Abstract
The circadian system controls a large array of physiological and metabolic functions. The molecular organization of the circadian clock is complex, involving various elements organized in feedback regulatory loops. Here we demonstrate that the RelB subunit of NFκB acts as a repressor of circadian transcription. RelB physically interacts with the circadian activator BMAL1 in the presence of CLOCK to repress circadian gene expression at the promoter of the clock-controlled gene Dbp. The repression is independent of the circadian negative regulator CRY. Notably, RelB −/− fibroblasts have profound alterations of circadian genes expression. These findings reveal a previously unforeseen function for RelB as an important regulator of the mammalian circadian system in fibroblasts.
Keywords: Dbp, Clock mutant, BMAL1, NFκB, RelB, circadian, fibroblasts, inflammation
Introduction
The clock system controls many physiological and metabolic functions in different organisms.1 In mammals, a clock-driven transcriptional machinery is responsible for regulating the circadian gene expression of 10–20% of the genes within most tissues.2-5 Aberrant regulation by the circadian machinery may lead to various pathological conditions, including neurodegeneration, insomnia, inflammation, obesity, diabetes and cancer.6-12 The transcription factors CLOCK and BMAL1 are central to the positive transcriptional loop: after heterodimerization they bind to E-box promoter elements in the regulatory regions of many clock-controlled genes (CCGs). Among the CCGs, there are the Per and Cry genes, which encode negative regulators of CLOCK:BMAL1. These interplays are responsible for the oscillation of circadian gene expression.13
Accumulating evidence shows the presence of bidirectional links between circadian regulation and inflammatory response.14-21 Previous studies have demonstrated that stimulation of fibroblasts with tumor necrosis factor-α (TNF-α) represses circadian transcription.22,23 Moreover, we have recently observed that circadian disruption is associated with acute bacterial infection in mice (unpublished data), whereas other reports indicate that circadian disruption is involved in the development of symptoms associated to the inflammatory state.24-26
The NFκB transcription factor plays a central role in the inflammatory response. It is composed by five different subunits that can homo- or hetero-dimerize to form a variety of transcriptionally active isoforms with widely different roles in the transcriptional activation or repression of inflammatory genes.27-30
Here we report on the interplay between the circadian clock and the NFκB transcriptional pathway. Cells with a disrupted clock system display an altered response to lipopolysaccharide (LPS) and aberrant levels of some specific components of the NFκB complex. We show physical and functional interaction between RelB and BMAL1. This results in the repression of CLOCK:BMAL1-driven transcription and in alteration of the circadian expression profile in mouse embryo fibroblasts lacking RelB. Our findings reveal a molecular link between two transcription pathways previously thought to be independent, providing a molecular framework to interpret the physiological relationship between the inflammatory response and circadian rhythms.
Results
Reduced inflammatory response in cells with a disrupted circadian clock
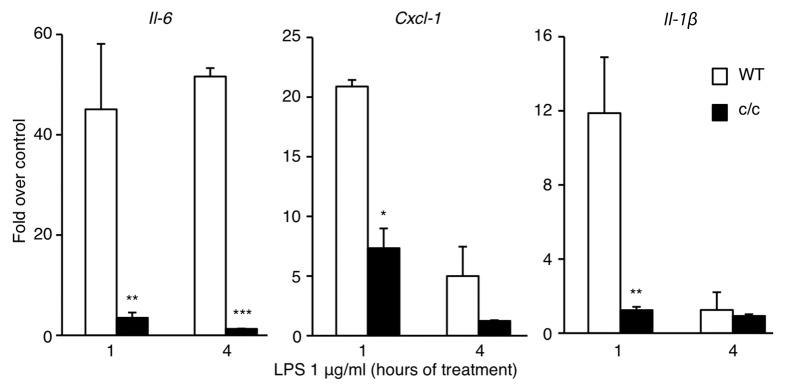
To explore whether the circadian clock could modulate the inflammatory response, we studied cultured cells with a disrupted clock system compared with their wild-type counterpart. We followed the timing of expression of various cytokines 1 h and 4 h after LPS stimulation of mouse embryonic fibroblasts (MEFs) derived from wild-type and Clock mutant mice (c/c), where a single point mutation determines a deletion of exon 19 within the CLOCK coding sequence,31-34 thus generating a dominant negative mutant CLOCK protein that renders the CLOCK:BMAL1 heterodimers functionally defective.35,36 The expression of the proinflammatory genes Il-6, Cxcl1 and Il-1β (Fig. 1) was drastically reduced in c/c MEFs compared with the wild-type cells. We also observed that these cells were only slightly responsive to stimulation with recombinant TNF-α (Fig. S1), thus confirming that the low responsivity was independent of the stimulus applied to the cells to induce the inflammatory response. We also monitored the expression of circadian genes after TNF-α stimulation (Fig. S1). As previously reported,22,23 TNF-α leads to a repressed expression of circadian genes in wild-type cells, while a constantly low level of Per2 and Dbp mRNAs was detected in c/c MEFs. Thus, a normally functioning circadian clock is necessary to obtain an efficient inflammatory response.
Figure 1.Clock mutant MEFs are less resposive to LPS stimulation. Time course of mRNA expression of different cytokines after LPS stimulation (1 μg/ml) of wt and Clock mutant (c/c) MEFs, measured by quantitative real time PCR. Shown are fold changes in gene expression compared with unstimulated cells. All the values are the mean +/− s.e.m. (n = 6); (*) p < 0.05, (**) p < 0.01, (***) p < 0.001.
