Abstract
Enriching target sequences in sequencing libraries via capture hybridization to bait/probes is an efficient means of leveraging the capabilities of next-generation sequencing for obtaining sequence data from target regions of interest. However, homologous sequences from non-target regions may also be enriched by such methods. Here we investigate the fidelity of capture enrichment for complete mitochondrial DNA (mtDNA) genome sequencing by analyzing sequence data for nuclear copies of mtDNA (NUMTs). Using capture-enriched sequencing data from a mitochondria-free cell line and the parental cell line, and from samples previously sequenced from long-range PCR products, we demonstrate that NUMT alleles are indeed present in capture-enriched sequence data, but at low enough levels to not influence calling the authentic mtDNA genome sequence. However, distinguishing NUMT alleles from true low-level mutations (e.g. heteroplasmy) is more challenging. We develop here a computational method to distinguish NUMT alleles from heteroplasmies, using sequence data from artificial mixtures to optimize the method.
INTRODUCTION
Next-generation sequencing (NGS) platforms can process hundreds of thousands to millions of DNA templates in parallel, thereby providing dramatically faster and cost-effective sequence throughput compared with traditional capillary sequencing (1). With this fast-evolving technology, whole-genome sequencing is enabled for most organisms and will likely be routine in the future. However, currently, it is not yet feasible to sequence large numbers of complex genomes, as the cost and time required are still prohibitive. Moreover, whole-genome sequencing is not necessary for many studies that focus on some specific target region(s) of interest (e.g. specific genes, exons, regulatory elements, etc.). In such cases, targeted sequencing is preferable as most of the sequencing capacity is then devoted to the genomic region(s) of interest. Although various target-enrichment methods have been developed (2,3), it is unclear how homologous sequences from non-target regions might influence the sequencing results. This is particularly the case for the capture-enrichment method (4,5), which makes use of the similarity between a bait/probe and the target. Investigating this issue is particularly important for clinical and forensic applications, because contamination from homologous sequences could be misidentified as lower level (heterogeneous) mutations.
Mitochondrial DNA (mtDNA) genome sequencing offers an excellent opportunity to evaluate the contamination from homologous sequences in capture-enriched sequence data. First, sequences homologous to the mtDNA genome are well-characterized in the nuclear genome (nuclear mitochondrial DNA inserts or NUMTs) (6–8). NUMTs vary in length and similarity to the mtDNA genome; thus, NUMTs may co-enrich with the mtDNA. Second, a rapid and cost-effective method for capture enrichment of mtDNA is available (9), which has been carried out on hundreds of samples in our laboratory (10; unpublished), providing substantial comparative data to work with. Third, mtDNA sequencing in general has been widely carried out and has important biomedical and forensic applications (11), so it is important to understand the impact of NUMTs on capture-enrichment methods for mtDNA. Finally, the availability of mitochondria-free cell lines (which have nuclei but lack mitochondria) provides a perfect negative control for assessing the impact of NUMTs on capture-enriched sequence data.
In this study, we first collected all known NUMTs through a computational search of the nuclear genome and found many mtDNA positions that could be potentially affected by NUMTs. We evaluated the impact of NUMTs in capture-enriched sequence data by analyzing such data from a mitochondria-free cell line and the parental cell line, and by comparing long-range PCR enriched sequence data (expected to be largely free of NUMTs) to capture-enriched sequence data for the same samples. Our results indicate that NUMTs are indeed captured along with the mtDNA genome. Furthermore, we evaluated methods for distinguishing true low-level mutations (i.e. heteroplasmy) from NUMTs by analyzing artificially mixed samples.
MATERIALS AND METHODS
NUMTs in silico
To predict all NUMTs that exist in the human nuclear genome, previously published criteria were applied (6). BLASTn was used to compare the revised Cambridge reference sequence for the human mtDNA genome with the human nuclear genome (HG19 excluding mtDNA) (12,13). All hits with e-score ≤0.0001 were kept as potential NUMTs (see a full list in Supplementary Table S1). A Perl script was written to retrieve all positions showing a difference (substitution or indel) between each NUMT and the mtDNA genome (we call these ‘NUMTs-affected positions’). The position on the mtDNA genome, counts of corresponding NUMTs, and nucleotides in the mtDNA genome and NUMTs were recorded.
All reported human mtDNA polymorphism data were downloaded from the MITOMAP website (www.mitomap.org) (14). For each identified NUMTs-affected position, we examined whether the same mutational difference (occurring between the mtDNA and nuclear DNA copies) was also observed as an mtDNA polymorphism in human populations. All of these data can be downloaded from http://sourceforge.net/projects/dmcrop/files/.
