells to wild-type cell numbers. These results indicate that down-regulation of ush during crystal cell lineage commitment is required for development of these cells. Together with the observed increase in crystal cell number in ush loss-of-function assays, these findings suggest that Ush functions during hematopoiesis to limit the number of hemocyte precursors that enter the crystal cell lineage.
Figure 4.
FOG transgenes repress crystal cell production. (a and b) WT, wild-type embryos. (c–j) UASFOG embryos expressing FOG transgenes in crystal cells under the control of the lzGal4 driver. Dorsal view of stage 13 (a, c, e, g, and i) and stage 16 (b, d, f, h, and j) embryos stained for β-galactosidase protein to assay lzlacZ reporter gene expression. Misexpression of UASUsh (c and d), UASFOG-1 (e and f), or UASFOG-2 (g and h) in crystal cells reduced the number of these cells compared with similarly staged wild-type controls (a and b). Expression of UASΔFOG-2 (i and j) in crystal cells had no effect on crystal cell production. Arrows indicate wild-type numbers of crystal cells; open arrows indicate reduced numbers of crystal cells.
Considering that Ush is structurally and functionally similar to murine FOG-1 and FOG-2 (7, 8, 11, 12, 17, 22, 23), we wanted to determine whether expression of UASFOG-1 and UASFOG-2 would repress crystal cell production. Forced expression of either of these proteins (Fig. 4 e–h) reduced the average number of cells in stage 13–16 UASFOG-1 and UASFOG-2 embryos by 50% overall compared with wildtype controls. As with UASUsh, we observed a range of phenotypes from wild-type cell numbers to embryos completely devoid of crystal cells. Approximately 23% of the UASFOG-1 and UASFOG-2 embryos had no detectable crystal cells. All members of the FOG family of proteins characterized to date have a consensus PXDL amino acid sequence, which binds to the transcriptional corepressor CtBP (11, 16). Forced expression of a mutant form of FOG-2 with the CtBP interaction domain deleted (UASΔFOG-2) had no effect on crystal cell production (Fig. 4 i and j). Taken together, these data suggest that the mechanism of crystal cell repression by FOG class proteins is conserved and likely requires a corepressor such as CtBP.
FOG Transgenes Inhibit Cardial Cell Production.
In Drosophila, forced mesodermal expression of ush inhibits cardial cell production, presumably by negatively regulating Pnr function (17, 24). Given that FOG-1 and FOG-2 repress crystal cell production, we wanted to determine the effect of forced mesodermal expression of UASFOG-1, UASFOG-2, and UASΔFOG-2 on cardial cell production. The myogenic differentiation factor D-MEF2 was used to assess the status of cardial cells (32). Late-stage wild-type embryos have two contiguous rows of 52 cells present in the forming or mature dorsal vessel (Fig. 5a). In contrast, we observed a reduction in the number of cardial cells in embryos with twiGal4-driven mesodermal expression of any one of the four FOG transgenes (UASUsh, UASFOG-1, UASFOG-2, or UASΔFOG-2; Fig. 5 b–e). An average of 40% of embryos had gaps in the heart tube ranging from 10 cells missing to a complete absence of cardial cells. All four transgenes produced embryo populations with a similar range and severity of cardial cell reduction. These results indicate that the inhibitory function of the FOG proteins in this heart-formation assay is conserved. Furthermore, FOG proteins inhibit cardial cell production in the absence of the CtBP interaction domain, suggesting either that this corepressor is not required or that different corepressors operate in the cardial cell and hemocyte lineages.
Figure 5.
FOG transgenes inhibit cardial cell production. (a) WT, wild-type stage 16 embryo. (b–e) UASFOG stage 16 embryos expressing FOG transgenes in cardial cells under the control of the twiGal4 driver. Embryos are stained for D-MEF2 protein. Forced pan mesodermal expression of FOG transgenes results in a reduced number of cardial cells in the forming dorsal vessel. Arrows point to missing cardial cells.
FOG Transgenes Perturb Eye Development.