Specific elelements of the NFκB pathway are overexpressed in Clock-mutant fibroblasts
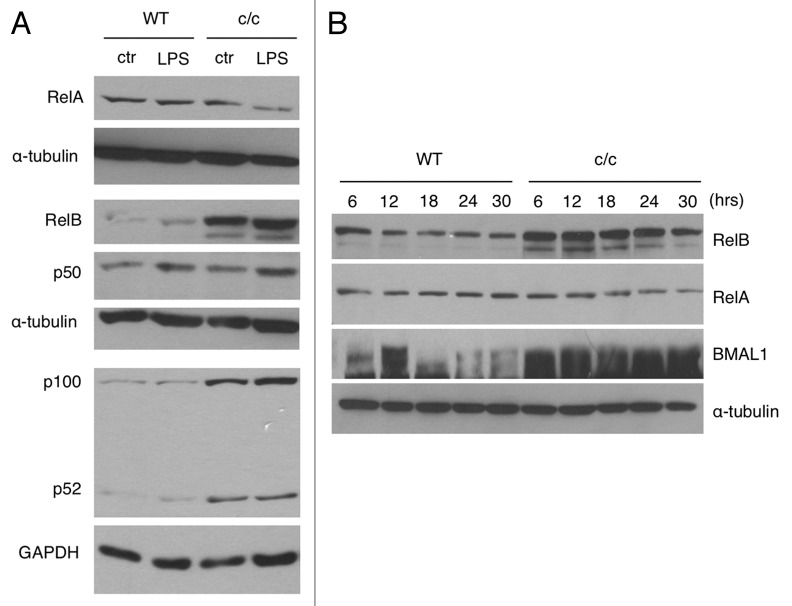
Based on the differential to LPS and TNF-α in cells with a disrupted clock (Fig. 1), we sought to explore the integrity of the NFκB signaling pathway. To do so, we monitored the protein levels of different NFκB subunits in wild-type and c/c MEFs, untreated or after LPS treatment. We observed a robust upregulation of the components of the non-canonical pathway RelB and p100/p52 in c/c fibroblasts as compared with isogenic wild-type cells. The upregulation appears independent from LPS stimulation (Fig. 2A). No differences in total levels of RelA and p50 were observed. The overexpression of RelB and p100/p52 is specific to c/c MEFs and not observed in cells carrying mutations in other clock components (Fig. S2).
Figure 2. Expression of NFκB subunits in c/c MEFs. (A) Endogenous expression of RelA, RelB, p50 and p100/p52 in wild type (WT) and Clock mutant (c/c) MEFs, treated for 1 h with LPS (1 μg/ml) or left untreated (ctr), was determined by western blot analysis. The α-tubulin and GAPDH were used as loading controls. (B) Wild-type and c/c MEFs were synchronized by 2 h serum-shock treatment. Total lysates were prepared at the indicated times (hrs, hours) post-synchronization and resolved by SDS-PAGE. Levels of RelB, RelA, BMAL1 and α-tubulin were detected by western blot analysis using specific antibodies.
These findings prompted us to investigate whether the expression of the transcriptionally active subunit RelB is regulated in a circadian manner. While the protein levels of RelB do not display a circadian oscillation in MEFs synchronized by serum shock, we observed a robust upregulation at all circadian time points in c/c MEFs compared with wild-type MEFs (Fig. 2B). Similarly, RelB mRNA showed only a marginal oscillation in both wild-type and c/c MEFs after serum-shock synchronization. Paralleling the protein levels (Fig. 2B), a significant upregulation of RelB transcript was present in c/c MEFs (Fig. S3).
Since RelB overexpression has been associated to repression of the inflammatory response in fibroblasts and induction of endotoxine tolerance,37-42 we concluded from these experiments that RelB expression is upregulated in c/c MEFs, and that this is likely to contribute to the altered response of these cells to an inflammatory stimulus (Fig. 1).
Physical interaction between RelB and BMAL1
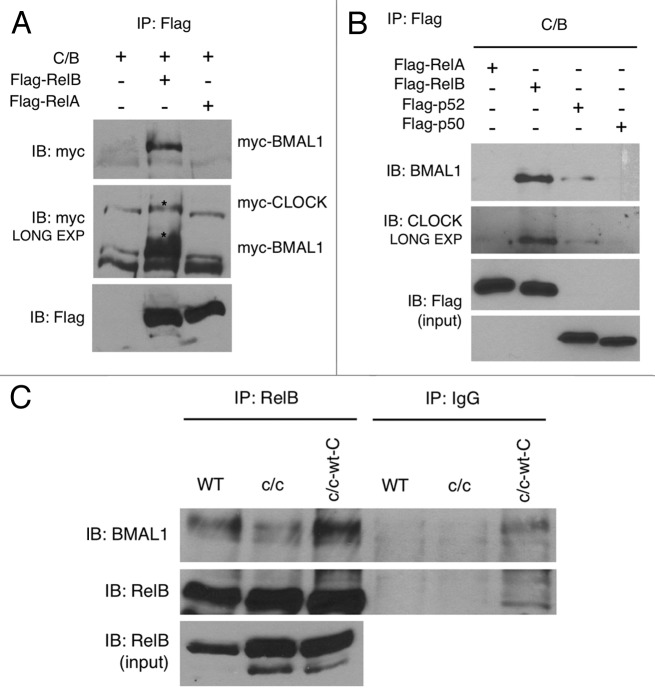
We sought to unravel the molecular mechanism by which the clock system may interplay with the NFκB pathway. Thus, we explored the possibility that one or several NFκB subunits could play a role in the regulation of circadian transcription by interacting with the components of the circadian machinery CLOCK and BMAL1. We ectopically overexpressed CLOCK and BMAL1, together with RelB, RelA, p50 or p52 in cultured cells. We found that RelB, but not RelA (Fig. 3A) or the other regolatory subunits (Fig. 3B), is able to efficiently interact with BMAL1, and weakly with CLOCK, thus suggesting a specific role of RelB in the circadian system.