Whole-mtDNA genome sequencing
DNA from 14 samples was extracted from cheek cell swabs/saliva/blood as described previously (15,16). MtDNA was enriched by an in-solution capture method and sequenced on the Illumina GAIIx platform (GAIIx; San Diego, CA, USA) via a multiplex sequencing protocol (9); the average sequencing coverage (in-target) was 930× with read length of 76 bp (single end). The same DNA samples were sequenced previously on the same platform using long-range PCR products to construct the sequencing libraries (15). Briefly, the mtDNA genome was amplified in two overlapping products of ∼9.7 and 7.3 kb and the average sequencing coverage was 1328× with read lengths of 36 and 76 bp (single-end) (15). Furthermore, whole-genome shotgun sequencing data from 13 samples (NA18501, NA18502, NA18504, NA18505, NA18507, NA18508, NA18510, NA18511, NA18517, NA18519, NA18520, NA18522, NA18523) was retrieved from the 1000 Genomes project (17); the average sequencing coverage for mtDNA was 1919× with read lengths of 36 bp (single end).
Quality control and genome assembly
First, the base quality score was recalibrated with the IBIS software using PhiX 174 sequencing data as the training dataset (18). Reads with more than five bases having a quality score <15 were removed from further analysis. The adapter sequences were trimmed, and reads were mapped to both the mtDNA reference genome (NC012920) and the entire genome (HG19) using the BWA assembler (19). Single-end reads with identical alignment positions were retained as most of them are derived from unique molecules (20). The resulting SAM output was further filtered by requiring not more than two mismatches and a minimum mapping quality score of 20. All bases with a quality score <20 were removed from the pileup file.
Mitochondria-free (rho zero) cell line
DNA from a mitochondria-free (rho zero) cell line and the parental cell line from which it was created were kindly provided by EMD-Millipore Inc. (San Diego, CA, USA). Genomic libraries were prepared and pooled with other libraries before hybridization capture (9). To avoid any possible contamination caused by jumping PCR or cluster misidentification, indexes were added at both ends of the template DNA (21), and all reads lacking the expected double indexes were discarded. These libraries were prepared in duplicate to investigate the reproducibility of the results.
Artificially mixed samples
Two mixtures were prepared from genomic DNA, one from two individuals differing at 34 positions in their mtDNA genomes, and the other from two individuals differing at 27 positions. Mixtures were prepared in the following proportions: 1:1, 1:3, 1:9, 1:19 and 1:39. DNA concentrations were assessed via qPCR (Mx3005PTM, Stratagene) using mtDNA-specific primers (22), and the DNA samples were diluted and mixed in the above proportions in triplicate, and then used for capture enrichment and Illumina GAIIx sequencing as described above.
Distinguishing NUMT alleles from heteroplasmy
To distinguish NUMT alleles from true heteroplasmic positions, we made use of the DREEP (Detecting low-level mutations by utilizing the re-sequencing error profile of the data) software that we developed previously to distinguish low-level mutations (i.e. heteroplasmy) from sequencing errors (20). This method assigns a quality score to the minor allele at each position, which is a Phred-like value that measures the deviation of the observed minor allele count from the expect error count derived from a reference panel. Here, NUMTs were regarded as a special type of sequencing error, and the quality score assigned by DREEP then reflects the deviation of the minor allele count from the expected minor allele count (caused by sequencing error and NUMTs), derived from a reference panel. The DREEP software is available at http://dmcrop.sourceforge.net.
RESULTS
NUMTs in silico
We identified 1077 NUMTs in silico from analysis of the HG19 human reference genome. HG19 is a composite of sequences from multiple individuals, and 28 of the identified NUMTs are located on unplaced sequences or alternative loci. The NUMTs range in length from 34 to 8798 bp, with an average of 240 bp. The percent sequence identity to human mtDNA ranged from 78% to 100%. By comparing the mtDNA reference sequence with the NUMTs, we found 8239 positions (49.7% of the mtDNA genome) that could be potentially affected by NUMT alleles in that the NUMTs possess one or more alternative nucleotides relative to the mtDNA reference sequence. The count of alternative NUMTs alleles mapped to the same mtDNA position ranges from 1 to 46, and >90% of them were identical (Supplementary Table S2). Moreover, the majority (74%) of alternative NUMT alleles were not found among modern human mtDNAs, suggesting that they are either mutations that arose in the NUMT after the insertion event or insertions of mtDNA sequences that are no longer present in modern human populations.
NUMTs in the mt-free (rho zero) cell line
The amount of endogenous mtDNA in the mt-free (rho zero) cell line was examined, by comparing the enrichment for reads mapping to the mtDNA genome after capture hybridization of sequencing libraries prepared from both the mt-free cell line and its parental cell line. A large fraction of the reads from both the mt-free cell line and its parental cell line mapped to the HG19 human reference genome (Table 1), while only 11.2% of the reads were inferred to be duplicate reads, indicating that there was sufficient endogenous DNA in the libraries for sequencing. However, ∼40% of the reads from the parental cell line mapped to the mtDNA genome, whereas <0.1% of the reads from the mt-free cell line mapped to mtDNA (Table 1). Thus, the mt-free cell line is indeed essentially devoid of mtDNA. The few reads from the mt-free cell line that do map to mtDNA could reflect a small amount of surviving mtDNA, contamination, artifacts such as jumping PCR, or NUMTs. If the mtDNA genome is used as the only mapping reference, more reads from the mt-free cell line would be mapped to the mtDNA (Table 1), 94.6% of which could also be mapped to NUMTs.
