Abstract
It is well known that remnants of partial or whole copies of mitochondrial DNA, known as Nuclear MiTochondrial sequences (NUMTs), are found in nuclear genomes. Since whole genome sequences have become available, many bioinformatics studies have identified putative NUMTs and from those attempted to infer the factors involved in NUMT creation. These studies conclude that NUMTs represent randomly chosen regions of the mitochondrial genome. There is less consensus regarding the nuclear insertion sites of NUMTs — previous studies have discussed the possible role of retrotransposons, but some recent ones have reported no correlation or even anti-correlation between NUMT sites and retrotransposons. These studies have generally defined NUMT sites using BLAST with default parameters. We analyze a redefined set of human NUMTs, computed with a carefully considered protocol. We discover that the inferred insertion points of NUMTs have a strong tendency to have high-predicted DNA curvature, occur in experimentally defined open chromatin regions and often occur immediately adjacent to A + T oligomers. We also show clear evidence that their flanking regions are indeed rich in retrotransposons. Finally we show that parts of the mitochondrial genome D-loop are under-represented as a source of NUMTs in primate evolution.
INTRODUCTION
Mitochondria are believed to share a common ancestor with α-proteobacteria (1). Mitochondria contain their own small genomes (mtDNA, around 16 000 bp in mammals), but rely on the nuclear genome to encode ∼99% of their proteins. Many of these proteins' genes are thought to have migrated from the mitochondrial to the nuclear genome in the distant past.
The process of inserting mtDNA into the nuclear genome continues to this day. Starting with the discovery of mtDNA-like sequences in the mouse nuclear genome (2), so-called Nuclear MiTochondrial sequences (NUMTs) have been found in a wide variety of Eukaryotic organisms (3,4).
The usual mechanism of NUMT insertion appears to be non-homologous end joining repair (5–7). mtDNA fragments are inserted and joined with nuclear DNA ends during nuclear double-strand break (DSB) repair. However the sequence characteristics (if any) of NUMT producing DSB sites is less clear.
Several studies have investigated general features of NUMT insertion sites. Ricchetti et al. (8) identified human NUMTs with BLASTN and claimed that human NUMTs preferentially insert into introns (rather than intergenic regions) of the genome, while Mishmar et al. (9) showed evidence that NUMTs tend to have low to moderate G+C content in their 100bp flanks (e.g. insert into L1–H1 isochores) and occur in areas which tend to have a different G+C content than their surroundings. Examining yeast NUMTs, Lenglez et al. (10) suggested they are associated with DNA replication origins (ORIs), due to the fragility of ORIs caused by the pausing of replication forks.
Several early studies hinted at a possible relationship between NUMTs and retrotransposons. Farrelly and Butow (11) found a yeast NUMT that in some strains is neighbored by a tandem pair of transposable (Ty) elements, Tsuzuki et al. (12) found two human NUMTs flanked by repetitive elements, Zullo et al. (13) found two rat NUMTs near a LINE element, and Ossorio et al. (14) found a NUMT flanked by direct or inverted repeats in the protist Toxoplasma gondii. Willet-Brozick et al. (15) described an individual human NUMT (not fixed in the general population) incorporated into a reciprocal translocation through a double-stranded break, in which one original breakpoint occurred in the 3′-end of an Alu repeat element and the other within an L1 repeat element.
Unfortunately, more systematic investigations have produced diverse and sometimes contradictory findings. Blanchard et al. (16) found only a couple of yeast NUMTs with flanking transposable elements, whose presence they attributed to chance. Mishmar et al. (9) analyzed 247 computationally (BLAST) identified human NUMTs and concluded that repetitive elements, especially LINEs and Alu's are significantly enriched in NUMT flanks. In another BLAST-based computational study, Gherman et al. (3) found repetitive elements to be under-represented in NUMT flanks, although they suggested this might be due to inaccurate NUMT boundary estimation. Qu et al. (17) also estimated the repetitive element content of the flanks of computationally identified human NUMTs, but did not draw any conclusion regarding their frequency. Jensen-Seaman et al. (18) claimed that repetitive elements are under-represented in human, but not chimpanzee, NUMTs.
As for the region of the mtDNA contributing to NUMTs, Mourier et al. (19) observed a deficiency of NUMTs from the mtDNA D-loop region, but attributed that to the difficultly of detecting NUMTs from this rapidly evolving region. Indeed most studies conclude that the mitochondrial source DNA and nuclear insertion sites of NUMTs are both randomly chosen (20,21).
In this study, we aimed to clear up the inconsistencies reported in the literature, by using careful methodology. Surprisingly, we not only succeeded in doing this, but also discovered completely new characteristics of NUMT insertion sites.
MATERIALS AND METHODS
Sequence data
We downloaded the nuclear genomes of human (hg18), rhesus (rheMac2), mouse (mm9) and rat (rn4), and the mitochondrial genomes of human (hg18), chimpanzee (panTro2), orangutan (ponAbe2), mouse (mm9), rat (rn4) and opossum (monDom5) from the UCSC web site. In addition, we obtained the mitochondrial genomes of rhesus (NC_005943), gorilla (NC_001645), gibbon (NC_002082), squirrel monkey (NC_012775), guinea pig (NC_000884), squirrel (NC_002369), rabbit (NC_001913) and hedgehog (NC_002080) from the NCBI Genome database.
NUMT detection
Alignment software
We used the LAST (22) program to perform local sequence alignment. We used LAST because BLAST cannot compute the E-values for the scoring system we used, and the E-value calculation of LAST is more accurate than BLAST (which assumes a 25% background probability for each base).
Substitution and gap scoring scheme
The best methodology for delineating NUMTs in nuclear genomes has not been carefully examined. In most previous studies, BLAST was used with its default settings (3,8,9,17–21). The default score matrix for DNA in BLAST is +1 for all matches, −3 for all mismatches, 1 for gap-open penalty and 1 for gap-extension penalty. Unfortunately, this scoring scheme is tuned for 99% identity alignments (23), rather than the low identity expected when aligning older NUMTs to mtDNA. Instead, we used the scoring scheme of +1 for matches, −1 for mismatches, 7 for gap-open penalty and 1 for gap-extension penalty, which is suitable for detecting distant homology (24).
Repeat masking
Repetitive and low complexity regions are not modeled by the BLAST statistics (25) (also used in LAST) and may sometimes achieve apparently significant E-value scores by chance. We therefore masked such regions in both the mitochondrial and nuclear genome with the recently developed repeat masking software tantan (26). We used so-called ‘soft-masking’ in which repetitive regions are disallowed as match ‘seeds’, but can be included in alignments during the final extension phase.
Sequence similarity E-value threshold
We empirically determined an E-value threshold which is as aggressive as possible, while still maintaining a low risk of producing false positives. To test the risk of false positives, we prepared two kinds of decoy tests.
Reversed mitochondrial genome
Random nuclear pseudo-genome
The reversed genome was obtained by simply reversing (but not complementing) the mitochondrial genome for each species (human, rhesus monkey, mouse and rat) examined. Since DNA sequences do not evolve by simple reversal, any match between this query and the nuclear genome can be considered to be spurious. The minimum E-value observed in this test was 2.23.
We produced nuclear pseudo-genomes by generating 1000 random sequences, each of the same length as the real genome. We used a first-order Markov model for this in order to retain the approximate dinucleotide content of the real genome. For each species, we queried the real mitochondrial genome against each of the 1000 nuclear pseudo-genomes. The minimum E-value obtained was 0.0912.
Considering these results and our desire to be conservative, we concluded that 10−3 should be a safe threshold, with very little risk of false positives. In fact, when we matched the real mitochondrial and nuclear genomes, we observed no hits with an E-value between 10−3 and 10−4, so even the worst E-value in our NUMTs set should be considered highly significant.
Treatment of circular mitochondrial genome
Since mtDNA is circular, we concatenated two linearized mtDNA sequences together, so that standard sequence comparison methods could be used without risk of losing hits due to boundary effects.
Detection of nuclear duplication of NUMTs
Some matches between the mitochondrial and nuclear genome are not the direct product of a NUMT insertion, but result from subsequent duplication in the nucleus. To investigate this, we divided the hits into two classes: those which can be unambiguously labeled as original NUMTs and those which may be the product of nuclear duplications. We performed this classification by comparing 200 bp flanking sequence similarities (cut-off >90%) and crosschecking the Segmental duplication (SD) database (27). Unfortunately, there is no assembly for mouse (mm9) available in the SD, so we looked for duplicated NUMTs ourselves. First, we extracted the 400 bp sequence obtained by concatenating the 200 bp upstream and downstream flanks of each NUMT; then, we aligned each pair of these and considered any pair with >90% identity as the likely product of a nuclear duplication event.
Consolidation of co-linear matches
As a final step, we chained together co-linear NUMT fragments which appeared to originate from a single NUMT consequently separated by insertions or deletions in the nuclear genome. Recall that we used LAST to compute matches of local similarity between the nuclear and mitochondrial genomes. To handle nuclear deletions, we considered mitochondrially co-linear matches separated by no more than 200 bp of mtDNA to be mitochondrially contiguous. To model insertions in the nuclear genome, we merged any mitochondrially contiguous matches within 10 kb. We tuned the values 200 bp and 10 kb empirically, based on manual inspection of several dozen matches.
NUMT insertion age estimation by phylogenetic analysis
For reference, we first computed a phylogenetic tree of the mitochondrial genomes, and then one additional tree for each NUMT.
To handle the circular mitochondrial genomes, we converted each mitochondrial genome to a single linear sequence starting at the D-loop origin. Next, we multiply aligned these sequences with ClustalW version 2.0.11 (28) and computed rooted phylogenetic trees with the RETREE program of Phylip version 3.68 (29). As expected, the obtained tree topology matched the known phylogenetic relationships of the organisms involved.
We computed a tree for each NUMT in a similar way; the NUMT sequence was multiply aligned (ClustalW) to the mitochondrial region of each species corresponding to where it matched the human mitochondrial genome, followed by phylogenetic tree inference with Phylip. All tree computations were bootstrapped 1000 times. Finally, we estimated the insertion age of each NUMT by manual inspection of its tree. For insertion age, we adopted seven periods defined by common ancestor divergence of a species with human: (o) before mouse (outgroup), (a) after mouse and before rhesus monkey, (b) after rhesus and before gibbon, (c) after gibbon and before orangutan, (d) after orangutan and before gorilla, (e) after gorilla and before chimpanzee and, (f) after chimpanzee (Supplementary Figure S1). Although we included squirrel monkey mtDNA in the phylogenetic trees, we did not define an ‘after squirrel monkey and before rhesus’ age. This is because we found it difficult to validate that age by multiple alignment (see ‘Results’ section, age estimation verification).