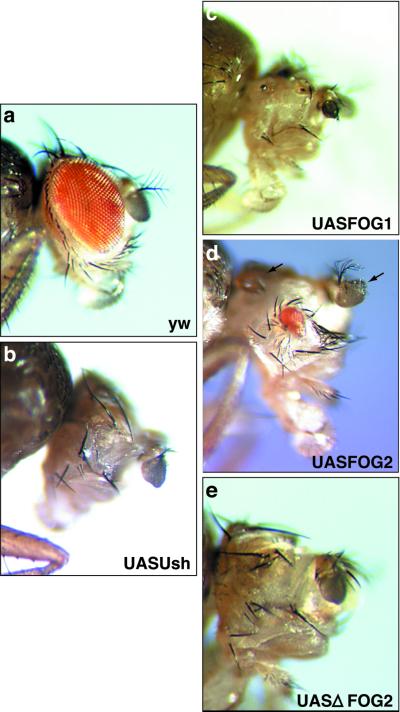
Pnr is also expressed at the dorsal margin of the eye imaginal disk and acts upstream of wingless to control proper eye and head capsule development (33). In the wild-type adult, the ommatidia have a distinct polarity and there is a single pair of antennae (Fig. 6a). eyGal4-driven expression of any one of the four FOG transgenes in the eye disk produces phenotypes comparable to those observed in pnr mutants (33). These phenotypes include ectopic production of antennae (Fig. 6d), reduction or loss of the eye (Fig. 6 b–e), disrupted ommatidial polarity, and death of ≈80% of the population before eclosion. These results are similar to those observed in animals with pnrGal4-driven misexpression of Ush (33). Our observations suggest that forced expression of FOG proteins in the eye disk inhibits the function of Pnr and that this perturbation of eye development does not require interaction with a protein that binds the PXDL sequence.
Figure 6.
FOG transgenes perturb eye development. (a) Adult progeny of a cross between eyGal4 virgins and y w males. (b–e) Adult flies with eyGal4-driven misexpression of any one of the four FOG transgenes (UASUsh, UASFOG-1, UASFOG-2, or UASΔFOG-2) in the eye imaginal disk showing reduction or loss of eye. Arrows indicate ectopic production of antennae.
Discussion
In this report we have shown that Ush is expressed in hemocyte precursors and plasmatocytes throughout embryogenesis and during larval development. Srp acts upstream of Ush during hematopoiesis, and Srp-positive plasmatocytes were observed in ush mutant embryos, indicating that Ush is not required for plasmatocyte specification and migration. Nonetheless, the observation that Ush is expressed in the plasmatocyte lineage suggests it is required for some aspect of plasmatocyte development. Ush is down-regulated during crystal cell lineage commitment, consistent with a role for the protein as a negative regulator of crystal cell production. Genetic studies demonstrated further that Ush limits crystal cell production and that this function may be conserved evolutionarily among the FOG proteins. Moreover, repression of crystal cell production by FOG proteins appears to require CtBP. In contrast, forced expression of the FOG factors repressed cardial cell production and disrupted eye development in the absence of the binding site for this transcriptional corepressor.
Lebestky et al. (21) recently reported that Srp acts upstream of two lineage-specific transcription factors, Gcm and Lz. In this model of Drosophila hematopoiesis, Srp-positive hemocyte precursors give rise to a large subpopulation of Gcm-positive cells (plasmatocyte lineage) and a smaller subpopulation of Lz-positive cells (crystal cell lineage). These authors reported that misexpression of Gcm in crystal cells can change their fate to that of the plasmatocyte lineage. However, they also reported that lz expression did not change, nor did the number of crystal cells increase, in gcm loss-of-function mutants (21). This result indicates that other factors are required to restrict the number of hemocyte precursors that enter crystal cell lineage commitment. Crystal cell numbers increased in ush mutant embryos and decreased in embryos with lz enhancer-directed misexpression of Ush. Thus, Ush may function to restrict the size of the crystal cell population by regulating crystal cell lineage commitment.