Table 1.
Mapping results for the mt-free cell line and the parental cell line
| Cell linea | Number of reads | percent mapped reads (HG19)b | percent mtDNA reads (HG19)c | percent mtDNA reads (MT)d |
|---|---|---|---|---|
| RHO1 | 459 888 | 87.72 | 0.007 (34) | 0.15 (692) |
| RHO2 | 219 470 | 74.12 | 0.017 (38) | 0.28 (620) |
| WT1 | 928 428 | 93.31 | 41.3 | 45.7 |
| WT2 | 1071 782 | 85.04 | 41.5 | 50.8 |
aBoth cell lines were sequenced twice (76-bp paired-end reads with double indexes), from independent sequencing libraries. RHO, mt-free cell line, WT, parental cell line.
bPercentage of reads mapped to the entire genome (nuclear DNA + mtDNA).
cPercentage of reads mapped to mtDNA when using the entire genome (nuclear DNA + mtDNA) as the mapping reference (number of reads in parentheses).
dPercentage of reads mapped to mtDNA when using only the mtDNA genome as the mapping reference (number of reads in parentheses).
Comparison of the mapping results for the mt-free cell line versus the parental cell line clearly shows evidence of NUMTs in the sequence data. When using mtDNA alone as the reference for mapping, we found hundreds of positions that differed between the reads mapping to the mtDNA genome from the mt-free cell line and the parental cell line (Table 2). Reads mapped to these positions in the mt-free cell line were thus unlikely to be derived from the surviving mitochondria as 87% of the mt-free cell line-specific alleles in such cases were included in the aforementioned NUMTs database. The count of these putative observed NUMT alleles in the database is higher than that of NUMT alleles that were not observed in the sequence data from the mt-free cell line (7.73 versus 4.62, P < 0.001 in Mann–Whitney U-test). Moreover, 88% of these putative NUMT alleles were also observed in the parental cell line as minor alleles, with a frequency between 0.02% and 0.93%. All the above evidence supports the interpretation that these are indeed reads coming from NUMTs.
Table 2.
Reads mapping to potential NUMTs in sequence data from the mt-free cell line and the parental cell line
| Major mt-free allele = Major parental allele |
Major mt-free allele ≠ Major parental allele |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Minor mt-free allele = Minor parental allele |
Minor mt-free allele ≠ Minor parental allele |
Major mt-free allele = Minor parental allele |
Major mt-free allele ≠ Minor parental allele |
||||||||
| Comparison | Ref | POSa | Presentb | Absent | Present | Absent | POS | Present | Absent | Present | Absent |
| RHO1 versus WT1 | MT | 9061 | 38 | 8 | 29 | 14 | 203 | 142 | 11 | 44 | 6 |
| HG19 | 540 | 0 | 0 | 0 | 0 | 7 | 5 | 1 | 1 | 0 | |
| RHO2 versus WT2 | MT | 8025 | 13 | 15 | 2 | 9 | 164 | 120 | 28 | 14 | 2 |
| HG19 | 898 | 1 | 1 | 0 | 0 | 17 | 1 | 11 | 5 | 0 | |
aNumber of positions in the mtDNA genome included in reads from the mt-free cell line.
bPresent means the mt-free allele is present in the NUMTs database and absent means the mt-free allele is not in the NUMTs database.
When comparing the discrepant positions between the mt-free cell line and the parental cell line given by two independent mt-free cell line sequencing libraries, ∼70% of them overlapped, of which 95% were included in the NUMTs database. The non-overlapping alleles could be either rare NUMT alleles (76% were included in the NUMTs database) or sequencing errors. We then created another database, called RHO94, that consists of 94 positions that: (i) differed between the true mtDNA genome sequence of the cell line and the reads obtained from the mt-free cell line; (ii) were observed in reads from both libraries from the mt-free cell line; and (iii) were also observed as minor alleles in the parental cell line sequencing data. This database is thus expected to contain NUMT alleles that are especially likely to occur in capture-enriched sequence data.
The choice of mapping (assembly) reference seems to be a crucial factor in influencing the amount of discrepant positions. When using the entire human genome sequence (HG19) as the mapping reference rather than just mtDNA, 90% of the discrepant positions disappeared (Table 2). However, mapping to the entire genome sequence causes other problems, as discussed below.