Detection limit of short, old NUMTs
We conducted two tests to estimate the detection limit of our methodology. In one test, we simulated short NUMTs by randomly extracting substrings of NUMTs of lengths 50–1000 in increments of 50. For this analysis, we used human NUMTs from age (a), because this age is relatively old and contains NUMTs of various lengths. For each simulated short NUMT length l, we randomly selected a real NUMT (of length >l) and a length l segment of that NUMT. We then blanked out the remainder of that NUMT with n's, and ran LAST. We repeated this 10 000 times to estimated the detection false-negative rate.
In another test, we randomly extracted 100 segments of length 100, 150, 200 and 500 bp from the mouse mtDNA D-loop region, randomly planted them somewhere in the human genome, and then searched for them with LAST as we did for real NUMTs.
NUMT insertion frequency in particular genomic contexts
Repetitive elements
From the UCSC web site, we downloaded the position of repetitive elements as defined by RepeatMasker and Tandem Repeats Finder, and used this information to compute the frequency of repetitive elements in NUMT flanks up to length 5000 bp. Note that unlike the analysis performed by Jensen-Seaman et al. (18), our computation does not involve ‘windows’ (see Discussion section). For each flank position (say 5 bp from the NUMT edge), we simply compute the fraction of NUMT flanks for which that position is found within a repetitive element. We also performed a control experiment using randomly generated pseudo-NUMTs.
Oligomer frequency
We computed the fraction of 10 bp NUMT flanks which contain each possible length 2–6 oligomer (corresponding to 9–5 possible starting points). For a background probability p, we also computed the fraction for all length 10 bp windows in the overall nuclear genome. We evaluated significance using a binomial test with p as the probability of success.
DNA bendability and curvature prediction in NUMT flanks
We extracted 200 bp NUMT flanks (upstream 100 bp + downstream 100 bp) and calculated bendability and curvature scores of the flanks with the bendability/curvature predictor ‘bend.it’ (30) (http://hydra.icgeb.trieste.it/dna/bend_it.html).
Open chromatin regions
To test whether NUMT insertion sites correlate with open chromatin regions, we downloaded open chromatin data defined by DNase-seq (31), and FAIRE-seq data produced with the FAIRE (31) protocol, from the UCSC web site and used the coordinates to compute the fraction of NUMT flanks annotated as open chromatin regions.
Correlation amongst NUMT insertion site features
A+T oligomers versus retrotransposons. When visualizing the occurrence of A+T-rich oligomers in NUMTs, we noticed an apparent weak correlation with the presence of retrotransposons (Supplementary Figure S12). Thus we used a Fisher's exact test to examine the correlation between the presence of retrotransposons and A+T-only k-mers for k = 2–6, in NUMT flanks of lengths {10, 50, 100, 200, 300, 400 and 500}bp. Only one combination, 6-mer with 300 bp flanks yielded a significant P-value (0.0258) and considering multi-testing effects, we do not regard this observation as statistically significant.
DNA bendability/curvature versus oligomers. To examine the relationship between NUMT flank oligomer frequency and predicted DNA curvature, we randomly chose 5000 sequences of length of 200 bp from the human nuclear genome and computed their DNA curvature scores. Then, we paired each NUMT flank with the sequence from this random sample with the closest curvature score—thus obtaining a random sample of nuclear DNA, under the constraint that the sample and NUMT flanks have a similar distribution of curvature scores. Finally, using this curvature normalized sample for oligomer background probabilities, we computed binomial test P-values for biased oligomer frequency (Supplementary Table S1).
Open chromatin regions vs DNA bendability/curvature. To examine the correlation of open chromatin and highly curved DNA regions, we cross-checked the highly curved background DNA collected from the above analysis with open chromatin regions and computed P-values by binomial test.
Retrotransposons versus open chromatin regions. To investigate whether retrotransposons and open chromatin regions co-occur at NUMT insertion sites, we calculated P-values with Fisher's exact test. For this calculation, we counted the number of NUMTs flanked by retrotransposons within +100/−100 bp and the number of NUMTs embedded in open chromatin regions.
RESULTS
NUMT dataset
Applying the sequence similarity search and post-processing procedure described in ‘Materials and Methods’ section to the genomes of human, rhesus monkey, mouse and rat, we obtained 724, 742, 162 and 97 NUMTs, covering 632,224, 622,584, 71,794 and 30,762 bp, respectively. The human and mouse datasets are given in tab separated field format in the supplementary material files hg18-numts.tsv and mm9-numts.tsv and the others are available upon request. To investigate the effect of alignment scoring parameters, we also computed NUMT datasets using the default BLAST alignment scoring parameters, yielding 391, 363, 93 and 34 NUMTs covering 483 689, 464 916, 48 709, and 6347 bp in the four species, respectively.
Statistical characteristics of NUMT insertion sites
Using our non-duplicated human NUMT set (610 loci), we investigated several genome features which we thought might correlate with NUMT insertion events.
Retrotransposons are highly enriched in NUMT flanks
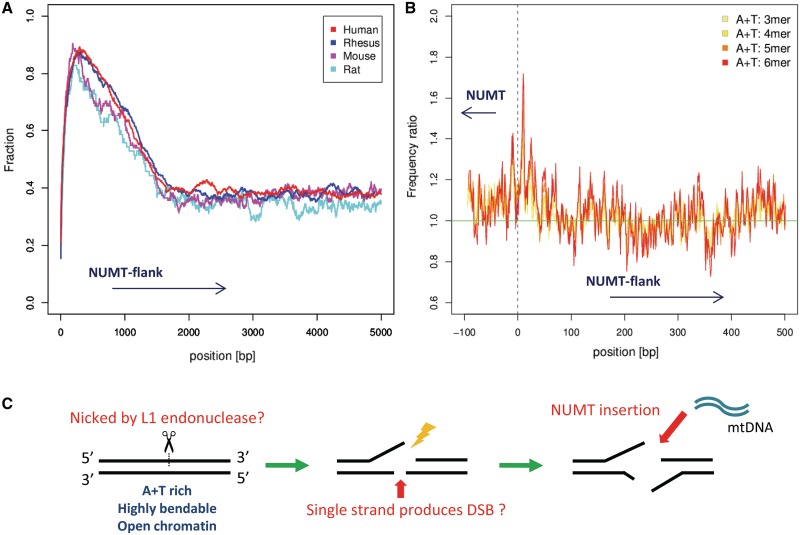
We used the output of RepeatMasker and Tandem Repeats Finder to investigate the frequency of various categories of repetitive elements. In direct contradiction to some recent reports (3,18) (see ‘Discussion’ section), we found a high density of retrotransposons in the flanking regions of human, rhesus, mouse and rat NUMTs (Figure 1A). More specifically, the fraction of bases masked as retrotransposons in the length 1000 bp genomic regions flanking NUMTs is much greater than the overall genome average: human 89.3% versus 40.6%, rhesus monkey 87.9% versus 39.8%, mouse 90.4% versus 37.8% and rat 82.85% versus 35.4%. This over-representation is statistically significant (binomial test, P-value < 0.001) in all species. We also checked if one particular class of retrotransposon (SINE, LINE or LTR) is especially over-represented, but this was not the case (results not shown). Finally, we checked the frequency of tandem repeats, which were not significantly over-represented (P-value ∼ 0.59).
Figure 1.
Features of NUMT insertion sites in the nuclear genome. (A) The proportion of NUMT flank bases covered by retrotransposons at each distance from 1 to 5000 bp is shown. (B) The relative ratio of 3–6 bp A+T-only oligomers in NUMT flanks is shown. The horizontal axis gives the position relative to the inferred NUMT boundary. The vertical axis gives the ratio of A+T-only oligomers centered at each flank position relative to the overall genome average. (C) Additional observations and discussion regarding NUMT insertion sites is depicted. We observe that the immediate flanking region of NUMTs tend to have A+T rich oligomers, high predicted DNA bendability and curvature and occur in open chromatin regions. From the enrichment of retrotransposons seen in (A) and other circumstantial evidence we speculate that L1 endonuclease nicking may have mediated many NUMT insertion events in primates.
Position and orientation of NUMT associated retrotransposons
We investigated the position and orientation of retrotransposons in or near NUMTs. Amongs human NUMTs acquired in recent evolutionary history, we found only 10 NUMTs inserted within a retrotransposon, but 547 NUMTs with at least one retrotransposon in one or both of their 1000 bp flanks. The 3′–5′-orientation of the retrotransposons (toward or away from their neighboring NUMT) was approximately equal (600 versus 587).
A+T-rich oligomers are enriched in NUMT flanks
Figure 1B shows an elevated frequency of A+T oligomers in human NUMT flanks and the bordering NUMT regions. Using the procedure described in ‘Materials and Methods’ section, we tested the statistical enrichment of length 2–6 oligomers in NUMT flanks; for human, rhesus, mouse and rat NUMTs. In the first 10 bp of NUMT flanks, each species showed a significant enrichment of A+T dinucleotides (AA, AT, TA and TT); human: p ≈ 0.00069, rhesus: p ∼ 0.0021, mouse: p∼0.0030 and rat: p ∼ 0.0032. As can be seen in Table 1, the most significant oligomers of length 3–6 are also A + T rich, with TAT attaining a P-value of 2.0 × 10−15. TTTTAA, the consensus oligomer recognized by L1-endonuclease (L1-EN), was also significantly enriched (P-value ∼ 0.00139).
Table 1.
Oligomers enriched in NUMT flanks
|
P-value in each species |
||||
|---|---|---|---|---|
| Oligomer | Human | Rhesus | Mouse | Rat |
| TAT | 2.0×10−15 | 5.8×10−8 | 9.2×10−6 | 0.11797 |
| TAC | 0.02822 | 6.1×10−4 | 3.8×10−4 | 0.00130 |
| TATA | 1.4×10−11 | 8.1×10−7 | 3.2×10−4 | 0.02857 |
| AAAC | 0.01485 | 0.00303 | 3.6×10−4 | 8.0×10−4 |
| TATAT | 1.7×10−6 | 1.3×10−6 | 0.00168 | 0.08631 |
| ATTAT | 9.6×10−4 | 7.9×10−8 | 0.03483 | 0.04502 |
| AAACT | 0.08377 | 0.03268 | 7.2×10−6 | 0.05856 |
| AAAAC | 0.00357 | 0.00326 | 0.00619 | 4.7×10−4 |
| TATATA | 4.2×10−4 | 3.6×10−6 | 0.06707 | 0.13717 |
| ATTATT | 0.00370 | 7.9×10−8 | 0.02320 | 0.00689 |
| AAACTT | 0.02436 | 0.09466 | 1.1×10−4 | 0.07799 |
| AATTTA | 0.00406 | 0.14369 | 0.02235 | 4.4×10−4 |
| TTTTAA | 0.00139 | 0.00722 | 0.21079 | 0.01762 |
The statistical enrichment (by binomial test) of the presence of various oligomers of length 3–6 in the 10 bp flanks of NUMTs is shown for four species. For each length, the most significant oligomer for each species is shown in bold, and underlined if it is the overall most significant for that species.