Figure 3. RelB interaction with BMAL1 and CLOCK. (A) HEK-293 cells were cotransfected with Myc-CLOCK and Myc-BMAL1 (C/B), without or with Flag-RelB or Flag-RelA. Flag-tagged proteins were immunoprecipitated by FLAG-Agar, and abundance of coimmunoprecipitated proteins was determined by western blotting with anti-Myc antibody and anti-Flag antibody. Asterisks indicate specific signals for BMAL1 and CLOCK proteins. (B) HEK-293 cells were cotransfected with Myc-CLOCK and Myc-BMAL1 (C/B) and series of expression vectors as described. Total lysates were prepared and subjected to immunoprecipitation using FLAG-Agar and coimmunoprecipitated proteins were detected by western blotting with anti-BMAL1 and anti-CLOCK antibodies. Lower panels show the expression of Flag-tagged proteins in total cell lysates as an input. (C) Cell extracts prepared from wild-type (WT), Clock mutant (c/c) and Clock mutant stably transfected with wild-type Myc-CLOCK (c/c-wt-C) were immunoprecipitated with RelB antibody or normal IgG, and immunoprecipitated BMAL1 and RelB were detected by probing with the BMAL1 and RelB antibody, respectively. Lower panel shows RelB expression in total cell lysates as an input.
Next, we determined whether native RelB interacts with BMAL1 and/or CLOCK expressed endogenously in cultured cells. A specific interaction was found with BMAL1 in wild-type MEFs (Fig. 3C), while we were not able to detect unequivocal interaction with CLOCK under the same conditions. To test whether CLOCK is dispensable for the interaction of RelB with BMAL1, we transfected CLOCK and BMAL1 alone or in combination, together with RelB. We found that RelB interacts with BMAL1 alone, and that the interaction is reinforced in the presence of CLOCK (Fig. S4A). No interaction with CLOCK alone was found (not shown).
We then explored whether mutations of the CLOCK protein may affect the RelB-BMAL1 interaction. We transfected full-length CLOCK or various deletion mutants, known to reduce or abolish CLOCK functions. RelB interaction with BMAL1 was more robust in the presence of a full-length CLOCK protein than in the presence of truncated forms of CLOCK, either with a deletion in the N terminal region or with deletion of exon 19, domains affecting protein-protein interaction and transcriptional activation, respectively (Fig. S4B). Based on these results, we then tested the RelB-BMAL1 interaction in wild-type and c/c MEFs. The interaction of RelB with BMAL1 was significantly reduced in c/c MEFs as compared with wild-type cells (Fig. 3C). Stable ectopic expression of CLOCK in c/c MEFs rescued the efficacy of RelB-BMAL1 interaction at levels similar than wild-type MEFs (Fig. 3C). From these experiments we conclude that RelB readily interacts with BMAL1, and that this interaction is reinforced and dependent on the presence of a functional full-length CLOCK protein.
RelB represses CLOCK:BMAL1-driven trancriptional activation
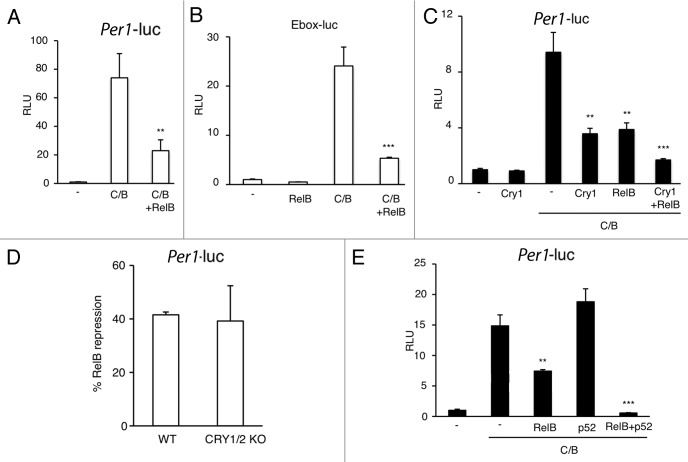
The specific BMAL1 interaction with RelB prompted us to explore whether it could be functionally relevant in modulating CLOCK:BMAL1-driven transcriptional activation. To address this question we performed luciferase assays on transiently transfected cultured cells. First, we observed a significant repression of CLOCK:BMAL1-driven transactivation on the Per1 promoter by RelB (Fig. 4A). Using a synthetic reporter vector carring only E-box promoter elements fused to the luciferase gene, we demonstrated that the repression was E-box-mediated (Fig. 4B). CLOCK:BMAL1-driven expression is known to be severely repressed by the circadian proteins CRYs.43 As comparison, RelB induces a repression of comparable extent than CRY1 (Fig. 4C), whereas co-expression of CRY1 and RelB leads to an additive repression (Fig. 4C). Importantly, RelB-mediated inhibition of CLOCK:BMAL1 is maintained in Cry1/Cry2-deficient cells (Fig. 4D), thus indicating that RelB repression is independent on the negative circadian regulators CRYs. Since RelB requires heterodimerization to be transcriptionally active,27-30 we asked whether the association with the regolatory subunit p52 could increase the repression. Interestingly, we observed increased repression in presence of p52 (Fig. 4E).