Comparison between long-rang PCR and capture-enriched sequence data
Fourteen samples, which were sequenced from long-range PCR products in our previous study (16), were sequenced again via the capture-enrichment method here. The expectation is that the sequence data from long-range PCR products should be largely free of NUMTs, as only single bands of the expected size were observed after gel electrophoresis of the PCR products. With mtDNA as the mapping reference, the two methods gave the same major allele at all positions, suggesting there were indeed more reads derived from mtDNA than that from the NUMTs with alternative alleles in the capture-enriched sequence data. Before comparing the minor alleles detected by the two methods, a quality filter was applied to remove sequencing errors, in which all minor alleles with frequency <1% on any strand were discarded. After quality filtering, the capture-enrichment method gave twice as many minor alleles as the long-range PCR method (406 versus 250, see details in Supplementary Table S3), with only 12 minor alleles detected by both methods. There were significantly more minor alleles in the NUMT database from the capture-enrichment method than from the long-range PCR method (50% versus 23%, P = 1.08 × 10−11, Fisher’s exact test), as well as in the RHO94 database (16% versus 3%, P = 7.44 × 10−8, Fisher’s exact test). This suggests that indeed NUMTs are enriched by the capture-enrichment method. If we exclude all minor alleles found in the NUMTs database, then the two methods give similar numbers of minor alleles (192 versus 204). Presumably these minor alleles reflect sequencing errors not removed by the quality filter, contamination, true NUMTs missing from the database and/or heteroplasmies. The average frequency of minor alleles from the capture-enrichment data that were found in the RHO94 database was 2.6 ± 3.3% (Supplementary Table S4).
The number of minor alleles detected in the reads from the shotgun library was much less than the number of minor alleles detected in reads from the long-range PCR or capture-enriched libraries (Table 3). This could reflect cross-contamination and/or index misidentification that occurred during handling/sequencing multiple samples in one library, as observed previously (21). Conversely, the shotgun library had the highest proportion of NUMT alleles among all minor alleles (85%, compared to 23.6–49.9% for the other methods), indicating the enrichment efficiency is indeed much higher for mtDNA than for NUMTs.
Table 3.
Minor allele profile in different sequencing libraries (after quality filter) mapped with either the mtDNA genome (MT) or entire genome (HG19) as the mapping reference
| Methods | Ref | Minor allele count | In NUMTs database | Not in NUMTs database | In RHO94 database |
|---|---|---|---|---|---|
| LR-PCR | MT | 250 | 58 | 192 | 7 |
| Capture | MT | 406 | 202 | 204 | 63 |
| Shotgun | MT | 33 | 28 | 5 | 8 |
| LR-PCR | HG19 | 278 | 61 | 217 | 1 |
| Capture | HG19 | 227 | 44 | 183 | 0 |
| Shotgun | HG19 | 4 | 2 | 2 | 0 |
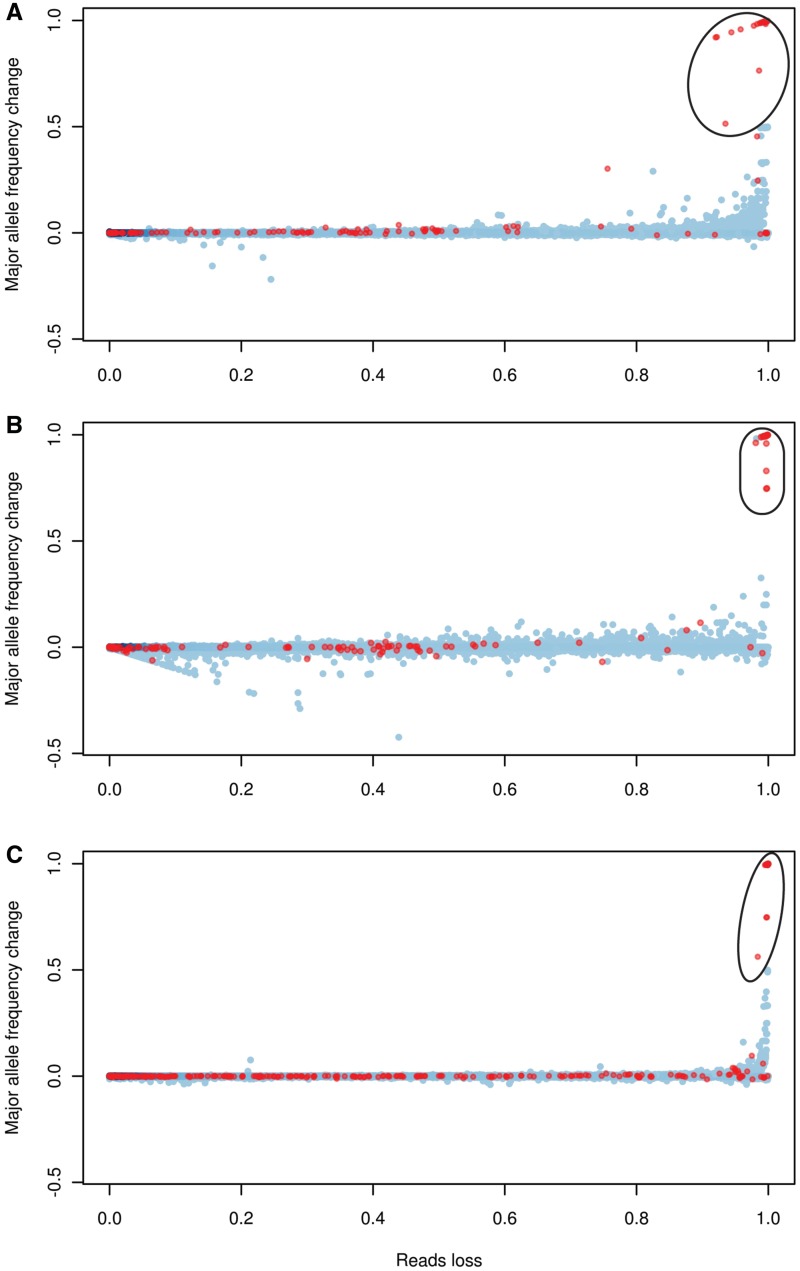
When using the entire genome as the mapping reference, the proportion of NUMT alleles in the capture-enriched data was significantly reduced from 50% to 19% (P = 1.74 × 10−14, Fisher’s exact test) and became equivalent to that of the long-range PCR method (19% versus 22%, P = 0.510, Fisher’s exact test). However, at the same time, there were 39 positions whose major alleles differed from the consensus sequence called when the mtDNA genome alone was used as the mapping reference. For 31 of these 39 positions, the consensus allele differed from the reference mtDNA but was identical to a NUMT. These positions have extremely low coverage (<10×), with >90% of the reads that mapped to mtDNA filtered out because they could be better mapped to the nuclear DNA genome. This problem was also observed with the long-range PCR method and the shotgun method (Figure 1). The positions with major alleles that changed depending on the reference genome tend to occur in several regions along the mtDNA, such as positions 4769, 8860 and 7256, all of which have very similar NUMTs.