One might consider the possibility that this is an artifact of poorly determined flank boundaries (i.e. hypothesize that we are accidentally treating mtDNA-derived DNA as flanks), but since human nuclear DNA has a higher A+T content (59.1%) than mtDNA (55.5%), we can safely rule out this possibility.
We also looked for statistically significant co-occurrence of oligomer pairs, but did not find any.
NUMT insertion sites typically have high DNA curvature and/or bendability
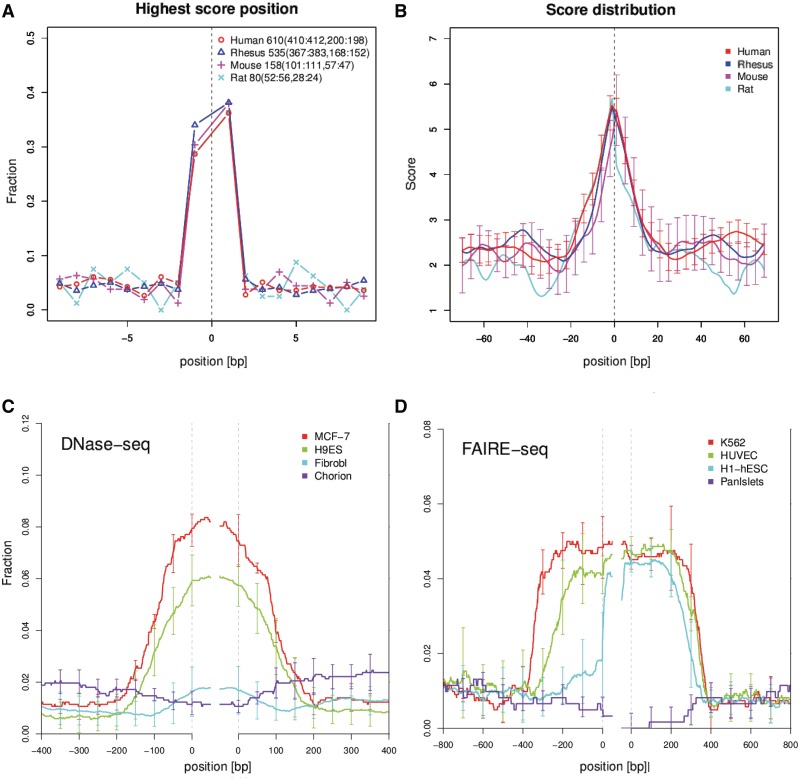
It is well known that A-tracts can cause bending in DNA (33), and it is thought that such bending plays important roles in several biological processes (e.g. gene regulation, packaging and DNA replication) (34,35). In light of our observation that NUMT flanks are rich in A+T oligomers, we decided to investigate the predicted DNA bendability and curvature of approximately reconstructed pre-NUMT insertion sequences, obtained by concatenating the upstream and downstream flanks of each NUMT (see ‘Materials and Methods’ section for details). As shown in Figure 2A and B, for all the organisms tested (human, rhesus, mouse and rat), the inferred NUMT insertion position is usually a local maximum of predicted curvature. A similar trend is seen for predicted bendability as well (Supplementary Figure S3), although the NUMTs which contribute to the high average bendability are not always the same ones which contribute to the high average curvature (Supplementary Figure S4).
Figure 2.
Predicted DNA curvature in NUMT flanks. (A) The horizontal axis gives the distance from the inferred NUMT insertion site. The vertical axis gives the fraction of human NUMTs which attain a local maximum (within the 20 bp window shown) in predicted DNA curvature. (B) The score distribution of DNA curvature in concatenated NUMT flanks is shown. The vertical bars represent standard error. Clear peaks of curvature are observed at inferred NUMT insertion sites. Distributions of experimentally identified open chromatin regions with DNase-seq (C) and FAIRE-seq (D) in NUMT flanks. (C,D): The horizontal axis shows the distance from the inferred NUMT insertion sites. The vertical axis shows the fraction of human NUMTs which have open chromatin at each position.
To see whether the over-representation of A+T rich oligomers in NUMTs flanks can be explained as a result of some selection for highly curved DNA, we computed the statistical over-representation of 10 bp NUMT flank oligomers against a background of 10 bp genomic regions chosen to have a similar distribution of predicted DNA curvature scores. However, use of this curvature score normalized background did not change the results dramatically (Supplementary Table S1).
NUMT insertion sites prefer open chromatin regions
Open chromatin regions are segments of the genomic DNA which are exposed to interacting molecules (36). Previous research has found that active retrotransposons, such as L1 tend to be newly integrated at open chromatin regions (37).
By crosschecking NUMT insertion sites with open chromatin regions defined by DNase-seq and FAIRE-seq, we found that NUMTs also correlate with open chromatin regions, as measured in most cell lines (binomial test, P-value ≪0.05, Figure 2C and D). In particular, NUMT insertion sites strongly correlate with open chromatin measured in the germ cell lines H9ES by DNase-seq, and H1-hESC by FAIRE-seq (P-value ∼1.3 × 10−25), for which the coverage of NUMT flanks within 10 bp of FAIRE-seq boundaries was 4.75%, seven times the genome-wide average of 0.647%.
Correlation of open chromatin data with other NUMT insertion site features
We investigated the co-occurrence of retrotransposons and open chromatin regions at NUMT insertion sites. As defined by FAIRE-seq, we observed some co-occurrence between open chromatin regions and retrotransposons at NUMT sites (the P-value for H1-hESC was 0.047, by Fisher's exact test). On the other hand, DNase hypersensitive sites did not tend to co-occur with retrotransposons at NUMT insertion sites (the P-value for H1-hESC was 0.79). The correlation between retrotransopons and NUMTs might be slightly higher for those in open chromatin, but is clear for both types of NUMTs (Supplementary Figure S9, FAIRE-seq data).
We also investigated the relationship between predicted DNA bendability (or curvature) and open chromatin. To do this we used the random sample of genomic sites, constrained to have a similar distribution of bendability (or curvature) scores as NUMT insertion sites, described in ‘Materials and Methods’ section. For the entire NUMT set, no significant correlation was found with either the FAIRE-seq or DNase-seq data. However, when considering only the most recent age (f) NUMTs, a strong correlation between high bendability and open chromatin was observed (binomial test; P-values: FAIRE-seq ∼3.0 × 10−10 and DNase-seq ∼1.8 × 10−12). Indeed the mean value of bendability scores for the age (f) FAIRE− NUMTs (5.68±0.25) was only somewhat elevated over the genomic background 4.84±0.54, while that for age (f) FAIRE+ NUMTs was highly elevated (7.73±0.30). Curvature also correlated positively with age (f) NUMTs, but only weakly (P-values: FAIRE-seq ∼0.028 and DNase-seq ∼0.040).
Finally, we investigated the correlation between the frequency of A+T rich oligomers and open chromatin properties in NUMT flanks. We calculated the statistical over-representation of oligomers in 10 bp NUMT flanks in open versus other chromatin regions, using the H1-hESC FAIRE-seq data. Similar oligomers were over-represented in both types of NUMTs, but the level of over-representation was stronger (especially for TTT) in open chromatin regions (Supplementary Table S2).
NUMTs do not cluster together
We checked whether NUMTs form clusters or exhibit insertion hotspots on specific chromosomes in human, rhesus monkey, mouse and rat. But no chromosome was significantly enriched (Fisher's exact test, P-value ∼0.64) nor were any hotspots evident in statistical tests (KS test, P-value∼0.54) nor upon visual inspection (Supplementary Figure S2).
Features showing no strong correlation to NUMTs
Chromosomal fragile sites. Chromosomal fragile sites are believed to produced breaks and gaps, due to replication delay in those regions (38–40). However we found that human NUMTs are not significantly enriched in FSs (binomial test, P-value ∼0.67). In this analysis, we used the chromosome bands (e.g. 10q23.3 and Xp22.31) of FSs listed in the review of Schwartz et al. (41) by converting the band information to the genome positions based on information found in the annotation file ‘cytoBand’ from the UCSC web site (42).
Long genes. DSBs can be produced by transcription associated recombination (43). One might speculate that the probability of DSBs may be higher in long genes due to transcriptional pausing (38). To investigate whether NUMTs preferentially insert into long genes, we computed the length distribution of genes found within 100 bp of NUMTs. However, we found no such correlation in our dataset (binomial test, P-value ∼0.78).
CpG islands. We checked whether human NUMTs tend to be embedded within CpG islands as defined by the ‘cpgIslandExt’ track in the UCSC Genome Browser. However, only 1 NUMT out of 610 non-duplicated NUMTs was contained in a CpG island, which is not significantly different than expected by chance (KS test, P-value ∼0.26).
The D-loop region seldom produces NUMTs
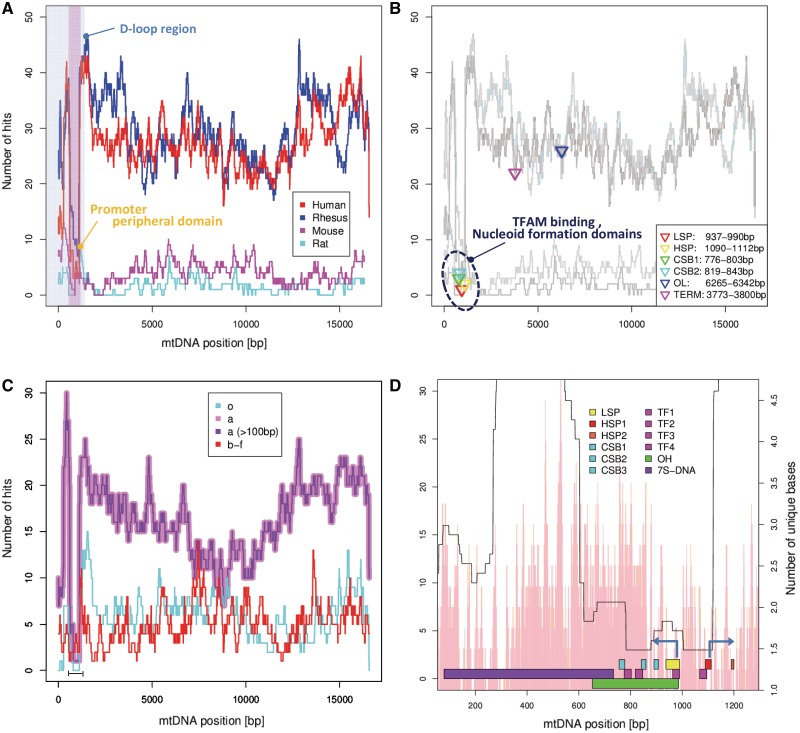
By crosschecking NUMT positions and their source mtDNA positions, we estimated the distribution of source mtDNA corresponding to human NUMTs. We found that the mitochondrial promoter region and its peripheral domains (600 bp–1120 bp from the D-loop start point) in the D-loop have seldom been transferred in human or rhesus monkey (Figure 3A). However, this trend is less clear for mouse and rat NUMTs, perhaps simply due to the small number and overall length of the rodent NUMTs.