Figure 4. RelB represses CLOCK:BMAL1 transactivation potential. (A) Effect of RelB on CLOCK:BMAL1 dependent transcription. Vectors expressing CLOCK and BMAL1 (C/B) were cotransfected with a construct containing the mPer1-luc promoter, with or without RelB. The total DNA amount was kept constant by adding carrier plasmid DNA. After normalization for transfection efficiency using β-galactosidase activity, reporter gene activity was expressed as relative luciferase units (RLU) (activity of the control transfected only with non-expressing plasmid was set to 1). All the values are the mean +/− SD (n = 3); (**) p < 0.01 (B) The E box promoter element mediates RelB repression of CLOCK:BMAL1. Experimental condition was as in A except that a reporter construct containing three copies of the Ebox consensus sequence was used (Ebox-luc). All the values are the mean +/− SD (n = 3); (***) p < 0.001. (C) Additive effect of RelB and cry1 repression on CLOCK:BMAL1 dependent transcription. Experimental conditions are as in A except that Cry1 was cotransfected with C/B or C/B + RelB. All the values are the mean +/− SD (n = 3); (**) p < 0.01, (***) p < 0.001. (D) RelB repression is CRY-independent. CLOCK and BMAL1 were cotransfected as in A in wild-type (WT) and CRY1/2 KO MEFs. % of RelB repression of CLOCK:BMAL1 transactivation is shown. All the values are the mean +/− SD (n = 3). (E) Effect of RelB and p52 on CLOCK:BMAL1 dependent transcription. Vectors expressing CLOCK and BMAL1 (C/B) were cotransfected with a construct containing the Per1-luc promoter, with or without RelB and p52, as described. After normalization for transfection efficiency using β-galactosidase activity, reporter gene activity was expressed as relative luciferase units (RLU) (activity of the control transfected only with non-expressing plasmid was set to 1). All the values are the mean +/− SD (n = 3); (**) p < 0.01, (***) p < 0.001.
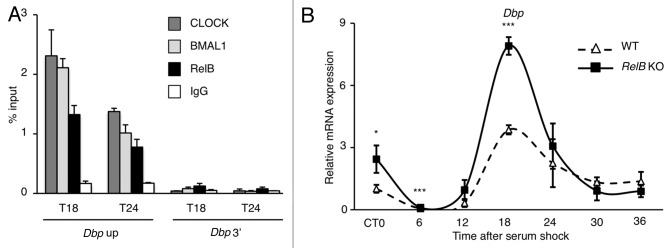
To better understand how the repression could take place, we performed a chromatin IP experiment at the promoter of the circadian gene Dbp in synchronized MEFs. Interestingly, RelB was immunoprecipitated together with CLOCK and BMAL1 on Dbp promoter transcriptionally active region (Fig. 5A) or on Per2 TSS promoter region (Fig. S5), while no enrichment was observed at the 3′ region of Dbp gene (Fig. 5A) or on Bmal1 promoter (Fig. S5). We then asked if the presence of RelB at the promoter of circadian genes was dependent on CLOCK and BMAL1. We performed the same experiment in BMAL1 KO MEFs, where the circadian transcription is abolished, and the CLOCK:BMAL1 complex is not present at the chromatin level, and we could not either detect any presence of RelB (Fig. S6). These results indicate that RelB is within a chromatin complex with CLOCK and BMAL1 at the promoter of circadian genes.
Figure 5. RelB negative regulation is required for proper circadian gene expression. (A) Cross-linked cell extracts were isolated at the indicated time points after serum shock from MEFs. The samples were subjected to ChIP assay with anti-CLOCK, anti-BMAL1, anti-RelB and anti-IgG, and analyzed by quantitative PCR with primers for Dbp promoter (Dbp Up and Dbp 3′). Control IgG and Dbp 3′UTR were used as control for immunoprecipitation and PCR, respectively. All the values are the mean +/− SD (n = 3). (B) Circadian Dbp mRNA expression profile in wild-type (WT) and RelB KO MEFs, after serum-shock synchronization, analyzed by quantitative PCR. The values are relative to those of β-actin mRNA levels at each circadian time (CT). Time 0 (unsynchronized cells, CT0) in wt cells was set to 1. All the values are the mean +/− s.e.m. (n = 3), (*) p < 0.05, (***) p < 0.001.
RelB controls the amplitude of circadian transcription
Our findings strongly suggested that RelB modulates CLOCK:BMAL1 and thereby contribute to the normal oscillation of circadian transcription. This notion was confirmed by using a chromatin immunoprecipitation approach, which demonstrated that RelB is present at the promoter of the clock-controlled gene Dbp in parallel with CLOCK and BMAL1 (Fig. 5A). To address the question, we used MEFs in which the RelB gene is deleted and compared to the profile of circadian gene expression with equivalent wild-type MEFs cells. We followed the expression of various circadian genes during a 24 h cycle after serum-shock synchronization. The transcription of Dbp was enhanced in asynchronous cells (CT0), and the amplitude of the oscillation during the circadian cycle was significantly increased in the absence of RelB (Fig. 5B), consistent with a repressive role. Similar results were obtained for the circadian-regulated genes Rev-erbα and Cry1 (Fig. S7). Interestingly, the transcription of the period genes was downregulated (Fig. S7), suggesting that RelB could play a more complex role in the maintainance of the functionality of the clock machinery.
Discussion
Previous reports showed as RelB expression is involved in the repression of the inflammatory response in fibroblasts.39-42,44,45 Different mechanisms have been considered, including the recruitment of chromatin modifier enzymes,37,46,47 thereby creating a repressive chromatin state, or stabilization of negative regulators, such as IkBα.38,40 Importantly, RelB was also considered as the major mediator of the endotoxin tolerance status in fibroblasts.42 This observation is of particular interest, since we observed increased levels of RelB in Clock-mutant MEFs compared with isogenic wild-type cells, and we also observed reduced induction of inflammatory genes expression in Clock-mutant macrophages (unpublished data) and MEFs (Fig. 1). We believe that these notions are important for future investigations.