Figure 1.
Correlation between read loss and major allele frequency change. Read loss was calculated as the percentage of reads that could be mapped when using the mtDNA genome as the reference (mapping quality score ≥20) but discarded when mapped to the entire genome (mapping quality score <20). Major allele frequency change was calculated as the frequency change of the correct allele (defined as the allele obtained when using mtDNA as the reference). Each dot represents one position in the mtDNA sequence of one of 14 samples (13 samples for the shotgun data). Blue dots indicate positions with consensus alleles that are not included in the NUMTs database; color intensity is proportional to the number of dots. Red dots represent positions with consensus alleles that differ from the reference mtDNA and are the same as a NUMT allele. The circled dots indicate positions whose major alleles changed when mapping to different reference genomes (mtDNA alone versus the entire genome). (A) long-range PCR data; (B) capture-enriched data; (C) shotgun data.
The reduction in reads not only can lead to the wrong consensus allele, but can also cause gaps and reduce the power to detect low-level mutations (heteroplasmy). With 36-bp single-end data, 36% of the reads would be removed in total, while 16% of the positions in the mtDNA genome would not have any mapped reads and 35% of the positions would lose more than half of the mapped reads. Longer reads and paired-end information does help to reduce the loss of reads (Supplementary Figure S1); for instance, no gap was observed when using 76-bp paired-end reads, but 7% of the reads were still discarded and 5% of the positions lost at least half of the mapped reads.
Inferring low-level mutations (heteroplasmy) from capture-enriched sequence data
Although most of the reads from capture-enriched libraries that mapped to mtDNA were authentic mtDNA reads, the existence of reads coming from NUMTs must be considered when attempting to infer heteroplasmy. To investigate difficulties that might arise in distinguishing heteroplasmy from NUMTs, we created a series of artificially mixed samples to mimic different levels of heteroplasmy (see details in ‘Materials and Methods’ section), which were then sequenced to an average coverage of 2824×.
First, different minor allele frequency (MAF) thresholds were applied to remove the NUMT alleles and sequencing errors. With a requirement of a minimum MAF of 0.01 on both strands, the false-negative error rate (FN) was 0.7% (6 out of 888 mixed positions were missed), while the false-positive rate (FP) was 11.1% (98 out of 882 detected mixed positions were false positives) when reads were mapped to the mtDNA genome. In contrast, mapping the reads to the entire genome increased the FN to 9.0%, while no change was observed in the FP (11.0%). However, the percentage of NUMT alleles in the FP was reduced from 57.1% with the mtDNA genome as the only mapping reference to 25.8% with the entire genome as the mapping reference (P = 0.006, Fisher’s exact test). Using the entire genome as the mapping reference thus helps eliminate reads mapping to NUMTs. In order to filter out all false positives, a minimum MAF of 0.055 on both strands is needed, which then results in FN = 38.9%, and 93.5% of the heteroplasmies with MAF of 2.5–5% would be lost. In contrast, when using the entire genome as the mapping reference, a minimum MAF of 0.02 would remove all false positives with FN = 15.9% and only 29.2% of the heteroplasmies with MAF of 2.5%–5% would be lost. However, as before, using the entire genome as the mapping reference results in gaps (4.7% of the mtDNA) due to loss of reads assigned to the mtDNA genome; 6.6% of the heteroplasmies (in each level) were lost as they were located in such gaps.