Recent studies have indicated that FOG proteins may function to regulate the commitment of several hematopoietic lineages. Ectopic expression of FOG proteins mFOG-1, mFOG-2, and xFOG (Xenopus FOG) early in Xenopus development represses red blood cell formation, possibly by down-regulating Gata-1 expression (16). These data suggest that FOG proteins may act to limit the differentiation of erythrocytes to prevent depletion of pluripotent stem cells (16). Furthermore, by using an in vitro avian hematopoietic differentiation system, Querfurth et al. (28) demonstrated that FOG-1 represses eosinophil-specific gene expression and that forced expression of FOG-1 in eosinophils produces a multipotent precursor phenotype. Thus, down-regulation of FOG-1 in multipotent hematopoietic precursors is an essential step in eosinophil differentiation (28). In addition to our findings that misexpressed Ush repressed crystal cell production, our data showing FOG-1 and FOG-2 also repressed crystal cell number indicate that the mechanism by which these proteins limit crystal cell number may be conserved. Taken together, studies using the Drosophila and vertebrate systems suggest that FOG proteins function to preserve the multipotent hemocyte precursor pool by controlling the lineage commitment of specific cell types.
An additional factor that may be required to control lineage commitment is CtBP. This transcriptional corepressor may interact with FOG-1 and FOG-2 to repress erythrocyte differentiation, because a mutant version of FOG-2 lacking the consensus PXDL sequence failed to repress erythrocyte differentiation when ectopically expressed during Xenopus development (16). We have shown that CtBP may be required for FOG protein repression of crystal cell production. It is noteworthy that CtBP likely functions during Drosophila hematopoiesis because a lacZ reporter gene inserted in the enhancer region of the CtBP gene is expressed in the larval plasmatocyte lineage (34). Thus, the FOG and CtBP class of transcriptional regulators may act together to control hemocyte lineage commitment in a pathway that is conserved evolutionarily.
FOG function involves binding to its GATA partner's N-terminal zinc finger. Srp is the only known hematopoietic GATA factor in Drosophila and reportedly contains a single C-terminal zinc finger (29). However, a survey of the srp genomic sequence shows an ORF within the third intron of the gene that putatively encodes an N-terminal zinc finger with 96% homology to that of Pnr (data not shown). This raises the possibility that Ush interacts with an alternatively spliced isoform of Srp during hematopoiesis.
Ush appears to negatively regulate the cardiogenic function of the GATA-4 homologue Pnr (17, 24), converting Pnr from a transcriptional activator to a repressor as observed during sensory bristle development (5, 35). As with Ush, forced mesodermal expression of FOG-1, FOG-2, and ΔFOG-2 also produced a diminution of cardial cells. These results demonstrate a functional conservation of the FOG proteins during Drosophila cardiogenesis, which most likely involves negative regulation of the cardiogenic activity of Pnr. In addition, forced expression of FOG proteins disrupted eye development-producing phenotypes that mimic pnr loss of function mutants, presumably by repressing Pnr activation of its downstream effector genes.
The disruption of eye development and the repression of cardial cell production by FOG proteins occurred in the absence of the CtBP-binding motif. This is consistent with the work of Svensson et al. (36), which showed that FOG-1 and FOG-2 repression of GATA-4 activation of cardiac promoters did not require the corepressor CtBP. Rather, conserved N-terminal regions of the murine FOG proteins were required for the repression of GATA-4 transcriptional activation, indicating that an alternative repressor mechanism may be used to negatively regulate GATA-4 (36). An emerging hypothesis suggests that CtBP may be a hematopoietic corepressor, and an alternative corepressor may be required during heart development. Our results showing that ΔFOG-2 does not repress crystal cell production but does repress cardial cell production is evidence for this dual mechanism of FOG gene regulation during heart development and hematopoiesis in a single experimental organism.
In conclusion, our data suggest that Ush and Lz function antagonistically during crystal cell lineage commitment and that Ush is required to limit the overproliferation of crystal cells. This demonstrates a possible intersection between the FOG and AML-1 gene pathways, which may prove important for understanding vertebrate hematopoiesis. Furthermore, this study expands the molecular characterization of the earliest events of hematopoiesis in Drosophila, identifying additional conserved genes that establish the fly as a model organism for hematopoiesis.