Figure 3.
The mitochondrial D-loop region tends not to form NUMTs. (A–D): Histograms of NUMT frequencies. The horizontal axis indicates position in the mitochondrial genome and the vertical axis the number of NUMTs whose inferred source mtDNA overlaps at each position. Note that human mtDNA is circular, so the right edge of plots A–C are conceptually joined to the left edge. A the distribution in several species. The vertically shaded region denotes the D-loop region, with the portion under-represented in NUMTs highlighted in a darker shade. (B) Superimposes the binding positions of several mtDNA binding proteins. (C) The distribution of human NUMTs inserted in different ages. The region under-represented in NUMTs is indicated with a bar. (D) A close-up of the NUMT depleted D-loop region (LSP, light strand promoter, HSP, heavy strand promoter, CSB, conserved sequence block, TF, TFAM binding region, OH, heavy strand replication origin and 7S DNA, displacement loop). The arrows represent direction of transcription from LSP and HSP1. The pink histogram-like bars show a rough estimate of local NUMT detection hardness due to sequence divergence—the number of unique characters (base or gap) in a multiple alignment of the mtDNA of each organism used in this study, averaged over a window of four positions.
We further investigated whether under-representation of D-loop region-derived NUMTs holds for NUMTs of all ages. As shown in Figure 3C and D; the scarcity of NUMTs from the D-loop region is striking in NUMTs from age (a) (inserted after humans diverged from mice and before divergence from rhesus monkeys). It also appears that D-loop NUMTs may be under-represented in older (o) age and younger (b–f) age NUMTs as well, although this is not as clear. We note that there are many more NUMTs from age (a) than the other ages, which may contribute to the clear trends observed for that age.
Validation and estimation of detection limits
Age estimation verification
The phylogenetic age estimation procedure yielded highly significant (≤ 0.001) bootstrap P-values in all cases and we confirmed that the addition of the NUMT sequence never produced an inferred tree topology which contradicts known phylogeny (e.g. with human mtDNA placed closer to gorilla mtDNA than chimpanzee mtDNA) for the mtDNA. However, a small fraction of NUMTs were placed off of the line to human mtDNA; in which case we treated them as coming from before the closest split in the tree leading to the human mtDNA. For example, 12 of the 138 human NUMTs we consider as age (o) were actually placed on the mouse side of the split between human and mouse. The number of human NUMTs identified for each age is listed in the Supplementary Material (Table S3).
To validate our age estimation procedure, we checked the ages of all NUMTs inferred to have inserted after the split between humans and rhesus monkeys (age b–f), by manually inspecting the UCSC provided genome alignment of the human genome with the closest genome not expected to contain the NUMT (for example NUMTs of age (c) should be found in the human, chimpanzee, gorilla and orangutan genomes, but not in the gibbon or rhesus genomes). Of the 120 NUMTs in age (b–f), we found 108 NUMTs to be present or absent in exactly the genomes predicted, 12 NUMTs showed unexpected results, for example present in human, chimpanzee and orangutan, but missing in the gorilla genome—which may indicate a gorilla-specific deletion of the NUMT or possible errors in the nuclear genomes or their alignment. No cases were observed for which the most parsimonious explanation is a simple mistake in age estimation, e.g. a NUMT estimated at age (c) that is found in the gibbon genome. Unfortunately, the squirrel monkey nuclear genome is not completely finished, and human and mouse are sufficiently evolutionarily distant to cause considerable uncertainty in the alignment of neutrally evolving sequences (44). Therefore, we did not extend this manual inspection to age (a) NUMTs.
To augment validation by manual inspection, we computed the average identity of matches to the mtDNA for each age. As expected, the average identity decreased with increasing age: f, 97.2±5.9; e, 94.4±5.2; d, 90.2±7.3; c, 86.7±6.5; b, 82.6±8.9; a, 77.4±6.5 o, 74.1±7.9).
NUMT detection limits
As described in ‘Materials and Methods’ section, we performed a simulation to estimate the detection limit of short, old NUMTs; in which we blanked out all but randomly chosen fixed length segments of NUMTs and checked if they could still be found. For the estimated age (a) NUMTs, we found that a 50 bp length segment was sufficient for detection in all of 10000 trials.
We also performed a separate simulation, designed to provide a lower bound on our ability to detect NUMTs derived from the D-loop of the mitochondrial genome. As detailed in Materials and Methods, our test involved randomly chosen segments of the mouse D-loop mtDNA region, randomly planted into the human nuclear genome. Our search procedure was able to detect all fragments of length 200 bp or more, 94/100 of length 150 and 86/100 of length 100. Since the D-loop evolves faster than other mtDNA regions and also faster than neutrally evolving nuclear DNA (45), 86% can serve as a conservative estimate of our ability to detect NUMTs of length 100 bp, inserted after the common ancestor of human and mouse.
NUMTs were inserted after their neighboring retrotransposons
While performing manual inspection for the age (b–f) NUMTs, we also checked whether any retrotransposons found in their flanks in humans appeared to predate the insertion of the NUMT. Of the 103 age (b–f) non-duplicated NUMTs with retrotransposons in their 500 bp flanks, the retrotransposon appeared before the NUMT in 93 cases, and at the ‘same time’ (e.g. both the NUMT and the retrotransposon first appear in the orangutan genome) in 10 cases. No cases were found in which the NUMT appeared before the retrotransposon.
Phylogenetically younger NUMTs showed stronger correlation with open chromatin regions
We computed the proportion of NUMT flanks near open chromatin regions, as measured by DNase-seq and FAIRE-seq separately for NUMTs of each age. This analysis revealed that NUMTs of the youngest age show a much stronger correlation than older NUMTs (Supplementary Figure S5 and S6).
DISCUSSION
Why our results differ from previous work
Despite the fact that several computational studies have investigated the features of human NUMTs (6,8,9,46), we present novel findings: over-representation of A+T oligomers and open chromatin, and high predicted DNA curvature and bendability at NUMT insertion flanks, and a new conclusion regarding the under-representation of D-loop mtDNA in NUMTs. Indeed, our results contradict the conclusions of some previous work (e.g. the over-representation of transposable elements in NUMT flanks) (3,7,18). In this section, we discuss the reasons for this.
Why we found more NUMTs
Our NUMT dataset covers 632 kb, which is larger than the 400–500 kb reported in previous studies (6,21). This difference can be explained by the use of the BLAST default alignment scoring parameters, which in our hands produces a NUMT dataset of 486 kb. The default BLAST scoring scheme, which penalizes mismatches very severely, is optimal for alignments with 99% identity. It is therefore is only well suited for the detection of very recently created NUMTs. Note that inappropriately strict scoring parameters not only increase the risk of missing NUMTs, they also tend to produce excessively short alignments (47), leading to false ‘NUMT flanks’ which are in fact part of the NUMT.
Why we found retrotransposons enriched in NUMT flanks
In an otherwise careful and informative study, Jensen-Seaman et al. concluded that retrotransposons are ‘under’-represented in NUMT flanks, which directly contradicts our results. The reason why they concluded that retrotransposons are under-represented seems to be due to an unfortunate artifact in the way they computed retrotransposon density. As in our study, they used RepeatMasker-based information to identify retrotransposon sequences, but they did this by applying RepeatMasker to non-overlapping 100bp windows, starting at each NUMT flank—conditions under which RepeatMasker will often miss retrotransposons which straddle the window boundaries. On the other hand, we used the rmsk track from the UCSC web site which includes retrotransposons as identified by RepeatMasker. As shown in Supplementary Figure S7A, using the rmsk track, retrotransposons are-found to be roughly equally enriched in the human-specific dataset of NUMTs from Jensen-Seaman et al. (18) as in our general human NUMT dataset. As shown in Supplementary Figure S8, we also obtain a similar curve when running RepeatMasker ourselves on concatenated NUMT flanks (approximating the nuclear sequence present before the NUMT was inserted).
To further assure ourselves that our result is not due to some mistake in our NUMT flank retrotransposons density computation method, we artificially generated a dataset of randomly inserting ‘NUMTs’ (see ‘Materials and Methods’) and computed their NUMT flank density. As expected, the retrotransposon density of these simulated NUMTs was equal to the genomic background at all distances from the NUMTs (results not shown).
Gherman et al. (3) also reported retrotransposons to be under-represented in NUMTs flanks. However, they suggest this may be due under-estimating the length of NUMTs, so that the perceived ‘NUMT flanks’ are in fact often part of the NUMT itself. This is consistent with our concern that the default BLAST scoring parameters will tend to underestimate the length of most NUMTs.
Why we found NUMT flanks are A+T oligomer rich
Several in silico studies on human NUMTs have been conducted to date (7,18,21), but none of them mention the over-representation of A+T rich oligonucleotides immediately flanking NUMT insertion sites.
To see whether this is due to different datasets. we evaluated flanking oligomers of NUMT sets produced by Jensen-Seaman et al. (18) and Hazkani-Covo et al. (7) and found that the NUMT flanks also showed an enrichment of A+T rich oligomers; Jensen-Seaman et al.: TAA, ATGC, CTTGA and CAAAAA, with P-values of 0.0033, 0.0015, 0.00033 and 0.00017, Hazkani et al.: ATA, AATA, ATTAT and ATATGG, with P-values of 0.0068, 0.00013, 0.00004 and 0.000018, respectively. The consensus oligomer recognized by L1-EN (TTTTAA) was also significantly enriched (Jensen-Seaman et al.: p∼0.0066, Hazkani et al.: p∼0.00475). So the enrichment of A+T rich oligomers also generally holds for their datasets. Their P-values are not as significant as ours but this is probably explainable by the smaller sample size, as those authors only analyzed NUMTs created after the split between humans and chimpanzees.
Why we found the D-loop region under-represented
Most previous studies have concluded that ‘randomly chosen’ mtDNA are copied as NUMTs (7,19,20). Differences in NUMT detection protocols may explain why we noticed the D-loop under-representation (Supplementary Figure S7B). We used a careful NUMT detection protocol designed to identify non-duplicated NUMTs as accurately as possible (see Supplementary Text and Figure S10). Interestingly, the only other study to identify the paucity of D-loop-derived NUMTs was Mourier et al. (19) who used the DBA program (48) to post-process BLAST hits by co-linear alignment merging.