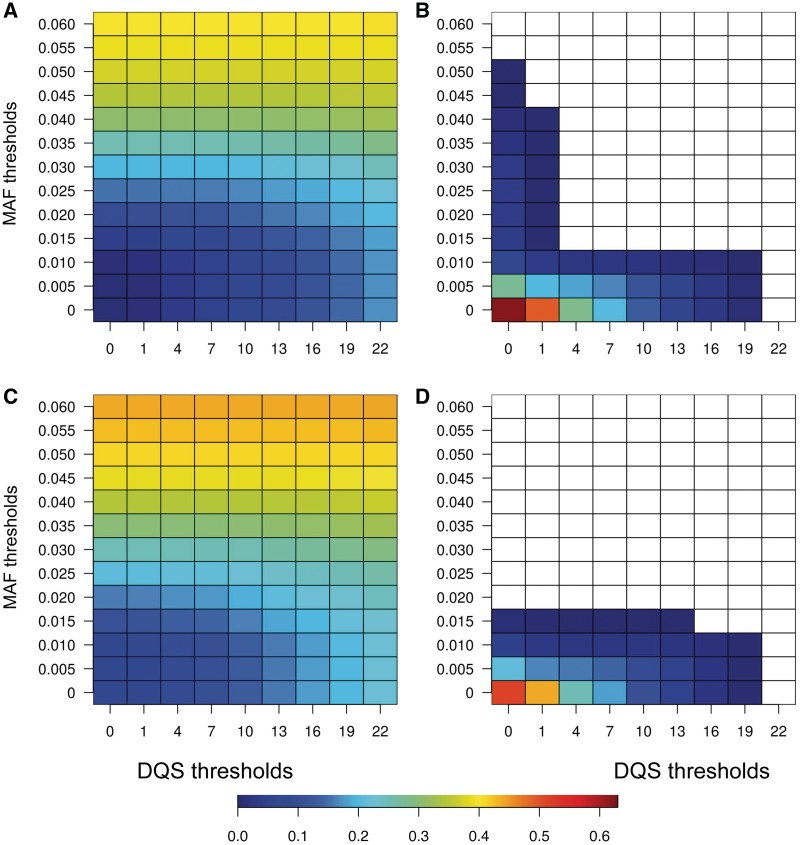
To filter out NUMT alleles for the analysis of heteroplasmy, a possible solution is to make use of the population sequencing data. The underlying principle is that data generated by the same capture-enrichment protocol from different individuals should be similarly influenced by NUMTs. Thus, NUMTs alleles should be observed more often and with higher frequencies than true heteroplasmies, and hence data from other samples may help to identify these NUMT alleles. Here, 2272 samples sequenced with the same protocol and on the same platform were used as the reference, and quality scores assigned by DREEP (DQS) were used to reflect the relative magnitude of the MAF compared with the reference panel. The FN and FP were calculated under different thresholds of DQS and MAF (Figure 2). When using mtDNA as the mapping reference, DQS performed better than MAF, as the DQS threshold that resulted in no false positives had lower FN values compared with that of the MAF threshold, whereas the opposite trend was observed when using the entire genome as the mapping reference (Figure 2). The best threshold combination (giving no false positives and the lowest FN) when using mtDNA alone as the reference (MAF≥0.015, DQS≥4) gave a much lower FN than obtained when using the entire genome as the reference (MAF≥0.02) (6.8% versus 15.9%). A lower FN was still obtained when a more stringent threshold was applied under the first mapping strategy (MAF≥0.02, DQS ≥ 10 gave FN=13.8%). Moreover, this strategy performed better than others for most mixture levels (Table 4).
Figure 2.
False-negative rates and false discovery rates under different thresholds of MAF, DQS. (A and C) False-negative rate; (B and D) false-positive rate. A and B are results when using mtDNA as the mapping reference; C and D are results when using the entire genome (HG19) as the mapping reference. Empty bins in B and D represent no false positives. Basic thresholds used here are as follows: coverage ≥100; minor allele count ≥3 on each strand; minor allele count (number of distinct reads) ≥3 on each strand; and position is not located in C-stretch or STR regions (303–315, 512–525, 16 181–16 195).
Table 4.
False-negative rates under different mapping strategies and different mixture levels
| Thresholdsa | Ref. | Mixture level |
|||||
|---|---|---|---|---|---|---|---|
| 0.5 | 0.25 | 0.1 | 0.05 | 0.025 | All levels | ||
| MAF ≥0.055 | MT | 0 | 0 | 0.022 | 0.885 | 0.984 | 0.389 |
| MAF ≥0.015, DQS ≥4 | MT | 0 | 0 | 0 | 0.005 | 0.322 | 0.068 |
| MAF ≥0.02, DQS ≥10 | MT | 0 | 0 | 0 | 0.033 | 0.639 | 0.138 |
| MAF ≥0.02 | HG19 | 0.066 | 0.066 | 0.066 | 0.082 | 0.508 | 0.159 |
aAll thresholds result in no false positives.
DQS: DREEP quality score.
DISCUSSION
Although genome sequencing is now feasible for most organisms, targeted sequencing of specific genomic regions is preferred by many studies (2,3), either because it is more cost-effective to sequence more samples, or because specific targets are of interest. Many methods have been developed to enrich desired target sequences from sequencing libraries (2,3,9); in general, these methods either make use of specific primers to amplify the targeted region, or the segments of interest are enriched by hybridization to complementary probes/baits.