Recent findings from another laboratory confirm that elements of the circadian clock may interplay with the NFκB pathway (M. Antoch, personal communication). Here we have shown that the NFκB subunit RelB directly partecipates in the regulation of circadian transcription, by interacting with the core clock factors BMAL1 and CLOCK at the chromatin level. Both interaction and presence of RelB at the promoter of circadian genes appears to be strictly dependent on a functional CLOCK:BMAL1 complex (Fig. S4 and 6). Moreover, RelB appears to directly control circadian transcription as shown by the use of RelB-deficient MEFs (Fig. 5; Fig. S7). It is still unclear whether RelB-induced repression of the circadian clock has a role during the inflammatory response. It is tempting to speculate that RelB might partecipate in mediating the repression of circadian genes during inflammation (Fig. S1), establishing an intriguing link between the clock system and the NFκB pathway. This possibility could be of great relevance during inflammatory responses and apoptosis and could establish the basis for the development of novel pharmacological strategies and therapeutic approaches.
Material and Methods
Plasmids
C-terminal Flag-tagged RelB pcDNA3, RelA pcDNA3, p52 pcDNA3 and p50 pcDNA3 were purchased from Addgene. N-terminal Myc-tagged plasmids Myc-mCLOCK/pSG5, Myc-mCLOCKΔ19/pSG5, Myc-mBMAL1/pCS2 and Myc-mCRY1 were previously described.48 Myc-mCLOCKΔC/pSG5 and mCLOCKΔN/pSG5 were made by deletion of a DNA fragment from Myc-mCLOCK/pSG5, encoding the C-terminal part (571–855) and the N-terminal part (1–434) of mCLOCK, respectively. All the Myc-tagged proteins contain six copies of Myc epitope at the N and carboxy termini. Plasmids expressing both β-galactosidase (pGL3-lacZ) for transfection control and luciferase (luc) for luminometry based expression (pGL3-mPer1-Luc promoter, pGL3-Ebox X3 luc) were described previously.49
Reagents and antibodies
LPS and mouse recombinant TNFα were purchased from Sigma-Aldrich. Antibodies against RelB for western blot and ChIP experiments were from Cell Signaling Technology and Santa Cruz Biotecnology, Inc., respectively. Antibodies against RelA, p50 and p100/p52 were from Cell Signaling Technology. Antibodies against CLOCK and rabbit IgG were from Santa Cruz Biotecnology; anti-BMAL1 was from Abcam; anti-Myc and anti-GAPDH were from Millipore, anti-flag and anti-α-tubulin from Sigma-Aldrich. Anti-Flag M2 affinity resin for immunoprecipitation experiments was from Sigma-Aldrich.
Cell culture
HEK-293 cells (ATCC) were mantained in DMEM (4.5 g/L glucose) supplemented with 10% newborn calf serum (NCS) and antibiotics and cultured at 37°C in 5% CO2. MEFs from c/c mutants, Cry1/2 −/− and Bmal1 −/− were cultured in DMEM supplemented with 10% FBS and antibiotics. MEFs c/c + CLOCK were previously generated.50 RelB +/+ and −/− were coltured with 10% BCS and antibiotics. Wild-type and RelB −/− MEFs were generous gift of Dr. Alexander Hoffmann. Wild-type and Cry −/− MEFs were generous gift of Dr. Bert van der Horst.
Transient transfection and luciferase assay
Cells were transfected with BioT (Bioland Scientific LLC) according to the manufacturer’s protocol. Cell extracts were subjected to a luminometry-based luciferase assay and luciferase activity was normalized by β-galactosidase activity.
RNA extraction and quantitative real-time RT-PCR
For circadian experiments, after 2 h of serum shock with media containing 50% horse serum, cells were incubated with serum-free medium for the indicated time. For gene expression analysis by real-time PCR, total RNA was extracted with TRIzol Reagent and processed according to the instructions of the manufacturer. Next, 2 μg of RNA from each sample was retrotranscribed (SuperScript II Reverse Transcriptase, Invitrogen) and 4-fold diluted cDNA was used for each real time reaction. For a 20 μl PCR, 50 ng of cDNA was mixed with primers to final concentration of 150 nM and 4 μl of RT2 SYBR Green Fluor Fast master mix (QIAGEN). The reaction was first incubated at 95°C for 3 min, followed by 40 cycles at 95°C for 30 sec, and 60°C for 1 min. Each quantitative real-time PCR was performed using the Chromo4 real time detection system (BIO-RAD). The primers for ChIP and real-time qPCR for murine Bmal1, Dbp, Cry1, Per1, Per2, Per3, Dbp up promoter region, Dbp 3′ UTR promoter region, Per2 TSS promoter region, Bmal TSS were obtained from refs. 51.51 The primers for RelB, Il-6, Cxcl-1, Il-1β, Rev-Erbα, β-actin and Gapdh are as follows: RelB: FW 5′-TGATCCACATGGAATCGAGA-3′ RV CAGGAAGGGATATGGAAGCA; Il-6: FW 5′ - TTCCATCCAGTTGCCTTCTT - 3′ RV 5′ -CAGAATTGCCATTGCACAAC - 3′; Cxcl-1: FW 5′ - TGCACCCAAACCGAAGTCAT - 3′ RV 5′ - TTGTCAGAAGCCAGCGTTCAC - 3′; Il-1β: FW 5′ - CTCTCCAGCCAAGCTTCCTTGTGC - 3′ RV 5′ - GCTCTCATCAGGACAGCCCAGGT - 3′ Rev-erbα: FW 5′ - GGGCACAAGCAACATTACCA - 3′ RV 5′ - CACGTCCCCACACACCTTAC - 3′; β-actin: FW 5′ - GGCTGTATTCCCCTCCATCG - 3′ RV 5′ - CCAGTTGGTAACAATGCCATGT - 3′; Gapdh: FW 5′-TGTAGACCATGTAGTTGAGGTCA-3′ RV 5′- AGGTCGGTGTGAACGGATTTG-3′.