D-loop-derived NUMTs are also rare in the gorilla genome
In the introduction to their article, Jensen-Seaman et al. (18) mention that ‘Within the great apes, it has been suggested that the frequency of NUMTs is increased in gorillas, although these observations are limited to the mitochondria D-loop’, citing PCR-based work (49–51), performed before the gorilla genome was available. With this in mind, we examined the mtDNA source location of gorilla NUMTs, but found that the under-representation of D-loop-derived NUMTs is equally apparent in gorilla as in human (Supplementary Figure S11). Note that in general, we detected somewhat fewer gorilla NUMTs, but this may be due to the fact that the gorilla reference genome is less complete than the human one.
The large number of primate NUMTs may relate to retrotransposon activity?
NUMTs have been identified in many eukaryotes, but their numbers differ widely—with some species possessing several hundred detectable NUMTs, but others fewer than 10 (4,46). It has been suggested that this variation in NUMTs frequency arises from different mtDNA copy number and length between species, but exceptions exist (4).
We suggest that the activity of retrotransposons in recent evolutionary history may be related to frequent NUMT creation. The number of detectable NUMT insertion events is much higher in human (610) and rhesus monkey (667) than in mouse (158). We speculate this may reflect the burst of SINE, especially Alu, activity which occurred during primate evolution (>40 million years ago: after the divergence of early primates from their common ancestor with mouse) (52,53); while the activity of retrotransposons in the mouse line has been relatively constant (for example, see Figure 18 in (54)). However, further support for this hypothesis will require analyses involving more species. An alternative hypothesis is that the difference in NUMT frequency could relate to a difference in DSB repair in the human versus mouse line of evolution; in which light, the observation that human nuclear extract has a much higher DSB binding activity than that of mouse is intriguing (55).
Causes behind the contemporaneous expansion of retrotransposons and NUMTs
Interestingly, the coincidence in evolutionary timing of the expansion of retrotransposons and NUMTs has been discussed at length by Gherman et al. (3), who use that observation to argue that this expansion is due to a population bottleneck, as opposed to retrotransposition activity as hypothesized by Liu et al. (56). Specifically, Gherman et al. (3) assume that the insertion of retrotransposons and NUMTs is unrelated, and therefore that the coincidence in the timing of their expansion is most easily explained by population size effects. Note however that their NUMT definition methodology was unable to detect the strong over-representation of retrotransposons near NUMTs reported here.
Unfortunately, a complete resolution of the question of whether the rapid expansion of retrotransposons was due to increased retrotransposition activity, or a population bottleneck, or both is beyond the scope of our results. However, the strong over-representation of retrotransposons near NUMTs is suggestive of some connection between retrotransposons and NUMT insertion, which weakens the argument of Gherman et al. (3) for an exclusive role of population size. Indeed, we are tempted to speculate that an increased activity of L1-EN may have played a role in the expansion of both retrotransposons and NUMTs.
Mitochondrial D-loop under-represented
Unlike most previous studies (7,20), Mourier et al. (19) observed that the D-loop region is under-represented in detectable human NUMTs. However, they suggested that fact simply reflects the difficulty in detecting NUMTs originating in this quickly evolving part of the mitochondrial genome (19). It is indeed true that, except for a few small conserved blocks (57), the D-loop evolves relatively quickly (45,58). Thus, we concede that the difficulty of detecting older D-loop-derived NUMTs may contribute to their under-representation in NUMT datasets.
However, if detection problems were the only cause, we would only expect to see under-representation of the D-loop in NUMTs old enough to be make detection difficult. In contrast, we find that D-loop-derived NUMTs are strongly under-represented in human NUMTs of inferred age (a) (Figure 3C), which is not the oldest age. Moreover, our validation experiments (See Materials and Methods) demonstrate that we are able to reliably detect age (a) NUMTs of length ≥100 bp. As shown by the purple curve in Figure 3C, the under-representation of D-loop-derived NUMTs is clearly evident even when NUMTs shorter than 100 bp are removed from the analysis.
Why is the D-loop under-represented?
As shown in Figure 3D, the mtDNA contains two distinct under-represented regions, one of just over 500 bp including the light and heavy strand promoters and one of about 300 bp, separated by a band of high NUMT frequency, also around 300 bp long, roughly centered on the 7S DNA region. The clear shape and the control features in this region seem suggestive, but unfortunately we were unable to come up with a well-supported explanation of the NUMT under-representation.
Our best guess is that perhaps TFAM binds these regions and this somehow prevents the region from forming NUMTs. The structure of TFAM affords both specific and non-specific DNA binding (59). On the one hand, TFAM is a highly abundant nucleoid protein covering most or all of the mtDNA (60). However, it can also bind DNA specifically with nano molar affinity (61), with known bindings sites just upstream of the light-and heavy-strand promoters (61) and near conserved sequence block 2 (62). One might expect that as TFAM drops in concentration as mtDNA leaves mitochondria, TFAM would tend to remain bound to its specific binding sites even after most of the mtDNA becomes unoccupied. Moreover, the non-specific binding mode of TFAM is cooperative in vitro (63) so it is plausible that mtDNA adjacent to specific TFAM binding sites might also tend to remain occupied. Interestingly, TFAM has been reported to be found in the nucleus in human (64,65) and rat (66) cancer-derived cells and in mouse testis (67).
Although our best guess, this scenario faces two difficulties in explaining our results. First, the left-hand valley in Figure 3D needs to be explained. Interestingly, the length and spacing of the observed valleys are consistent with the DNA loop structures observed at low TFAM concentration in vitro (63), so in principle the two could be bound together by TFAM. Even so, one must postulate that TFAM also has an unreported, relatively high affinity site in the left-hand valley. This is not clearly supported by methylation studies (68), although part of the left-hand valley is outside of the region measured in that study. A recent mtDNA genome-wide DNase I protection assay (69) shows some sign of protective protein binding in each valley, but this non-TFAM-specific assay shows protection in many areas of the mtDNA. The second difficulty is to account for the sharp transitions seen at the ends of the valleys, which seems to require more than just cooperative binding of TFAM to explain. The placement of the sharpest transition directly after HSP1 seems suggestive but could also be coincidental.
Other theoretically possible explanations of the paucity of NUMTs derived from particular mtDNA regions include: (i) proteins other than TFAM protect those regions, (ii) those regions are preferentially degraded and (iii) they are strongly negatively selected against in the nuclear genome. Unfortunately, without a better understanding of the state of the mitochondria under conditions leading to germ-line NUMT creation, all hypotheses will remain speculative.
Retrotransposons seldom straddle NUMTs
As noted in the ‘Results’ section, out of 557 non-duplicated NUMTs within 1000 bp of a retrotransposon, only 10 were found within retrotransposons. Considering that Alu's are ∼300 bp and LINE's can be much longer, with random insertion in the vicinity of retrotransposons, the random expectation should be that around 13% (300/2300) of NUMTs within 1000 bp of a retrotransposon should fall within the retrotransposon. With this rough estimate, the P-value of only finding 10 or less out of 557 is around 10−21 (binomial test). Since RepeatMasker masks ∼40% of the genome, NUMTs within retrotransposons are also under-represented relative to random insertion into any position in the genome.
This under-representation of NUMT straddling retrotransposons is consistent with the conclusions of Mishmar et al. (9). However, to be careful, we considered the possibility that it is an artifact of retrotransposon identification by RepeatMasker; being concerned that perhaps the insertion of a NUMT would sometimes cause RepeatMasker to miss one side of the divided retrotransposon. Therefore, we ran RepeatMasker after artificially splicing out the NUMT (i.e. on a reconstructed approximation of the sequence present before the NUMT was inserted), to see if more retrotransposons would be found straddling the insertion point. However, the results of this test were identical to using the UCSC web site retrotransposon track — only 10 retrotransposons straddled the insertion point.
Retrotransposons do not noticeably tend to insert near NUMTs
Of the 103 age b–f NUMTs with retrotransposons in one of their 500 bp flanks, comparative analysis shows that in 93 cases the retrotransposon was there first, while 10 cases are indeterminate and in no cases did the retrotransposon clearly insert in or near an existing NUMT. However, this lack of recent retrotransposons inserting into existing NUMTs is not necessarily surprising, due to the slow rate of retrotransposon expansion in recent evolution (corresponding to ages b–f in this study) (52) and the small number of NUMTs. According to Repbase (70), the number of age b–f retrotransposons is roughly 10 000, so for random insertion, the expected number of insertions within 500 bp of our 610 NUMTs is only about 0.002. Thus, the lack of recent retrotransposons insertions into NUMTs is not surprising.
Summary of the characteristics of mammalian NUMT insertion sites
To facilitate the discussion, it is useful to list these observations:
NUMTs tend to insert near retrotransposons, but with no preference for the orientation of the retrotransposon.
NUMTs tend not to insert inside retrotransposons.
NUMTs tend to insert in regions with high local DNA curvature and/or bendability.
NUMTs tend to insert in regions with high A+T rich oligomers, especially TAT.
NUMTs tend to insert into open chromatin regions.
Observation 1 tempts us to hypothesize that L1-EN is involved. Retrotransposon-encoded endonucleases are known to create breaks in regions which contain A+T rich oligomers such as TTTTAA (71–73). Moreover, LINE-encoded endonuclease expression has been shown to create numerous DSBs in human HeLa, MCF7 and mouse HIH 3T3 cells (74).
This hypothesis is weakened by the fact that in recent NUMT insertion sites reconstructed by multiple alignment of the human, chimpanzee and gorilla genomes (18), the L1-EN consensus sequence TTTTAA is not frequently seen; even though it is consistently observed in the insertion sites of disease causing (therefore recent) L1-EN-dependent retrotransposons (75).
However, on the basis of structural considerations and retrotransposition assays, it has been suggested that target DNA bending, rather than the consensus sequence, may be the main determinant of L1-endonuclease recognition (72,76). We might imagine that the initial DNA nicking by L1-EN is largely determined by DNA bending or curvature and many NUMT creation events involve the resulting DSBs; while downstream steps of L1 formation (e.g. initiation of reverse transcription) are what cause the apparent strong preference for the TTTTAA consensus.
For discussion, we note that Observation 2 may conceivably be explained in terms of Observations 3–5. If the high predicted DNA curvature and abundance of A+T rich oligomers reflect some requirement for local DNA structure that Alu's and LINE's lack, then they may be less likely to form DSBs which lead to NUMT insertion. However, this is speculation for which we did not find corroborating evidence. Retrotransposons do contain A+T rich oligomers and regions of high predicted DNA curvature, and the presence of retrotransposons in NUMT flanks does not highly correlate with the DNA curvature or presence of A+T oligomers (results not shown).