Acknowledgments
We thank M. Brennen, Y. Kim, and J. Pollock for providing genetic and molecular reagents used in this study. We also thank R. Grenda for assistance with figures and S. Wang and P. Reid for assistance with confocal microscopy. This research was supported by grants from the National Institutes of Health (HL59151) and American Heart Association (0150007N) to R.A.S.
Abbreviations
- Bc
Black cell
- CtBP
C-terminal-binding protein
- FOG
Friend of GATA
- Lz
Lozenge
- Pnr
Pannier
- Srp
Serpent
- UAS
upstream activation sequence
- Ush
U-shaped
References
- 1.Orkin S H. Curr Opin Genet Dev. 1996;6:597–602. doi: 10.1016/s0959-437x(96)80089-x. [DOI] [PubMed] [Google Scholar]
- 2.Parmacek M S, Leiden J M. In: Heart Development. Harvey R P, Rosenthal N, editors. San Diego: Academic; 1999. pp. 291–306. [Google Scholar]
- 3.Tsang A P, Visvader J E, Turner C A, Fujiwara Y, Yu C, Weiss M J, Crossley M, Orkin S H. Cell. 1997;90:109–119. doi: 10.1016/s0092-8674(00)80318-9. [DOI] [PubMed] [Google Scholar]
- 4.Cubadda Y, Heitzler P, Ray R P, Bourois M, Ramain P, Gelbart W, Simpson P, Haenlin M. Genes Dev. 1997;11:3083–3095. doi: 10.1101/gad.11.22.3083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Haenlin M, Cubadda Y, Blondeau F, Heitzler P, Lutz Y, Simpson P, Ramain P. Genes Dev. 1997;11:3096–3108. doi: 10.1101/gad.11.22.3096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tevosian S G, Deconinck A E, Cantor A B, Rieff H I, Fujiwara Y, Corfas G, Orkin S H. Proc Natl Acad Sci USA. 1999;96:950–955. doi: 10.1073/pnas.96.3.950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Svensson E C, Tufts R L, Polk C E, Leiden J M. Proc Natl Acad Sci USA. 1999;96:956–961. doi: 10.1073/pnas.96.3.956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lu J R, McKinsey T A, Xu H, Wang D Z, Richardson J A, Olson E N. Mol Cell Biol. 1999;19:4495–4502. doi: 10.1128/mcb.19.6.4495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tsang A P, Fujiwara Y, Hom D B, Orkin S H. Genes Dev. 1998;12:1176–1188. doi: 10.1101/gad.12.8.1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Crispino J D, Lodish M B, MacKay J P, Orkin S H. Mol Cell. 1999;3:219–228. doi: 10.1016/s1097-2765(00)80312-3. [DOI] [PubMed] [Google Scholar]
- 11.Turner J, Crossley M. EMBO J. 1998;17:5129–5140. doi: 10.1093/emboj/17.17.5129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fox A H, Kowalski K, King G F, Mackay J P, Crossley M. J Biol Chem. 1998;273:33595–33603. doi: 10.1074/jbc.273.50.33595. [DOI] [PubMed] [Google Scholar]
- 13.Nibu Y, Zhang H, Levine M. Science. 1998;280:101–104. doi: 10.1126/science.280.5360.101. [DOI] [PubMed] [Google Scholar]
- 14.Tevosian S G, Deconinck A E, Tanaka M, Schinke M, Litovsky S H, Izumo S, Fujiwara Y, Orkin S H. Cell. 2000;101:729–739. doi: 10.1016/s0092-8674(00)80885-5. [DOI] [PubMed] [Google Scholar]
- 15.Svensson E C, Huggins G S, Lin H, Clendenin C, Jiang F, Tufts R, Dardik F B, Leiden J M. Nat Genet. 2000;25:353–356. doi: 10.1038/77146. [DOI] [PubMed] [Google Scholar]
- 16.Deconinck A E, Mead P E, Tevosian S G, Crispino J D, Katz S G, Zon L I, Orkin S H. Development (Cambridge, UK) 2000;127:2031–2040. doi: 10.1242/dev.127.10.2031. [DOI] [PubMed] [Google Scholar]