Chromatin immunoprecipitation (ChIP) assay
Dual crosslinking ChIP assay52 was used. Briefly, after 2 h of serum shock with media containing 50% horse serum, cells were incubated with serum-free medium for the indicated time. Then, cells were washed three times with room temperature PBS and PBS with 1 mM MgCl2 was added. Disuccinimidyl Glutarate (DSG, Pierce) was added to a final concentration of 2 mM for crosslinking and incubated 45 min at room temperature, formaldehyde was added to a final concentration of 1% (v/v) and cells incubated for 15 min for dual crosslinking, and glycine was added to a final concentration of 0.1 M and incubated for 10 min to quench formaldehyde cross-linking. After harvesting, cells were lysed in 500 µL ice-cold cell lysis buffer (50 mM Tris/HCl pH 8.0, 85 mM KCl, 0.5% NP40, 1 mM PMSF, 1x protease inhibitor cocktail (Roche) for 10 min on ice. Nuclei were precipitated by centrifugation (3,000 g for 5 min) resuspended in 600 µL ice-cold RIPA buffer (50 mM Tris/HCl pH 8.0, 150 mM NaCl, 1 mM EDTA pH 8.0, 1% Triton X-100, 0.1% SDS, 0.1% sodium deoxycolate, 1 mM PMSF, 1x protease inhibitor cocktail) and incubated on ice for 30 min. Sonication was performed to obtain DNA fragments 100–600 bp in length.
Supplementary Material
Acknowledgments
We thank Dr. Eugenio Mora, Dr. Benedetto Grimaldi, Dr. Gioacchino Natoli and all P.S.-C. laboratory members for help, reagents and discussions. M.M.B. was in part supported by postdoctoral fellowships from AIRC (Associazione Italiana per la Ricerca sul Cancro) and from Della Martin Foundation. This work was supported by the National Institute of Health and the Institut National de la Sante et de la Recherche Medicale.
Glossary
Abbreviations:
- CCGs
clock-controlled genes
- c/c
Clock mutant
- MEFs
mouse embryonic fibroblasts
- LPS
lipopolysaccharide
- TNF-α
tumor necrosis factor-α
- TSS
transcription start site
Footnotes
Previously published online: www.landesbioscience.com/journals/cc/article/21669
References
- 1.Schibler U, Sassone-Corsi P. A web of circadian pacemakers. Cell. 2002;111:919–22. doi: 10.1016/S0092-8674(02)01225-4. [DOI] [PubMed] [Google Scholar]
- 2.Akhtar RA, Reddy AB, Maywood ES, Clayton JD, King VM, Smith AG, et al. Circadian cycling of the mouse liver transcriptome, as revealed by cDNA microarray, is driven by the suprachiasmatic nucleus. Curr Biol. 2002;12:540–50. doi: 10.1016/S0960-9822(02)00759-5. [DOI] [PubMed] [Google Scholar]
- 3.Duffield GE, Best JD, Meurers BH, Bittner A, Loros JJ, Dunlap JC. Circadian programs of transcriptional activation, signaling, and protein turnover revealed by microarray analysis of mammalian cells. Curr Biol. 2002;12:551–7. doi: 10.1016/S0960-9822(02)00765-0. [DOI] [PubMed] [Google Scholar]
- 4.Panda S, Antoch MP, Miller BH, Su AI, Schook AB, Straume M, et al. Coordinated transcription of key pathways in the mouse by the circadian clock. Cell. 2002;109:307–20. doi: 10.1016/S0092-8674(02)00722-5. [DOI] [PubMed] [Google Scholar]
- 5.Storch KF, Lipan O, Leykin I, Viswanathan N, Davis FC, Wong WH, et al. Extensive and divergent circadian gene expression in liver and heart. Nature. 2002;417:78–83. doi: 10.1038/nature744. [DOI] [PubMed] [Google Scholar]
- 6.Antoch MP, Gorbacheva VY, Vykhovanets O, Toshkov IA, Kondratov RV, Kondratova AA, et al. Disruption of the circadian clock due to the Clock mutation has discrete effects on aging and carcinogenesis. Cell Cycle. 2008;7:1197–204. doi: 10.4161/cc.7.9.5886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Antoch MP, Kondratov RV, Takahashi JS. Circadian clock genes as modulators of sensitivity to genotoxic stress. Cell Cycle. 2005;4:901–7. doi: 10.4161/cc.4.7.1792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bellet MM, Sassone-Corsi P. Mammalian circadian clock and metabolism - the epigenetic link. J Cell Sci. 2010;123:3837–48. doi: 10.1242/jcs.051649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cutolo M, Seriolo B, Craviotto C, Pizzorni C, Sulli A. Circadian rhythms in RA. Ann Rheum Dis. 2003;62:593–6. doi: 10.1136/ard.62.7.593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lin JD, Liu C, Li S. Integration of energy metabolism and the mammalian clock. Cell Cycle. 2008;7:453–7. doi: 10.4161/cc.7.4.5442. [DOI] [PubMed] [Google Scholar]
- 11.Sahar S, Sassone-Corsi P. Circadian clock and breast cancer: a molecular link. Cell Cycle. 2007;6:1329–31. doi: 10.4161/cc.6.11.4295. [DOI] [PubMed] [Google Scholar]
- 12.Sahar S, Sassone-Corsi P. Metabolism and cancer: the circadian clock connection. Nat Rev Cancer. 2009;9:886–96. doi: 10.1038/nrc2747. [DOI] [PubMed] [Google Scholar]
- 13.Reppert SM, Weaver DR. Coordination of circadian timing in mammals. Nature. 2002;418:935–41. doi: 10.1038/nature00965. [DOI] [PubMed] [Google Scholar]