Regarding Observation 3, DNA bendability or flexibility is an important factor in the recognition or action of some proteins involved in DNA cleavage (e.g. DNase I (77,78)) and DSB repair (e.g. Topoisomerase IIα (79,80), DNA-PKcs (81), and PNKP (82), which bind to single-stranded nicks in dsDNA). Interestingly, L1-EN was found to have much higher in vitro nicking activity when presented with super-coiled versus relaxed DNA (72).
As reported in Materials and Methods, Observation 4 does not appear to simply be a consequence of Observation 3. Some explanations we considered for the A+T rich oligomers were: (i) they are the poly-A tails of neighboring retrotransposons, (ii) they may be related to the consensus sequence of L1-EN, (iii) they are copies of the poly-A tails from processed mtDNA transcripts, and (iv) they are related to Observation 5. The first explanation seems unlikely because retrotransposons in NUMT flanks are equally enriched in each orientation (5′ toward or away from the NUMT). The second explanation is not well supported, as oligomers containing TAT are more over-represented than the L1-EN consensus sequence TTTTAA. To test the third explanation, we mapped NUMTs to the mtDNA and compared their ends with the position of mitochondrial poly-A sites determined by high-throughput sequencing (69), but found no correspondence. The fourth explanation has some support from the increased over-represented of A+T rich oligomers in open chromatin NUMT insertion sites (Supplementary Table S2) and the moderate sequence preferences exhibited by nucleosomes (83).
Observation 5 supports accessibility as an important factor determining NUMT insertion sites, but not to the exclusion of a possible role for L1-EN. In fact, even for the most recent age (f) NUMTs (for which the chromatin context at the time of insertion should be closest to measurements on modern human cells), most NUMTs appear next to retrotransposons regardless of their observed chromatin state (Supplementary Figure S9). Interestingly, open chromatin has been reported to be more susceptible to nicking by L1-EN (37). The observation that repair of DSBs is less efficient and uses different pathways in heterochromatin versus euchromatin (84,85) may also be relevant.
We suspect that Observation 5 may partially explain the high predicted DNA bendability, but not curvature, observed at inferred NUMT insertion sites. DNA bendability is considered to be associated with open chromatin (86) and showed a strong correlation with FAIRE-seq data in age (f) NUMTs.
NUMTs as markers of chromatin structure in the past
The correlation between open chromatin measurements and very recent NUMTs is striking, but drops precipitously for NUMTs older than the split of humans and chimpanzee. Our interpretation of this is that open chromatin regions are poorly conserved around the regions of the genome (e.g. intergenic) in which we observed NUMTs, and thus for older NUMTs, the FAIRE-seq and DNase-seq data measurements on human cells are a poor indication of the environment at the time of their insertion.
What effect might selection have on our results?
The set of NUMTs we observe in the human genome is not a random sample of NUMT insertion events, because NUMT insertions which are significantly deleterious are unlikely to become fixed in the population. However, we expect this effect is unlikely to qualitatively affect the conclusions we draw regarding features which correlate with NUMT insertion sites—to do so, deleterious NUMTs would need to: (i) constitute a significant fraction of all NUMT insertion events, and (ii) exhibit anti-correlation with the features we observe, so that when taken together the correlations cancel each other out. We doubt condition (i) holds, but rather believe that most of the human genome can accept NUMT insertions without deleterious effects. This belief is consistent with recent estimates that only around 10–15% of the human genome is ‘functional’ (87) and also hinted at by the large number of new Alu and repetitive elements accepted in human evolution (52). Moreover, condition (ii) seems unlikely for the NUMT insertion site features we listed. First of all, it is hard to image why a lack of A+T rich oligomers, or lack of high DNA bendability would correlate strongly with sites under selection pressure. While for open chromatin regions, our intuition is that if there is a correlation with selection pressure, it is likely to be ‘positive’. Regarding retrotransposons, it does seem plausible that functional sites might be less likely to border retrotransposons; and perhaps even show a correlation over distance similar in shape to the inverse of that seen Figure 1A. However, for the magnitude of this effect to explain the observed fraction of retrotransposons found near extant NUMTs (peak at ∼0.8 versus a genome average of ∼0.4), a large fraction of the genome would have to be under strong selection.
Related observations in the yeast literature
Lenglez et al. (10) found that Schizosaccharomyces pombe NUMTs tend to be inserted immediately next to ORIs, and hypothesized that this is due to delay of replication at ORIs (10). On the other hand, ORIs tend to contain A+T rich oligomers (88,89) and Behrens et al. (90) noted that the yeast transposon Tf1 (transposon of fission yeast 1) is densely packed next to replication origins. Thus, although based on circumstantial evidence, we are tempted to speculate that retrotransposons might play a role in NUMT insertion in yeast as well as mammals.
Relevance beyond NUMTs?
Hazkani-Covo et al. (7) reconstructed the approximate pre-insertion sequence of 37 human-specific NUMTs by multiply aligning them with their flanks to human mtDNA and the chimpanzee nuclear genome. They observed that deletion of nuclear DNA did not occur in 54% of those events, in contrast to a nearly 100% rate observed in two experimental systems using V(D)J (91,92) and I-SceI-induced DSBs (93–96). In light of this, they hypothesized that NUMTs serve as filler DNA which mitigates the tendency of short stretches of nuclear DNA to be lost during DSB repair.
We do not present evidence to contradict their hypothesis, but note the logical possibility that repair without deletion may not be caused by the use of filler DNA, but rather that the kind of DSBs which commonly occur in primate germ line cells may have an inherent tendency to allow repair without deletion of nuclear material. In other words, the perceived connection to NUMTs may simply be due to the fact that the DSBs which create NUMTs afford reconstructing the details of their repair. Thus, the characteristics of NUMT insertion sites observed here may reflect mammalian germ-line DSBs in general.
CONCLUSION
By careful alignment methodology we have defined a highly reliable set of human NUMTs and inferred original NUMT insertion sites. Using the inferred insertion sites of non-duplicated NUMTs, we have shown conclusive evidence that retrotransposons are enriched in NUMT insertion site flanks (suggesting that NUMTs preferentially insert near them) and that A+T rich oligomers and regions of open chromatin and high local DNA curvature are enriched in the immediate vicinity (10 bp) of NUMT insertion sites. Finally, we show that the mitochondrial D-loop region has been under-represented in primate NUMT insertion events.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Tables 1–3, Supplementary Figures 1–12 and Supplementary References [4,18,19].
FUNDING
The Global Center of Excellence program “Deciphering Biosphere from Genome Big Bang” (to J.T.). Funding for open access charge: General research funds of AIST, a national lab in Japan.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Mircea Pacurar, for kindly providing us with a standalone version of DNA bendability/curvature predictors. We thank Toutai Mitsuyama, for helping to build NUMT tracks on the UCSC Genome Browser. An anonymous reviewer for suggesting we consider open chromatin data and Dmitrij Frishman and Stefanie Kaufmann for helpful discussion of this topic.
REFERENCES
- 1.Sagan L. On the origin of mitosing cells. J. Theol. Biol. 1967;14:255–274. doi: 10.1016/0022-5193(67)90079-3. Author later changed her name to Lynn Margulis. [DOI] [PubMed] [Google Scholar]
- 2.du Buy H, Riley F. Hybridization between the nuclear and kinetoplast DNA's of Leishmania enriettii and between nuclear and mitochondrial DNA's of mouse liver. Proc. Natl Acad. Sci. USA. 1967;57:790–797. doi: 10.1073/pnas.57.3.790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gherman A, Chen PE, Teslovich TM, Stankiewicz P, Withers M, Kashuk CS, Chakravarti A, Lupski JR, Cutler DJ, Katsanis N. Population bottlenecks as a potential major shaping force of human genome architecture. PLoS Genet. 2007;3:e119. doi: 10.1371/journal.pgen.0030119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hazkani-Covo E, Zeller RM, Martin W. Molecular Poltergeists: Mitochondrial DNA Copies (numts) in Sequenced Nuclear Genomes. PLoS Genet. 2010;6:e1000834. doi: 10.1371/journal.pgen.1000834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ricchetti M, Fairhead C, Dujon B. Mitochondrial DNA repairs double-strand breaks in yeast chromosomes. Nature. 1999;402:96–100. doi: 10.1038/47076. [DOI] [PubMed] [Google Scholar]
- 6.Woischnik M, Moreas CT. Pattern of organization of human mitochondrial pseudogenes in the nuclear genome. Genome Res. 2002;12:885–893. doi: 10.1101/gr.227202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hazkani-Covo E, Covo S. Numt-mediated double-strand break repair mitigates deletions during primate genome evolution. PLoS Genet. 2008;4:e1000237. doi: 10.1371/journal.pgen.1000237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ricchetti M, Tekaia F, Dujon B. Continued colonization of the human genome by mitochondrial DNA. PLoS Biol. 2004;2:e273. doi: 10.1371/journal.pbio.0020273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mishmar D, Ruiz-Pesini E, Brandon M, Wallacen DC. Mitochondrial DNA-like sequences in the nucleus (NUMTs): insights into our african origins and the mechanism of foreign DNA integration. Hum. Mutat. 2004;23:125–133. doi: 10.1002/humu.10304. [DOI] [PubMed] [Google Scholar]