Both enrichment strategies have been applied in mtDNA genome sequencing. Compared to shotgun libraries, mtDNA sequences were enriched 391-fold by long-range PCR and 155-fold by the in-solution capture method (Supplementary Table S5). Although PCR has long been regarded as the gold standard due to its higher specificity and reproducibility, when sample quality is an issue (e.g. with degraded DNA templates), the capture-hybridization method is much faster, cheaper, easier to use and more efficient. However, a concern of the capture-hybridization method is that NUMTs could be co-enriched with the authentic mtDNA sequences, whereas NUMTs are not expected to amplify in the long-range PCR protocol (23). Indeed, the results of this study demonstrated the existence of NUMTs in the capture-enriched sequence data, as identified by comparison to the NUMTs database that we constructed. It is quite probable that additional NUMTs are actually represented in the sequence data, as there could exist polymorphic NUMTs absent in the reference sequence, or NUMTs present in the human reference genome that were not detected by our in silico analysis. Despite the existence of NUMT-derived sequences in the capture sequence data, by examining the alternative NUMTs allele frequency in the mt-free and parental cell lines and in sequence data for 14 samples enriched by both long-range PCR and by capture hybridization, we found that NUMT alleles usually had a frequency <5%. Thus all capture-enriched sequence data gave the same mtDNA genome consensus sequences as that given by the long-range PCR products, and in general, we do not expect NUMTs to interfere with determining the authentic mtDNA genome sequence from capture-enriched data. However, as shown by the mt-free cell line data (Table 2), NUMTs could have a significant influence on the consensus sequence calling when the mitochondria become extremely rare. It would be interesting to test various types of samples (with different DNA concentrations, degradation status and ratio of mtDNA/nuclear DNA), to obtain an overall view of the impact of NUMTs on calling consensus mtDNA sequences.
Moreover, a bigger challenge is distinguishing NUMT alleles from true low-level mutations (heteroplasmy), as NUMT alleles and true heteroplasmies have the same mapping profile. To distinguish them, the most straightforward way would be to discard all minor alleles included in the NUMTs database. However, we showed in this study that not all putative NUMT alleles are included in the database; moreover, some putative NUMT alleles could nonetheless be true heteroplasmies. Another approach would be to use the entire genome as the reference for mapping, as reads from NUMTs would be mapped to both mtDNA and the nuclear genome and could thus be filtered out by requiring a minimum mapping quality score. However, we showed that authentic mtDNA reads would be discarded by this approach and result in sequencing gaps and even inaccurate calling of the consensus mtDNA genome sequence. A third approach would be to apply a higher MAF threshold to remove the NUMT alleles, but this results in a high false-negative rate: in our artificially mixed samples (with coverage of 2824×), a MAF of 5.5% is needed to filter out all the false positives, but 38.9% of the mixed positions would be missed by this criterion.
We therefore developed and investigated a method that makes use of a statistic that reflects the magnitude of the observed MAF relative to the expected frequency distribution (obtained from a reference panel). The DQS quality score given by DREEP was chosen for this purpose, as it represents the likelihood of the observation given the reference minor allele distribution. By applying thresholds to both MAF and DQS to the sequencing data from the artificial mixtures, we could achieve a much lower false-negative rate (6.8%) with no false positives. This method performed better than any others tested (Table 4). With this method, we can detect almost all mixtures with MAF ≥5% and half of the mixtures with MAF = 2.5%, with no false positives; the sensitivity could be further improved with higher sequencing depth.
In conclusion, we have demonstrated that NUMTs are present in capture-enriched sequence data, but not at a high enough level to interfere with calling accurate consensus mtDNA genome sequences. However, NUMTs could interfere with accurate detection of heteroplasmic mutations, and we have developed and tested a method for distinguishing NUMTs from true heteroplasmic positions in mtDNA sequence data. Given that accurate detection of heteroplasmies or other low-level mutations (e.g. arising from mixed samples) is important for some clinical or forensic applications of mtDNA analysis, we advocate that NUMT detection should be part of the data processing in such studies.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Tables 1–5 and Supplementary Figure 1.
DATA ACCESSION
The sequencing data are publicly available from the European Nucleotide Archive Sequence Read Archive (http://www.ebi.ac.uk/ena/) through accession number ERP001200.
FUNDING
Funding for open access charge: The Max Planck Society.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank EMD-Millipore Inc. (San Diego, CA, USA) for kindly providing the rho zero and parental cell lines, T. Maricic for help discussion and comments on the manuscript, and the MPI-EVA sequencing group and M. Kircher for technical support.