- 14.Bollinger T, Leutz A, Leliavski A, Skrum L, Kovac J, Bonacina L, et al. Circadian clocks in mouse and human CD4+ T cells. PLoS ONE. 2011;6:e29801. doi: 10.1371/journal.pone.0029801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gibbs JE, Blaikley J, Beesley S, Matthews L, Simpson KD, Boyce SH, et al. The nuclear receptor REV-ERBα mediates circadian regulation of innate immunity through selective regulation of inflammatory cytokines. Proc Natl Acad Sci USA. 2012;109:582–7. doi: 10.1073/pnas.1106750109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hayashi M, Shimba S, Tezuka M. Characterization of the molecular clock in mouse peritoneal macrophages. Biol Pharm Bull. 2007;30:621–6. doi: 10.1248/bpb.30.621. [DOI] [PubMed] [Google Scholar]
- 17.Hrushesky WJ, Langevin T, Kim YJ, Wood PA. Circadian dynamics of tumor necrosis factor alpha (cachectin) lethality. J Exp Med. 1994;180:1059–65. doi: 10.1084/jem.180.3.1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Keller M, Mazuch J, Abraham U, Eom GD, Herzog ED, Volk HD, et al. A circadian clock in macrophages controls inflammatory immune responses. Proc Natl Acad Sci USA. 2009;106:21407–12. doi: 10.1073/pnas.0906361106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Logan RW, Sarkar DK. Circadian nature of immune function. Mol Cell Endocrinol. 2011 doi: 10.1016/j.mce.2011.06.039. [DOI] [PubMed] [Google Scholar]
- 20.Marpegan L, Leone MJ, Katz ME, Sobrero PM, Bekinstein TA, Golombek DA. Diurnal variation in endotoxin-induced mortality in mice: correlation with proinflammatory factors. Chronobiol Int. 2009;26:1430–42. doi: 10.3109/07420520903408358. [DOI] [PubMed] [Google Scholar]
- 21.Silver AC, Arjona A, Walker WE, Fikrig E. The circadian clock controls toll-like receptor 9-mediated innate and adaptive immunity. Immunity. 2012;36:251–61. doi: 10.1016/j.immuni.2011.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cavadini G, Petrzilka S, Kohler P, Jud C, Tobler I, Birchler T, et al. TNF-alpha suppresses the expression of clock genes by interfering with E-box-mediated transcription. Proc Natl Acad Sci USA. 2007;104:12843–8. doi: 10.1073/pnas.0701466104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Petrzilka S, Taraborrelli C, Cavadini G, Fontana A, Birchler T. Clock gene modulation by TNF-alpha depends on calcium and p38 MAP kinase signaling. J Biol Rhythms. 2009;24:283–94. doi: 10.1177/0748730409336579. [DOI] [PubMed] [Google Scholar]
- 24.Castanon-Cervantes O, Wu M, Ehlen JC, Paul K, Gamble KL, Johnson RL, et al. Dysregulation of inflammatory responses by chronic circadian disruption. J Immunol. 2010;185:5796–805. doi: 10.4049/jimmunol.1001026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hashiramoto A, Yamane T, Tsumiyama K, Yoshida K, Komai K, Yamada H, et al. Mammalian clock gene Cryptochrome regulates arthritis via proinflammatory cytokine TNF-alpha. J Immunol. 2010;184:1560–5. doi: 10.4049/jimmunol.0903284. [DOI] [PubMed] [Google Scholar]
- 26.Taraborrelli C, Palchykova S, Tobler I, Gast H, Birchler T, Fontana A. TNFR1 is essential for CD40, but not for lipopolysaccharide-induced sickness behavior and clock gene dysregulation. Brain Behav Immun. 2011;25:434–42. doi: 10.1016/j.bbi.2010.11.001. [DOI] [PubMed] [Google Scholar]
- 27.Bonizzi G, Karin M. The two NF-kappaB activation pathways and their role in innate and adaptive immunity. Trends Immunol. 2004;25:280–8. doi: 10.1016/j.it.2004.03.008. [DOI] [PubMed] [Google Scholar]
- 28.Hoffmann A, Baltimore D. Circuitry of nuclear factor kappaB signaling. Immunol Rev. 2006;210:171–86. doi: 10.1111/j.0105-2896.2006.00375.x. [DOI] [PubMed] [Google Scholar]
- 29.Li Q, Verma IM. NF-kappaB regulation in the immune system. Nat Rev Immunol. 2002;2:725–34. doi: 10.1038/nri910. [DOI] [PubMed] [Google Scholar]
- 30.Natoli G, Saccani S, Bosisio D, Marazzi I. Interactions of NF-kappaB with chromatin: the art of being at the right place at the right time. Nat Immunol. 2005;6:439–45. doi: 10.1038/ni1196. [DOI] [PubMed] [Google Scholar]
- 31.Antoch MP, Song EJ, Chang AM, Vitaterna MH, Zhao Y, Wilsbacher LD, et al. Functional identification of the mouse circadian Clock gene by transgenic BAC rescue. Cell. 1997;89:655–67. doi: 10.1016/S0092-8674(00)80246-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.King DP, Vitaterna MH, Chang AM, Dove WF, Pinto LH, Turek FW, et al. The mouse Clock mutation behaves as an antimorph and maps within the W19H deletion, distal of Kit. Genetics. 1997;146:1049–60. doi: 10.1093/genetics/146.3.1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.King DP, Zhao Y, Sangoram AM, Wilsbacher LD, Tanaka M, Antoch MP, et al. Positional cloning of the mouse circadian clock gene. Cell. 1997;89:641–53. doi: 10.1016/S0092-8674(00)80245-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vitaterna MH, King DP, Chang AM, Kornhauser JM, Lowrey PL, McDonald JD, et al. Mutagenesis and mapping of a mouse gene, Clock, essential for circadian behavior. Science. 1994;264:719–25. doi: 10.1126/science.8171325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gekakis N, Staknis D, Nguyen HB, Davis FC, Wilsbacher LD, King DP, et al. Role of the CLOCK protein in the mammalian circadian mechanism. Science. 1998;280:1564–9. doi: 10.1126/science.280.5369.1564. [DOI] [PubMed] [Google Scholar]