- 10.Lenglez S, Hermand D, Decottignies A. Genome-wide mapping of nuclear mitochondrial DNA sequences links DNA replication origins to chromosomal double-strand break formation in Schizosaccharomyces pombe. Genome Res. 2010;20:1250–1261. doi: 10.1101/gr.104513.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Farrelly F, Butow RA. Rearranged mitochondrial genes in the yeast nuclear genome. Nature. 1983;301:296–301. doi: 10.1038/301296a0. [DOI] [PubMed] [Google Scholar]
- 12.Tsuzuki T, Nomiyama H, Setoyama C, Maeda S, Shimada K. Presence of mitochondrial-DNA-like sequences in the human nuclear DNA. Gene. 1983;25(2–3):223–229. doi: 10.1016/0378-1119(83)90226-3. [DOI] [PubMed] [Google Scholar]
- 13.Zullo S, Sieu L, Slightom J, Hadler H, Eisenstadt J. Mitochondrial D-loop sequences are integrated in the rat nuclear genome. J. Mol. Biol. 1991;221:1223–1235. [PubMed] [Google Scholar]
- 14.Ossorio P, Sibley L, Boothroyd J. Mitochondrial-like DNA sequences flanked by direct and inverted repeats in the nuclear genome ofToxoplasma gondii. J. Mol. Biol. 1991;222:525–536. doi: 10.1016/0022-2836(91)90494-q. [DOI] [PubMed] [Google Scholar]
- 15.Willett-Brozick JE, Savul SA, Richey LE, Baysal BE. Germ line insertion of mtDNA at the breakpoint junction of a reciprocal constitutional translocation. Hum. Genet. 2001;109:216–223. doi: 10.1007/s004390100564. [DOI] [PubMed] [Google Scholar]
- 16.Blanchard JL, Schmidt GW. Mitochondrial DNA migration events in yeast and humans: integration by a common end-joining mechanism and alternative perspectives on nucleotide substitution patterns. Mol. Biol. Evol. 1996;13:537–548. doi: 10.1093/oxfordjournals.molbev.a025614. [DOI] [PubMed] [Google Scholar]
- 17.Qu H, Ma F, Li Q. Comparative analysis of mitochondrial fragments transferred to the nucleus in vertebrate. J. Genet. Genomics. 2008;35:485–490. doi: 10.1016/S1673-8527(08)60066-1. [DOI] [PubMed] [Google Scholar]
- 18.Jensen-Seaman MI, Wildschutte JH, Soto-Calderon ID, Anthony NM. A comparative approach shows differences in patterns of numt insertion during hominoid evolution. J. Mol. Evol. 2009;68:688–699. doi: 10.1007/s00239-009-9243-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mourier T, Hansen AJ, Willerslev E, Arctander P. The human genome project reveals a continuous transfer of large mitochondrial fragments to the nucleus. Mol. Biol. Evol. 2001;18:1833–1837. doi: 10.1093/oxfordjournals.molbev.a003971. [DOI] [PubMed] [Google Scholar]
- 20.Tourmen Y, Baris O, Dessen P, Jacques C, Malthiery Y, Reynier P. Structure and chromosomal distribution of human mitochondrial pseudogenes. Genomics. 2002;80:71–77. doi: 10.1006/geno.2002.6798. [DOI] [PubMed] [Google Scholar]
- 21.Hazkani-Covo E, Graur D. A comparative analysis of numt evolution in human and chimpanzee. Mol. Biol. Evol. 2007;24:13–18. doi: 10.1093/molbev/msl149. [DOI] [PubMed] [Google Scholar]
- 22.Kielbasa SM, Wan R, Sato K, Horton P, Frith MC. Adaptive seeds tame genomic sequence comparison. Genome Res. 2011;21:487–493. doi: 10.1101/gr.113985.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Korf I, Yandell M, Bedell J. BLAST. Sebastopol, CA, USA: O'Reilly & Associates, Inc.; 2003. [Google Scholar]
- 24.Frith MC, Hamada M, Horton P. Parameters for accurate genome alignment. BMC Bioinformatics. 2010;11:80. doi: 10.1186/1471-2105-11-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Frith MC. A new repeat-masking method enables specific detection of homologous sequences. Nucleic Acids Res. 2010;39:e23. doi: 10.1093/nar/gkq1212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bailey JA, Gu Z, Clark RA, Reinert K, Samonte RV, Schwartz S, Adams MD, Myers EW, Li PW, Eichler EE. Recent segmental duplication in the human genome. Science. 2002;297:1003–1007. doi: 10.1126/science.1072047. [DOI] [PubMed] [Google Scholar]
- 28.Larkin M, Blackshields G, Brown N, Chenna R, McGettigan P, McWilliam H, Valentin F, Wallace I, Wilm A, Lopez R, et al. Clustal W and clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 29.Felsenstein J. PHYLIP—phylogeny inference package (Version 3.2) Cladistics. 1989;5:164–166. [Google Scholar]
- 30.Vlahoviček K, Kaján L, Pongor S. DNA analysis servers: plot.it, bend.it. Nucleic Acids Res. 2003;31:3686–3687. doi: 10.1093/nar/gkg559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Crawford G, Holt IE, Whittle J, Webb BD, Tai D, Davis S, Margulies EH, Chen Y, Bernat JA, Ginsburg D, et al. Genome-wide mapping of DNase hypersensitive sites using massively parallel signature sequencing (MPSS) Genome Res. 2006;16:123–131. doi: 10.1101/gr.4074106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Giresi PG, Kim J, McDaniell RM, Iyer VR, Lieb JD. FAIRE (Formaldehyde-Assisted Isolation of Regulatory Elements) isolates active regulatory elements from human chromatin. Genome Res. 2007;17:877–885. doi: 10.1101/gr.5533506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Koo HS, Wu HM, Crothers DM. DNA bending at adenine·thymine tracts. Nature. 1986;320:501–506. doi: 10.1038/320501a0. [DOI] [PubMed] [Google Scholar]
- 34.Hizver J, Rozenberg H, Frolow F, Rabinovich D, Shakked Z. DNA bending by an adenine-thymine tract and its role in gene regulation. Proc. Natl. Acad. Sci. USA. 2001;98:8490–8495. doi: 10.1073/pnas.151247298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gabrielian A, Simoncsits A, Pongor S. Distribution of bending propensity in DNA sequences. FEBS Lett. 1996;393:124–130. doi: 10.1016/0014-5793(96)00837-x. [DOI] [PubMed] [Google Scholar]
- 36.Song L, Zhang Z, Grasfeder LL, Boyle AP, Giresi PG, Lee B-K, Sheffield NC, Gräf S, Huss M, et al. Open chromatin defined by DNaseI and FAIRE identifies regulatory elements that shape cell-type identity. Genome Res. 2011;21:1757–1767. doi: 10.1101/gr.121541.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cost GJ, Golding A, Schlissel MS, Boeke JD. Target DNA chromatinization modulates nicking by L1 endonuclease. Nucleic Acids Res. 2001;29:573–577. doi: 10.1093/nar/29.2.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Helmrich A, Stout-Weider K, Hermann K, Schrock E, Heiden T. Common fragile sites are conserved features of human and mouse chromosomes and relate to large active genes. Genome Res. 2006;16:1222–1230. doi: 10.1101/gr.5335506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Glover TW, Arlt MF, Casper AM, Durkin SG. Mechanisms of common fragile site instability. Hum. Mol. Genet. 2005;14:197–205. doi: 10.1093/hmg/ddi265. [DOI] [PubMed] [Google Scholar]
- 40.Zlotorynski E, Rahat A, Skaug J, Ben-Porat N, Ozeri E, Hershberg R, Levi A, Scherer SW, Margalit H, Kerem B. Molecular basis for expression of common and rare fragile sites. Mol. Cell Biol. 2003;23:7143–7151. doi: 10.1128/MCB.23.20.7143-7151.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schwartz M, Zlotorynski E, Kerem B. The molecular basis of common and rare fragile sites. Cancer Lett. 2006;232:13–26. doi: 10.1016/j.canlet.2005.07.039. [DOI] [PubMed] [Google Scholar]
- 42.Furey TS, Haussler D. Integration of the cytogenetic map with the draft human genome sequence. Hum. Mol. Genet. 2003;12:1037–1044. doi: 10.1093/hmg/ddg113. [DOI] [PubMed] [Google Scholar]
- 43.Aguilera A, Gomez-Gonzalez B. Genome instability: a mechanistic view of its causes and consequences. Nat. Rev. Genet. 2008;9:204–217. doi: 10.1038/nrg2268. [DOI] [PubMed] [Google Scholar]
- 44.Schwartz S, Kent WJ, Smit A, Zhang Z, Baertsch R, Hardison RC, Haussler D, Miller W. Human-mouse alignments with BLASTZ. Genome Res. 2003;13:103–107. doi: 10.1101/gr.809403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lopez J, Culver M, Stephens JC, Johnson W, O'Brien S. Rates of nuclear and cytoplasmic mitochondrial DNA sequence divergence in mammals. Mol. Biol. Evol. 1997;14:277–286. doi: 10.1093/oxfordjournals.molbev.a025763. [DOI] [PubMed] [Google Scholar]
- 46.Richly E, Leister D. NUMTs in sequenced eukaryotic genomes. Mol. Biol. Evol. 2004;21:1081–1084. doi: 10.1093/molbev/msh110. [DOI] [PubMed] [Google Scholar]
- 47.Frith MC, Park Y, Sheetlin SL, Spouge JL. The whole alignment and nothing but the alignment: the problem of spurious alignment flanks. Nucleic Acids Res. 2008;36:5863–5871. doi: 10.1093/nar/gkn579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jareborg N, Birney E, Durbin R. Comparative analysis of noncoding regions of 77 orthologous mouse and human gene pairs. Genome Res. 1999;9:815–824. doi: 10.1101/gr.9.9.815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jensen-Seaman MI, Sarmiento EE, Deinard AS, Kidd KK. Nuclear integrations of mitochondrial DNA in gorillas. Am. J. Primatol. 2004;63:139–147. doi: 10.1002/ajp.20047. [DOI] [PubMed] [Google Scholar]
- 50.Thalmann O, Hebler J, Poinar HN, Paabo S, Vigilant L. Unreliable mtDNA data due to nuclear insertions: a cautionary tale from analysis of humans and other great apes. Mol. Ecol. 2004;13:321–335. doi: 10.1046/j.1365-294x.2003.02070.x. [DOI] [PubMed] [Google Scholar]
- 51.Anthony NM, Clifford SL, Bawe-Johnson M, Abernethy KA, Bruford MW, Wickings EJ. Distinguishing gorilla mitochondrial sequences from nuclear integrations and PCR recombinants: guidelines for their diagnosis in complex sequence databases. Mol. Phylogenet. Evol. 2007;43:553–566. doi: 10.1016/j.ympev.2006.09.013. [DOI] [PubMed] [Google Scholar]
- 52.Batzer MA, Deininger PL. Alu repeats and human genomic diversity. Nat. Rev. Genet. 2002;3:370–379. doi: 10.1038/nrg798. [DOI] [PubMed] [Google Scholar]
- 53.Cordaux R, Batzer MA. The impact of retrotransposons on human genome evolution. Nat. Rev. Genet. 2009;10:691–703. doi: 10.1038/nrg2640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Consortium IHGS. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 55.Lorenzini A, Johnson FB, Oliver A, Tresini M, Smith JS, Hdeib M, Sell C, Cristofalo VJ, Stamato TD. Significant correlation of species longevity with DNA double strand break recognition but not with telomere length. Mech. Ageing Dev. 2009;130:784–792. doi: 10.1016/j.mad.2009.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Liu G, Zhao S, Bailey JA, Sahinalp SC, Alkan C, Tuzun E, Green ED, Eichler EE. Analysis of primate genomic variation reveals a repeat-driven expansion of the human genome. Genome Res. 2003;13:358–368. doi: 10.1101/gr.923303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chang DD, Clayton DA. Priming of human mitochondrial DNA replication occurs at the light-strand promoter. Proc. Natl. Acad. Sci. USA. 1985;82:351–355. doi: 10.1073/pnas.82.2.351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Parsons TJ, Muniec DS, Sullivan K, Woodyatt N, Alliston-Greiner R, Wilson MR, Berry DL, Holland KA, Weedn VW, Gill P. A high observed substitution rate in the human mitochondrial DNA control region. Nat. Genet. 1997;15:363–368. doi: 10.1038/ng0497-363. [DOI] [PubMed] [Google Scholar]