REFERENCES
- 1.Shendure J, Ji H. Next-generation DNA sequencing. Nat. Biotechnol. 2008;26:1135–1145. doi: 10.1038/nbt1486. [DOI] [PubMed] [Google Scholar]
- 2.Mamanova L, Coffey AJ, Scott CE, Kozarewa I, Turner EH, Kumar A, Howard E, Shendure J, Turner DJ. Target-enrichment strategies for next-generation sequencing. Nat. Methods. 2010;7:111–118. doi: 10.1038/nmeth.1419. [DOI] [PubMed] [Google Scholar]
- 3.Mertes F, Elsharawy A, Sauer S, van Helvoort JM, van der Zaag PJ, Franke A, Nilsson M, Lehrach H, Brookes AJ. Targeted enrichment of genomic DNA regions for next-generation sequencing. Brief Funct. Genomics. 2012;10:374–386. doi: 10.1093/bfgp/elr033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Meldrum C, Doyle MA, Tothill RW. Next-generation sequencing for cancer diagnostics: a practical perspective. Clin. Biochem. Rev. 2011;32:177–195. [PMC free article] [PubMed] [Google Scholar]
- 5.Mason VC, Li G, Helgen KM, Murphy WJ. Efficient cross-species capture hybridization and next-generation sequencing of mitochondrial genomes from noninvasively sampled museum specimens. Genome Res. 2011;21:1695–1704. doi: 10.1101/gr.120196.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hazkani-Covo E, Zeller RM, Martin W. Molecular poltergeists: mitochondrial DNA copies (numts) in sequenced nuclear genomes. PLoS Genet. 2010;6:e1000834. doi: 10.1371/journal.pgen.1000834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Woischnik M, Moraes CT. Pattern of organization of human mitochondrial pseudogenes in the nuclear genome. Genome Res. 2002;12:885–893. doi: 10.1101/gr.227202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Simone D, Calabrese FM, Lang M, Gasparre G, Attimonelli M. The reference human nuclear mitochondrial sequences compilation validated and implemented on the UCSC genome browser. BMC Genomics. 2011;12:517. doi: 10.1186/1471-2164-12-517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Maricic T, Whitten M, Paabo S. Multiplexed DNA sequence capture of mitochondrial genomes using PCR products. PLoS One. 2010;5:e14004. doi: 10.1371/journal.pone.0014004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Barbieri C, Whitten M, Beyer K, Schreiber H, Li M, Pakendorf B. Contrasting maternal and paternal histories in the linguistic context of Burkina Faso. Mol. Biol. Evol. 2012;29:1213–1223. doi: 10.1093/molbev/msr291. [DOI] [PubMed] [Google Scholar]
- 11.Pakendorf B, Stoneking M. Mitochondrial DNA and human evolution. Annu. Rev. Genomics Hum. Genet. 2005;6:165–183. doi: 10.1146/annurev.genom.6.080604.162249. [DOI] [PubMed] [Google Scholar]
- 12.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 13.Andrews RM, Kubacka I, Chinnery PF, Lightowlers RN, Turnbull DM, Howell N. Reanalysis and revision of the Cambridge reference sequence for human mitochondrial DNA. Nat. Genet. 1999;23:147. doi: 10.1038/13779. [DOI] [PubMed] [Google Scholar]
- 14.Ruiz-Pesini E, Lott MT, Procaccio V, Poole JC, Brandon MC, Mishmar D, Yi C, Kreuziger J, Baldi P, Wallace DC. An enhanced MITOMAP with a global mtDNA mutational phylogeny. Nucleic Acids Res. 2007;35:D823–D828. doi: 10.1093/nar/gkl927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schonberg A, Theunert C, Li M, Stoneking M, Nasidze I. High-throughput sequencing of complete human mtDNA genomes from the Caucasus and West Asia: high diversity and demographic inferences. Eur. J. Hum. Genet. 2011;19:988–994. doi: 10.1038/ejhg.2011.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li M, Schonberg A, Schaefer M, Schroeder R, Nasidze I, Stoneking M. Detecting heteroplasmy from high-throughput sequencing of complete human mitochondrial DNA genomes. Am. J. Hum. Genet. 2010;87:237–249. doi: 10.1016/j.ajhg.2010.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.The 1000 Genomes Project Consortium. A map of human genome variation from population-scale sequencing. Nature. 2010;467:1061–1073. doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kircher M, Stenzel U, Kelso J. Improved base calling for the Illumina Genome Analyzer using machine learning strategies. Genome Biol. 2009;10:R83. doi: 10.1186/gb-2009-10-8-r83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li H, Durbin R. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li M, Stoneking M. A new approach for detecting low-level mutations in next-generation sequence data. Genome Biol. 2012;13:R34. doi: 10.1186/gb-2012-13-5-r34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kircher M, Sawyer S, Meyer M. Double indexing overcomes inaccuracies in multiplex sequencing on the Illumina platform. Nucleic Acids Res. 2012;40:e3. doi: 10.1093/nar/gkr771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Walker JA, Hedges DJ, Perodeau BP, Landry KE, Stoilova N, Laborde ME, Shewale J, Sinha SK, Batzer MA. Multiplex polymerase chain reaction for simultaneous quantitation of human nuclear, mitochondrial, and male Y-chromosome DNA: application in human identification. Anal. Biochem. 2005;337:89–97. doi: 10.1016/j.ab.2004.09.036. [DOI] [PubMed] [Google Scholar]
- 23.Yao YG, Kong QP, Salas A, Bandelt HJ. Pseudomitochondrial genome haunts disease studies. J. Med. Genet. 2008;45:769–772. doi: 10.1136/jmg.2008.059782. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.