- 36.Jin X, Shearman LP, Weaver DR, Zylka MJ, de Vries GJ, Reppert SM. A molecular mechanism regulating rhythmic output from the suprachiasmatic circadian clock. Cell. 1999;96:57–68. doi: 10.1016/S0092-8674(00)80959-9. [DOI] [PubMed] [Google Scholar]
- 37.Chen X, El Gazzar M, Yoza BK, McCall CE. The NF-kappaB factor RelB and histone H3 lysine methyltransferase G9a directly interact to generate epigenetic silencing in endotoxin tolerance. J Biol Chem. 2009;284:27857–65. doi: 10.1074/jbc.M109.000950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chen X, Yoza BK, El Gazzar M, Hu JY, Cousart SL, McCall CE. RelB sustains IkappaBalpha expression during endotoxin tolerance. Clin Vaccine Immunol. 2009;16:104–10. doi: 10.1128/CVI.00320-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.McCall CE, El Gazzar M, Liu T, Vachharajani V, Yoza B. Epigenetics, bioenergetics, and microRNA coordinate gene-specific reprogramming during acute systemic inflammation. J Leukoc Biol. 2011;90:439–46. doi: 10.1189/jlb.0211075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Xia Y, Chen S, Wang Y, Mackman N, Ku G, Lo D, et al. RelB modulation of IkappaBalpha stability as a mechanism of transcription suppression of interleukin-1alpha (IL-1alpha), IL-1beta, and tumor necrosis factor alpha in fibroblasts. Mol Cell Biol. 1999;19:7688–96. doi: 10.1128/mcb.19.11.7688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Xia Y, Pauza ME, Feng L, Lo D. RelB regulation of chemokine expression modulates local inflammation. Am J Pathol. 1997;151:375–87. [PMC free article] [PubMed] [Google Scholar]
- 42.Yoza BK, Hu JY, Cousart SL, Forrest LM, McCall CE. Induction of RelB participates in endotoxin tolerance. J Immunol. 2006;177:4080–5. doi: 10.4049/jimmunol.177.6.4080. [DOI] [PubMed] [Google Scholar]
- 43.Kume K, Zylka MJ, Sriram S, Shearman LP, Weaver DR, Jin X, et al. mCRY1 and mCRY2 are essential components of the negative limb of the circadian clock feedback loop. Cell. 1999;98:193–205. doi: 10.1016/S0092-8674(00)81014-4. [DOI] [PubMed] [Google Scholar]
- 44.Madge LA, May MJ. The NFκB paradox: RelB induces and inhibits gene expression. Cell Cycle. 2011;10:6–7. doi: 10.4161/cc.10.1.14291. [DOI] [PubMed] [Google Scholar]
- 45.McCall CE, Yoza BK. Gene silencing in severe systemic inflammation. Am J Respir Crit Care Med. 2007;175:763–7. doi: 10.1164/rccm.200610-1436CP. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.El Gazzar M, Yoza BK, Hu JY, Cousart SL, McCall CE. Epigenetic silencing of tumor necrosis factor alpha during endotoxin tolerance. J Biol Chem. 2007;282:26857–64. doi: 10.1074/jbc.M704584200. [DOI] [PubMed] [Google Scholar]
- 47.Liu TF, Yoza BK, El Gazzar M, Vachharajani VT, McCall CE. NAD+-dependent SIRT1 deacetylase participates in epigenetic reprogramming during endotoxin tolerance. J Biol Chem. 2011;286:9856–64. doi: 10.1074/jbc.M110.196790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Travnickova-Bendova Z, Cermakian N, Reppert SM, Sassone-Corsi P. Bimodal regulation of mPeriod promoters by CREB-dependent signaling and CLOCK/BMAL1 activity. Proc Natl Acad Sci USA. 2002;99:7728–33. doi: 10.1073/pnas.102075599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yujnovsky I, Hirayama J, Doi M, Borrelli E, Sassone-Corsi P. Signaling mediated by the dopamine D2 receptor potentiates circadian regulation by CLOCK:BMAL1. Proc Natl Acad Sci USA. 2006;103:6386–91. doi: 10.1073/pnas.0510691103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Doi M, Hirayama J, Sassone-Corsi P. Circadian regulator CLOCK is a histone acetyltransferase. Cell. 2006;125:497–508. doi: 10.1016/j.cell.2006.03.033. [DOI] [PubMed] [Google Scholar]
- 51.Nakahata Y, Kaluzova M, Grimaldi B, Sahar S, Hirayama J, Chen D, et al. The NAD+-dependent deacetylase SIRT1 modulates CLOCK-mediated chromatin remodeling and circadian control. Cell. 2008;134:329–40. doi: 10.1016/j.cell.2008.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nowak DE, Tian B, Brasier AR. Two-step cross-linking method for identification of NF-kappaB gene network by chromatin immunoprecipitation. Biotechniques. 2005;39:715–25. doi: 10.2144/000112014. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.