- 59.Hallberg BM, Larsson NG. TFAM forces mtDNA to make a U-turn. Nat. Struct. Mol. Biol. 2011;18:1179–1181. doi: 10.1038/nsmb.2167. [DOI] [PubMed] [Google Scholar]
- 60.Kukat C, Wurm CA, Spåhr H, Falkenberg M, Larsson NG, Jakobs S. Super-resolution microscopy reveals that mammalian mitochondrial nucleoids have a uniform size and frequently contain a single copy of mtDNA. Proc. Natl Acad. Sci. USA. 2011;108:13534–13539. doi: 10.1073/pnas.1109263108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Fisher RP, Topper JN, Clayton DA. Promoter selection in human mitochondria involves binding of a transcription factor to orientation-independent upstream regulatory elements. Cell. 1987;50:247–258. doi: 10.1016/0092-8674(87)90220-0. [DOI] [PubMed] [Google Scholar]
- 62.Suissa S, Wang Z, Poole J, Wittkopp S, Feder J, Shutt TE, Wallace DC, Shadel GS, Mishmar D. Ancient mtDNA genetic variants modulate mtDNA transcription and replication. PLos Genet. 2009;5:e1000474. doi: 10.1371/journal.pgen.1000474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kaufman BA, Durisic N, Mativetsky JM, Costantino S, Hancock MA, Grutter P, Shoubridge EA. The mitochondrial transcription factor TFAM coordinates the assembly of multiple DNA molecules into nucleoid-like structures. Mol. Biol. Cell. 2007;18:3225–3236. doi: 10.1091/mbc.E07-05-0404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Pastukh V, Shokolenko I, Wang B, Wilson G, Alexeyev M. Human mitochondrial transcription factor A possesses multiple subcellular targeting signals. FEBS J. 2007;274:6488–6499. doi: 10.1111/j.1742-4658.2007.06167.x. [DOI] [PubMed] [Google Scholar]
- 65.Han B, Izumi H, Yasuniwa Y, Akiyama M, Yamaguchi T, Fujimoto N, Matsumoto T, Wu B, Tanimoto A, Sasaguri Y. Human mitochondrial transcription factor A functions in both nuclei and mitochondria and regulates cancer cell growth. Biochem. Biophys. Res. Commun. 2011;408:45–51. doi: 10.1016/j.bbrc.2011.03.114. [DOI] [PubMed] [Google Scholar]
- 66.Dong X, Ghoshal K, Majumder S, Yadav SP, Jacob ST. Mitochondrial transcription factor A and its downstream targets are up-regulated in a rat hepatoma. J. Biol. Chem. 2002;277:43309–43318. doi: 10.1074/jbc.M206958200. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 67.Larsson NG, Garman JD, Oldfors A, Barsh GS, Clayton DA. A single mouse gene encodes the mitochondrial transcription factor A and a testis-specific nuclear HMG-box protein. Nat. Genet. 1996;13:296–302. doi: 10.1038/ng0796-296. [DOI] [PubMed] [Google Scholar]
- 68.Rebelo AP, Williams SL, Moraes CT. In vivo methylation of mtDNA reveals the dynamics of protein-mtDNA interactions. Nucleic Acids Res. 2009;37:6701–6715. doi: 10.1093/nar/gkp727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Mercer TR, Neph S, Dinger ME, Crawford J, Smith MA, Shearwood AM, Haugen E, Bracken CP, Rackham O, Stamatoyannopoulos JA, et al. The human mitochondrial transcriptome. Cell. 2011;146:645–658. doi: 10.1016/j.cell.2011.06.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Jurka J. Repbase update: a database and an electronic journal of repetitive elements. TRENDS Genet. 2000;16:418–420. doi: 10.1016/s0168-9525(00)02093-x. [DOI] [PubMed] [Google Scholar]
- 71.Toda Y, Saito R, Tomita M. Characteristic sequence pattern in the 5- to 20-bp upstream region of primate Alu elements. J. Mol. Evol. 2000;50:232–237. doi: 10.1007/s002399910026. [DOI] [PubMed] [Google Scholar]
- 72.Repanas K, Zingler N, Layer LE, Schumann GG, Perrakis A, Weichenrieder O. Determinants for DNA target structure selectivity of the human LINE-1 retrotransposon endonuclease. Nucleic Acids Res. 2007;35:4914–4926. doi: 10.1093/nar/gkm516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ichiyanagi K, Nishihara H, Duvernell DD, Okada N. Acquisition of endonuclease specificity during evolution of L1 retrotransposon. Mol. Biol. Evol. 2007;24:2009–2015. doi: 10.1093/molbev/msm130. [DOI] [PubMed] [Google Scholar]
- 74.Gasior SL, Wakeman TP, Xu B, Deininger PL. The human LINE-1 Retrotransposon creates DNA double-strand breaks. J. Mol. Biol. 2006;357:1383–1393. doi: 10.1016/j.jmb.2006.01.089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Chen JM, Stenson PD, Cooper DN, Ferec C. A systematic analysis of LINE-1 endonuclease-dependent retrotranspositional events causing human genetic disease. Hum. Genet. 2005;117:411–427. doi: 10.1007/s00439-005-1321-0. [DOI] [PubMed] [Google Scholar]
- 76.Weichenrieder O, Repanas K, Perrakis A. Crystal structure of the targeting endonuclease of the human LINE-1 retrotransposon. Structure. 2004;12:975–986. doi: 10.1016/j.str.2004.04.011. [DOI] [PubMed] [Google Scholar]
- 77.Hogan ME, Roberson MW, Austin RH. DNA flexibility variation may dominate DNase I cleavage. Proc. Natl Acad. Sci. USA. 1989;86:9273–9277. doi: 10.1073/pnas.86.23.9273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Heddi B, Abi-Ghanem J, Lavigne M, Hartmann B. Sequence-dependent DNA flexibility mediates DNaseI cleavage. J. Mol. Biol. 2010;395:123–133. doi: 10.1016/j.jmb.2009.10.023. [DOI] [PubMed] [Google Scholar]
- 79.Hardin AH, Sarkar SK, Seol Y, Liou GF, Osheroff N, Neuman KC. Direct measurement of DNA bending by type IIα topoisomerases: implications for non-equilibrium topology simplification. Nucleic Acids Res. 2011;39:5729–5743. doi: 10.1093/nar/gkr109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Lee S, Jung SR, Heo K, Byl JA, Deweese JE, Osheroff N, Hohng S. DNA cleavage and opening reactions of human topoisomerase IIα are regulated via Mg2+-mediated dynamic bending of gate-DNA. Proc. Natl Acad. Sci. USA. 2012;109:2925–2930. doi: 10.1073/pnas.1115704109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Dip R, Naegeli H. More than just strand breaks: the recognition of structural DNA discontinuities by DNA-dependent protein kinase catalytic subunit. FASEB J. 2005;19:704–715. doi: 10.1096/fj.04-3041rev. [DOI] [PubMed] [Google Scholar]
- 82.Garces F, Pearl LH, Oliver AW. The structural basis for substrate recognition by mammalian polynucleotide kinase 3′ phosphatase. Mol. Cell. 2011;44:385–396. doi: 10.1016/j.molcel.2011.08.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Valouev A, Johnson SM, Boyd SD, Smith CL, Fire AZ, Sidow A. Determinants of nucleosome organization in primary human cells. Nature. 2011;474:516–520. doi: 10.1038/nature10002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Goodarzi AA, Jeggo P, Lobrich M. The influence of heterochromatin on DNA double strand break repair: getting the strong, silent type to relax. DNA Repair (Amst) 2010;9:1273–1282. doi: 10.1016/j.dnarep.2010.09.013. [DOI] [PubMed] [Google Scholar]
- 85.Jakob B, Splinter J, Conrad S, Voss KO, Zink D, Durante M, Löbrich M, Taucher-Scholz G. DNA double-strand breaks in heterochromatin elicit fast repair protein recruitment, histone H2AX phosphorylation and relocation to euchromatin. Nucleic Acids Res. 2011;39:6489–6499. doi: 10.1093/nar/gkr230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Vinogradov AE. DNA helix: the importance of being GC-rich. Nucleic Acids Res. 2003;31 doi: 10.1093/nar/gkg296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Ponting CP, Hardison RC. What fraction of the human genome is functional? Genome Res. 2011;21:1769–1776. doi: 10.1101/gr.116814.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Gomez M, Antequera F. Organization of DNA replication origins in the fission yeast genome. EMBO J. 1999;18:5683–5690. doi: 10.1093/emboj/18.20.5683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Nieduszynski CA, Knox Y, Donaldson AD. Genome-wide identification of replication origins in yeast by comparative genomics. Genes Dev. 2006;20:1874–1879. doi: 10.1101/gad.385306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Behrens R, Hayles J, Nurse P. Fission yeast retrotransposon Tf1 integration is targeted to 5′ ends of open reading frames. Nucleic Acids Res. 2000;28:4749–4716. doi: 10.1093/nar/28.23.4709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Lieber MR, Ma Y, Pannicke U, Schwarz K. The mechanism of vertebrate nonhomologous DNA end joining and its role in V(D)J recombination. DNA Repair (Amst) 2004;3:817–826. doi: 10.1016/j.dnarep.2004.03.015. [DOI] [PubMed] [Google Scholar]
- 92.Ramsden D, Paige C, Wu G. Kappa light chain rearrangement in mouse fetal liver. J. Immunol. 1994;153:1150–1160. [PubMed] [Google Scholar]
- 93.Capp JP, Boudsocq F, Besnard AG, Lopez BS, Cazaux C, Hoffmann JS, Canitrot Y. Involvement of DNA polymerase μ in the repair of a specific subset of DNA double-strand breaks in mammalian cells. Nucleic Acids Res. 2007;35:3551–3560. doi: 10.1093/nar/gkm243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Guirouilh-Barbat J, Huck S, Bertrand P, Pirzio L, Desmaze C, Sabatier L, Lopez BS. Impact of the KU80 pathway on NHEJ-induced genome rearrangements in mammalian cells. Mol. Cell. 2004;14:611–623. doi: 10.1016/j.molcel.2004.05.008. [DOI] [PubMed] [Google Scholar]
- 95.Honma M, Sakuraba M, Koizumi T, Takashima Y, Sakamoto H, Hayashi M. Non-homologous end-joining for repairing I-Sel-induced DNA double strand breaks in human cells. DNA Repair (Amst) 2007;6:781–788. doi: 10.1016/j.dnarep.2007.01.004. [DOI] [PubMed] [Google Scholar]
- 96.Rebuzzini P, Khoriauli L, Azzalin CM, Magnani E, Mondello C, Giulotto E. New mammalian cellular systems to study mutations introduced at the break site by non-homologous end-joining. DNA Repair (Amst) 2004;4:546–555. doi: 10.1016/j.dnarep.2004.12.011. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.