- 17.Fossett N, Zhang Q, Gajewski K, Choi C Y, Kim Y, Schulz R A. Proc Natl Acad Sci USA. 2000;97:7348–7353. doi: 10.1073/pnas.97.13.7348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mathey-Prevot B, Perrimon N. Cell. 1998;92:697–700. doi: 10.1016/s0092-8674(00)81396-3. [DOI] [PubMed] [Google Scholar]
- 19.Dearolf C R. Biochim Biophys Acta. 1998;1377:M13–M23. doi: 10.1016/s0304-419x(97)00031-0. [DOI] [PubMed] [Google Scholar]
- 20.Rizki T M. In: Genetics and Biology of Drosophila. Ashburner M, Novitski E, editors. 2B. London: Academic; 1978. pp. 398–451. [Google Scholar]
- 21.Lebestky T, Chang T, Hartenstein V, Banerjee U. Science. 2000;288:146–149. doi: 10.1126/science.288.5463.146. [DOI] [PubMed] [Google Scholar]
- 22.Liew C K, Kowalski K, Fox A H, Newton A, Sharpe B K, Crossley M, Mackay J P. Struct Fold Des. 2000;8:1157–1166. doi: 10.1016/s0969-2126(00)00527-x. [DOI] [PubMed] [Google Scholar]
- 23.Matthews J M, Kowalski K, Liew C K, Sharpe B K, Fox A H, Crossley M, MacKay J P. Eur J Biochem. 2000;267:1030–1038. doi: 10.1046/j.1432-1327.2000.01095.x. [DOI] [PubMed] [Google Scholar]
- 24.Gajewski K, Fossett N, Molkentin J D, Schulz R A. Development (Cambridge, UK) 1999;126:5679–5688. doi: 10.1242/dev.126.24.5679. [DOI] [PubMed] [Google Scholar]
- 25.Cripps R M, Black B L, Zhao B, Lien C L, Schulz R A, Olson E N. Genes Dev. 1998;12:422–434. doi: 10.1101/gad.12.3.422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hu J. Ph.D. dissertation. Louisville, KY: Univ. of Louisville; 1995. [Google Scholar]
- 27.Rizki T M, Rizki R M, Grell E H. Roux Arch Dev Biol. 1980;188:91–99. doi: 10.1007/BF00848799. [DOI] [PubMed] [Google Scholar]
- 28.Querfurth E, Schuster M, Kulessa H, Crispino J D, Doderlein G, Orkin S H, Graf T, Nerlov C. Genes Dev. 2000;14:2515–2525. doi: 10.1101/gad.177200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rehorn K P, Thelen H, Michelson A M, Reuter R. Development (Cambridge, UK) 1996;122:4023–4031. doi: 10.1242/dev.122.12.4023. [DOI] [PubMed] [Google Scholar]
- 30.Sam S, Leise W, Hoshizaki D K. Mech Dev. 1996;60:197–205. doi: 10.1016/s0925-4773(96)00615-6. [DOI] [PubMed] [Google Scholar]
- 31.Brand A H, Perrimon N. Development (Cambridge, UK) 1993;118:401–415. doi: 10.1242/dev.118.2.401. [DOI] [PubMed] [Google Scholar]
- 32.Lilly B, Zhao B, Ranganayakulu G, Paterson B M, Schulz R A, Olson E N. Science. 1995;267:688–693. doi: 10.1126/science.7839146. [DOI] [PubMed] [Google Scholar]
- 33.Maurel-Zaffran C, Treisman J E. Development (Cambridge, UK) 2000;127:1007–1016. doi: 10.1242/dev.127.5.1007. [DOI] [PubMed] [Google Scholar]
- 34.Braun A, Lemaitre B, Lanot R, Zachary D, Meister M. Genetics. 1997;147:623–634. doi: 10.1093/genetics/147.2.623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Garcia-Garcia M J, Ramain P, Simpson P, Modolell J. Development (Cambridge, UK) 1999;126:3523–3532. doi: 10.1242/dev.126.16.3523. [DOI] [PubMed] [Google Scholar]
- 36.Svensson E C, Huggins G S, Dardik F B, Polk C E, Leiden J M. J Biol Chem. 2000;275:20762–20769. doi: 10.1074/jbc.M001522200. [DOI] [PubMed] [Google Scholar]